Abstract
The Foxo transcription factors (Foxo1, Foxo3, Foxo4) modulate cell fate decisions in diverse systems. Here we show that Foxo1-dependent gene expression was critical at multiple stages of B cell differentiation. Early deletion of Foxo1 caused a severe block at the pro-B cell stage, due to a failure to express interleukin 7 receptor α (IL-7Rα). Foxo1 inactivation in late pro-B cells resulted in an arrest at the pre-B cell stage due to a reduction in Rag1 and Rag2 expression. Deletion of Foxo1 in peripheral B cells led to fewer lymph node B cells due to reduced L-selectin expression, and failed class switch recombination due to impaired Aicda upregulation. Thus, Foxo1 regulates a transcriptional program that is essential for early B cell development and peripheral B cell function.
Introduction
B cell commitment, survival, and immunoglobulin assembly are all products of a carefully orchestrated network of transcription factors guiding gene expression1. In hematopoietic stem cells (HSCs), graded expression of the transcription factors PU.1 and Ikaros direct development away from the myeloid lineage and into common lymphoid progenitors (CLPs)2, 3. Deletion of either PU.1 or Ikaros in mice leads to a block in HSC differentiation into CLPs and a lack of expression of downstream B cell specific transcription factors. While the microenvironmental cues that direct early B cell commitment and differentiation are not entirely understood, IL-7R {http://www.signaling-gateway.org/molecule/query?afcsid=A001267} and Flt-3 expression are critical1. The transcription factors E2A, EBF and Pax5 are each required for the transition from CLPs to the pro-B cell stage4–6. With respect to the transcriptional hierarchy, E2A regulates expression of EBF and Pax5, as ectopic expression of EBF can rescue B cell-specific gene expression in E2A-deficient cells7. B cells reaching the pre-B cell stage require the functions of transcription factors IRF4 and IRF8 to cease expression of pre-BCR components and to promote Rag1 and Rag2 expression8, 9. A better understanding of the early B cell transcriptional network has evolved over recent years, but the inability of known transcription factors to completely rescue the developmental block(s) observed in gene-ablation models suggests that additional factors and feedback mechanisms exist.
In addition to their roles in early B cell development, many of these same regulatory factors appear to function in peripheral B cell differentiation and activation. Forced expression of PU.1, which is expressed at low amounts in mature B cells, reduces the ability of peripheral B cell to switch to immunoglobulin G1 (IgG1) upon in vitro stimulation10. Similarly, activated B cells overexpressing a major E2A antagonist, Id3, fail to induce expression of activation-induced cytidine deaminase (AID), suggesting that E2A regulates the Aicda gene11. Lastly, it has also been shown that IRF4 plays a dual role in peripheral B cell differentiation by promoting Aicda expression and regulating plasma cell generation12, 13.
The phosphatidylinositol-3-kinase (PI(3)K) pathway is important in both B cell development and peripheral B cell maturation and function. Deletion of either the catalytic or the regulatory subunits of PI(3)K in B cells leads to impairments in B cell development and reductions in peripheral B cell subsets (follicular, marginal zone and B-1)14. Conversely, inactivation of the inositol phosphatase Pten, which is the enzymatic antagonist of PI(3)K, results in an expansion of B-1 and marginal zone B cells15, 16. PI(3)K is activated by the BCR and other receptors on B cells, resulting in the transient production of phosphatidylinositol-3,4,5-trisphosphate (PI(3,4,5)P3). PI(3,4,5)P3 binds to the pleckstrin homology (PH) domain of various effector proteins (for example, Akt), resulting in plasma membrane recruitment and activation. Transcriptional regulation downstream of PI(3)K is accomplished in part by members of the evolutionarily conserved Daf-16-related Forkhead O (Foxo) family consisting of Foxo1 {http://www.signaling-gateway.org/molecule/query?afcsid=A000944}, Foxo3 {http://www.signaling-gateway.org/molecule/query?afcsid=A000945}, and Foxo4 {http://www.signaling-gateway.org/molecule/query?afcsid=A002832}. Phosphorylation of the Foxo proteins by Akt at three conserved sites induces their nuclear export and consequent inactivation of transcriptional activity17. Known targets of Foxo factors include genes encoding the cell cycle regulators, p27 and cyclinD2, pro-apoptotic factors, Bim and FasL, and regulators of reactive oxygen species (ROS), MnSOD and GADD45 (reviewed in17). Deletion of all Foxo factors in HSCs leads to a severe block in the generation of CLPs due to impaired protection from ROS18. From the analysis of the Foxo transcriptome in various lineages, it is clear that the Foxo factors regulate their many targets in a highly context-specific manner with virtually no overlap from one cell type to the next18, 19; thus underscoring the need for direct analysis of Foxo actions in specific physiological settings. Of relevance to the current study, it is notable that while all three Foxo factors are expressed in B cells, their unique and redundant roles in B cell development and activation have not been elucidated.
Here we employed multiple gene targeting approaches to determine the unique and redundant roles of Foxo1 and Foxo3 in B cell development and activation. Germline inactivation of Foxo3 did not impair early B cell generation, formation of Ig-positive B cells, or peripheral B cell differentiation or antibody responses. Conversely, selective deletion of Foxo1 at discrete stages of B cell development revealed specific functions for Foxo1 at multiple B cell checkpoints. Inactivation of Foxo1 in the early pro-B, pre-B, or transitional B cells compartments resulted in impaired B cell differentiation associated with reduced expression of Il7rα, Rag1, Rag2, L-selectin and Aicda. These findings establish Foxo1 as a critical member of the transcription factor network directing early B cell development and peripheral immune function.
Results
Foxo1 is required for IL-7Rα and Rag expression
Although signaling via the PI(3)K pathway is critical for B cell generation and the antibody response, little is known about the control of transcription by PI(3)K signaling effectors. In recent years, the Foxo factors, which are negatively regulated by Akt phosphorylation, have emerged as critical downstream regulators controlling diverse physiological processes such as cell cycle progression, metabolism, apoptosis, and oxidative defense. To determine the role of Foxo1 in B cell development and function, mice with loxP sites inserted into the Foxo1 gene (Foxo1L/L) were crossed with mice that express Cre-recombinase (Cre) under the control of the mb1 promoter (mb1Cre)20. Mb1Cre mice express Cre at an early B lymphoid progenitor stage, mediating deletion of loxP-flanked (floxed) genes at the earliest pro-B cell stage of development. The Foxo1L/Lmb1Cre mice were compared to wild-type (Foxo1+/+mb1Cre) littermate controls as well as to Foxo3 null (Foxo3−/−) mice21.
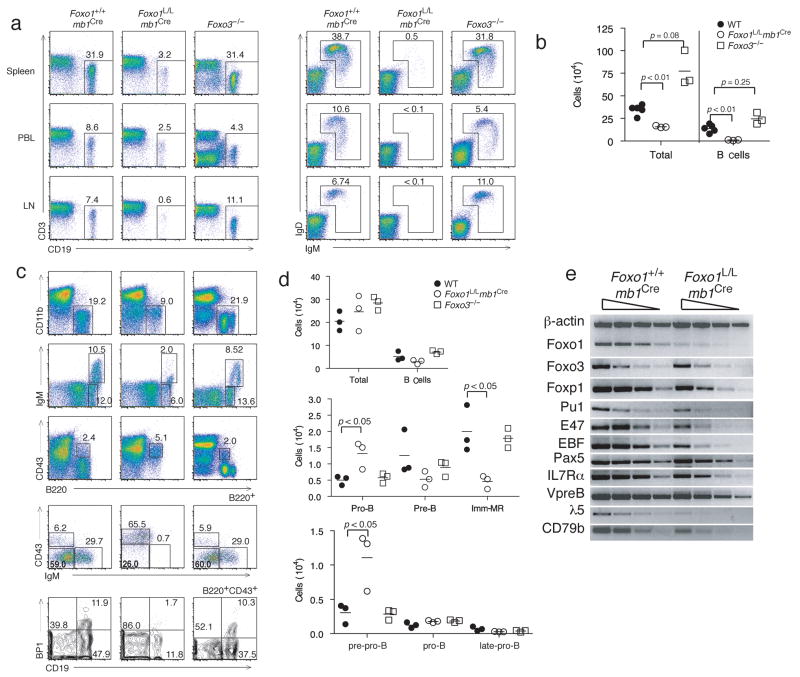
Peripheral lymphoid organs were analyzed by flow cytometry to enumerate B cell subsets. In the spleen, lymph node and peripheral blood, Foxo1L/Lmb1Cre mice contained 10% of the number of B cells that wild-type or Foxo3−/− mice contain (Fig. 1a). In addition, Foxo1-deficient B cells present in the periphery did not express surface IgM or IgD (Fig. 1a). As has been reported22, Foxo3−/− mice had an increase in total cell numbers; however, B cell numbers were not significantly increased compared to wild-type littermate controls (Fig. 1b). Due to the sharp decrease in peripheral B cells in Foxo1L/Lmb1Cre mice, we examined the B cell compartment of the bone marrow from Foxo1L/Lmb1Cre mice to determine if B cell development was blocked at an early stage. Indeed, Foxo1L/Lmb1Cre mice showed a decreased percentage of total B cells in the bone marrow as compared to wild-type mice (Fig. 1c), and consistent with efficient deletion of Foxo1 in early B cells (Supplementary Fig. 1a online). A close analysis of the bone marrow also revealed a marked decrease in the percentages of B220+IgM+ immature and mature B cells, and B220+CD43−IgM− pre-B cells compared to wild-type and Foxo3−/− mice (Fig. 1c). In addition, the Foxo1L/Lmb1Cre mice had a 2-fold increase in the percentage of B220+CD43+ pro-B cells (Fig. 1c). Based on the differential display of cell surface markers, pro-B cells can be further subdivided into 3 stages of development known as pre-pro-B, early-pro-B, and late-pro-B cells. Foxo1L/Lmb1Cre mice had a significant reduction in the percentages of pro-B cells that were CD19+BP1− (early-pro-B) and CD19+BP1+ (late-pre-B), but an increase in the percentage of CD19−BP1− (pre-pro-B) cells (Fig. 1c,d). Semiquantitative RT-PCR analysis of cDNA revealed a general reduction in the expression of genes known to be critical for early B cell differentiation in pro-B cells from Foxo1L/Lmb1Cre mice (Fig. 1e, Supplementary Fig. 2 online). Whether these genes are direct or indirect targets of Foxo1 will require further investigation. Together, these results suggest that B cell development in Foxo1L/Lmb1Cre mice is partially blocked at the transition from pre-pro-B to the early-pro-B stage and that very few B cells progress beyond the cycling pre-B stage.
Figure 1. Foxo1 directs pro-B cell development.
(a) Representative FACS profiles of n = 3–5 mice per group of spleen, peripheral blood, and lymph node from Foxo1+/+mb1Cre, Foxo1L/Lmb1Cre, and Foxo3−/− mice indicating B cell (CD19+) percentages (left) and percentage of cells expressing IgM and IgD (right). (b) Absolute numbers (symbol) and averages number (dash) of splenocytes and splenic B cells from n=3–5 mice per group. (c) Representative FACS profiles of n = 3–5 mice per group showing percentages of bone marrow B cells, and subsets of immature and mature B cells, pro-B cells, pre-B cells, and subcompartments of pre-pro-B (CD19−BP1−), early-pro-B (CD19+BP1−) and late-pro-B (CD19+BP1+) cells. (d) Absolute number (symbols) and average numbers (dashes) of bone marrow cells and B lineage cells subcompartments. (e) Representative semi-quantitative PCR of 3 independent experiments of cDNA obtained from Foxo1+/+mb1Cre pro B cells (IgM−IgD−CD2−CD25−B220+) and Foxo1L/Lmb1Cre pro B cells (B220+).
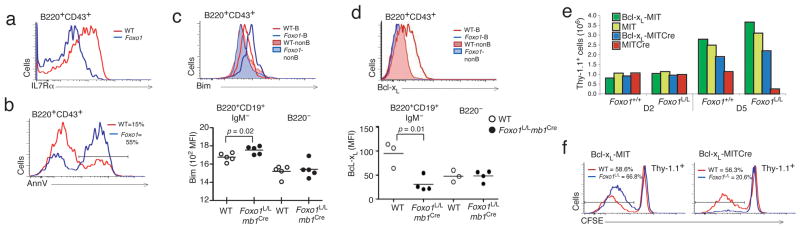
A pro-B cell block in development can occur due to two primary types of defects. One defect is marked by failed IL-7R signaling, resulting in increased cell death and reduced proliferation23, 24. The second defect, such as that observed in Rag2−/− mice, is attributed to failed pre-BCR assembly and signaling. The earliest stages of B cell development are dependent on IL-7 signaling, with a gradual transition to pre-BCR-dependent proliferation and differentiation. In Foxo1L/Lmb1Cre mice, pro-B cells failed to expand in IL-7 conditioned cultures, indicating that these cells could present defects in cell proliferation and or survival (data not shown). Because IL-7Rα is necessary for IL-7 signaling at the pro-B cell stage, IL-7Rα expression was analyzed by flow cytometry. While wild-type pro-B cells exhibited high expression of IL-7Rα, Foxo1L/Lmb1Cre mice did not up-regulate IL-7Rα at the pro-B cell stage (Fig. 2a). To test whether the lack of IL-7R in Foxo1-deficient pro-B cells had an effect on pro-B cell survival, Annexin V expression was analyzed on pro-B cells from Foxo1L/Lmb1Cre and wild-type mice. While wild-type pro-B cells had relatively few (15%) Annexin V+ apoptotic cells, these cells represented the majority (55%) of the pro-B cells in Foxo1L/Lmb1Cre mice, indicating that Foxo1-deficient B cells are prone to apoptosis at this stage of development (Fig. 2b).
Figure 2. Foxo1 regulates IL-7Rα expression and pro-B cell survival.
(a) IL-7Rα expression on pro-B cells from Foxo1+/+mb1Cre (red) and Foxo1L/Lmb1Cre (blue) mice. (b) Ann V expression on late-pro-B cells from Foxo1+/+mb1Cre (red) and Foxo1L/Lmb1Cre (blue) mice. (c) Intracellular Bim expression in pro-B (line) and non-B (shaded) cells and MFI analysis of Bim expression from pro-B and non-B cells (5 mice of each group indicated with symbols, and average MFI indicated with dashes). (d) Intracellular Bcl-xL expression of pro-B (line) and non-B (shaded) cells and MFI of Bcl-xL expression of pro-B and non-B cells (3–4 mice of each group). (e) Foxo1+/+ and Foxo1L/L B220+ bone marrow cells were grown in culture with IL-7 for 2 days, infected with MSCV-Bcl-xL or left uninfected, followed by infection with MIT or MIT-Cre. MIT-Cre-transduced (Thy1.1+) B cells were enumerated by FACS on day 2 and day 5. (f) Bcl-xL infected cells superinfected with either MIT or MIT-Cre were labeled with CFSE and cultured for 4 days. FACS analysis of proliferation was determined by CFSE division. FACS profiles are representative of 3–5 independent experiments.
To gain insight into the mechanisms of Foxo1L/Lmb1Cre pro-B cell death, we analyzed the expression of Bim and Bcl-xL, pro- and anti-apoptotic members of the Bcl-2 family, respectively. Flow cytometric analysis of freshly isolated pro-B cells from Foxo1L/Lmb1Cre mice were found to express increased amounts of Bim and decreased amounts of Bcl-xL (Fig. 2c,d). Overexpression of either Bcl-2 or Bcl-xL has been shown to attenuate apoptosis, and the Bcl-xL transgene allows for the expansion and survival of pro-B cells25. To determine if early B cell development could be rescued by protecting Foxo1-deficient B cells from apoptosis, Foxo1F/F pro-B cells that do not express Cre were expanded in IL-7 then transduced with a mouse stem cell based retrovirus encoding Bcl-xL (MSCV-Bcl-xL). Two days later, secondary infections were performed with either empty virus (MIT) or Cre-expressing virus (MIT-Cre) to eliminate Foxo1. While induced deletion of Foxo1 in the absence of exogenous Bcl-xL resulted in rapid cell death, Foxo1L/L cells expressing both Bcl-xL and Cre survived in culture, but did not proliferate (Fig. 2e,f). Thus, promoting the survival of Foxo1-deficient pro-B cells is necessary but not sufficient to rescue the developmental block in Foxo1L/Lmb1Cre mice.
In addition to the defect in survival, Foxo1L/Lmb1Cre pro-B cells failed to express normal amounts of intracellular μ heavy chain (Fig. 3a). Foxo1L/Lmb1Cre and Rag2−/− mice showed similar staining patterns for μ heavy chain at the pro B cell stage (Fig. 3a). To determine if Foxo1L/Lmb1Cre cells can undergo heavy chain Ig rearrangements, genomic DNA from purified pro-B cells was analyzed by PCR followed by Southern blot hybridization. While DH to JH rearrangement was normal, Foxo1L/Lmb1Cre pro-B cells showed impaired VH to (D)JH rearrangement (Fig. 3b). This result is consistent with a defect in IL-7R signaling, since IL-7 has been implicated in promoting the accessibility of the VH gene locus26, 27. It has also been shown that Rag enhancer regions (Erag) bear conserved forkhead binding sites28, and hence Foxo1 could directly regulate Rag expression. Indeed, we found that Foxo1L/Lmb1Cre pro-B cells had decreased transcription of Rag1 and Rag2 (Fig. 3c). A similar decrease in Rag1 and Rag2 expression was observed in cultured pre-B cells upon acute deletion of Foxo1 with Cre-expressing virus (Supplementary Fig. 3 online). To determine if Foxo1 could also bind to the Erag region, chromatin was isolated from primary pre-B cells propagated in IL-7 and subjected to chromatin immunoprecipitation (ChIP) analysis. Interestingly, Foxo1 bound to the same conserved forkhead binding sites as Foxp140, suggesting that they co-regulate Rag expression in part by binding to the Erag enhancer (Fig. 3d). Additionally, ChIP data from peripheral T cells suggests that Foxo1 does not bind to the Rag enhancer region when Rag is not expressed (Supplementary Fig. 4).
Figure 3. Foxo1 regulates RAG expression to induce immunoglobulin gene rearrangement.
(a) Representative FACS profile of n=3 mice per group of intracellular μ heavy chain expression in pro-B cells from Foxo1+/+mb1Cre (red), Foxo1L/Lmb1Cre (blue), and Rag−/− (shaded gray) bone marrow. (b) Southern blot analysis of distal VH-DJH rearrangements (VHJ558), proximal VH-DJH rearrangements (VH7183), D-J rearrangements, and control (Cμ) in pro-B cells. (c) Pro-B cells were analyzed by semi-quantitative RT-PCR for expression of Rag1, Rag2, Foxo1, and Actb expression. (d) Foxo1 binding to the Erag enhancer region was analyzed by ChIP on chromatin from pre-B cells from IL-7 cultures using antibodies for Foxo1, E47, and an isotype control, followed by PCR of three regions of the Erag enhancer region. All 3 regions contain putative forkhead binding sites while Erag1 and Erag2 regions contain E-box binding sites. Southern blot, RT-PCR, and ChiP analyses are representative of 3 independent experiments.
Foxo1 regulates kappa rearrangement in pre-B cells
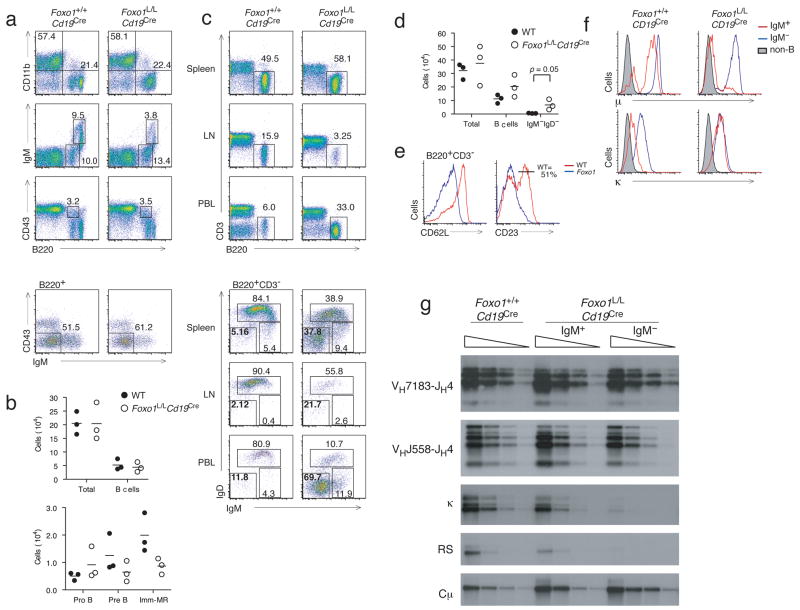
To determine if Foxo1 is required beyond the pro-B cell stage, Foxo1L/L mice were crossed with mice expressing Cre under the control of the Cd19 promoter (Cd19Cre)29. Cd19 expression is induced by Pax5 and EBF at the pro-B cell stage, conferring substantial yet partial Cd19Cre-mediated deletion in pre-B and immature B cells, and nearly complete deletion by the mature B cell stage29 (Supplementary Fig. 1a,b). The Foxo1L/Lmb1Cre mice had normal pro-B and pre-B cell compartments with a decrease in the percentage of immature and mature B cells relative to wild-type (Foxo1+/+Cd19Cre; Fig. 4a,b). In the periphery, Foxo1L/LCd19Cre mice showed an increase in the percentage of B cells in the spleen and peripheral blood and a sharp reduction in the percentage of B cells in lymph nodes relative to wild-type (Fig. 4c). Intriguingly, a prominent IgM−IgD− B cell population was present in the peripheral blood and spleen of Foxo1L/LCd19Cre mice that was not observed in wild-type mice (Fig. 4c,d). These IgM− B cells also exhibited low expression of the B cell maturation markers CD21 and CD23 (Fig. 4e), suggesting they are not mature B cells. To determine if the IgM−IgD− B cells present in the Foxo1L/LCd19Cre mice are pre-B cells, splenic B cells were stained for surface IgM and IgD and either intracellular μ-heavy chain or κ-light chain. While a high proportion of IgM−IgD− wild-type B cells expressed intracellular κ-light chain, suggesting that they are class switched B cells, very few IgM−IgD− Foxo1L/LCd19Cre B cells expressed intracellular κ- or λ light chains, but expressed abundant amounts intracellular μ-heavy chain (Fig. 4f, Supplementary Fig. 5 online). Consistent with these data, IgM−IgD− Foxo1L/LCd19Cre B cells also showed a decrease in the amount of Vκ-Jκ rearrangement, as well as reduced excision of the Vκ-Jκ locus via downstream rearrangement with the recombination signal (RS) element (Fig. 4g). IgM−IgD− Foxo1L/LCd19Cre B cells were found to be negative for expression of pre-BCR components VpreB1 and λ5 (data not shown), suggesting that they are small resting pre-B cells that transited to the periphery. These results are consistent with decreased Rag expression and heavy chain gene rearrangement seen at the pro-B cell stage in Foxo1L/Lmb1Cre mice. Foxo1L/LCd19Cre mice were also crossed onto the Foxo3−/− background. The B cell compartment of these mice was indistinquishable from Foxo1L/LCd19Cre counterparts (Supplementary Fig. 6 online), indicating that the functions we have ascribed to Foxo1 are unaffected by the presence or absence of Foxo3.
Figure 4. Foxo1 regulates kappa chain gene rearrangement in pre-B cells.
(a) Representative FACS profiles from Foxo1+/+Cd19Cre and Foxo1L/LCd19Cre mice indicating percentages of total B cells, immature and mature B cells, pro-B cells, and pre-B cells. (b) Quantitation of bone marrow B cell subsets from n=3 mice per group (indicated by symbols) and average numbers of each group (indicated by dashes). (c) Representative FACS profiles indicating percentage of B cells and IgM and IgD expression on B220+ B cells from the spleen, peripheral blood, and lymph node. (d) Quantitation of splenic B cell subsets from n=3 mice per group (indicated by symbols) and average numbers of each group (indicated by dashes). (e) Expression of CD21 and CD23 maturation markers on splenic B cells from Foxo1+/+Cd19Cre and Foxo1L/LCd19Cre mice. (f) Expression of intracellular μ heavy chain and κ light chain in splenic IgM+ (red), IgM− (blue) B cells, and non-B cells (shaded gray). (g) Representative southern blot analysis of 3 independent experiments of proximal and distal VH(DH)JH rearrangement, Vκ-Jκ rearrangement, Vκ to RS rearrangement, and cμ control in IgM+ and IgM− B cells. FACS plots are representative of 3 mice/group.
Foxo1 is required for AID and CD62L expression
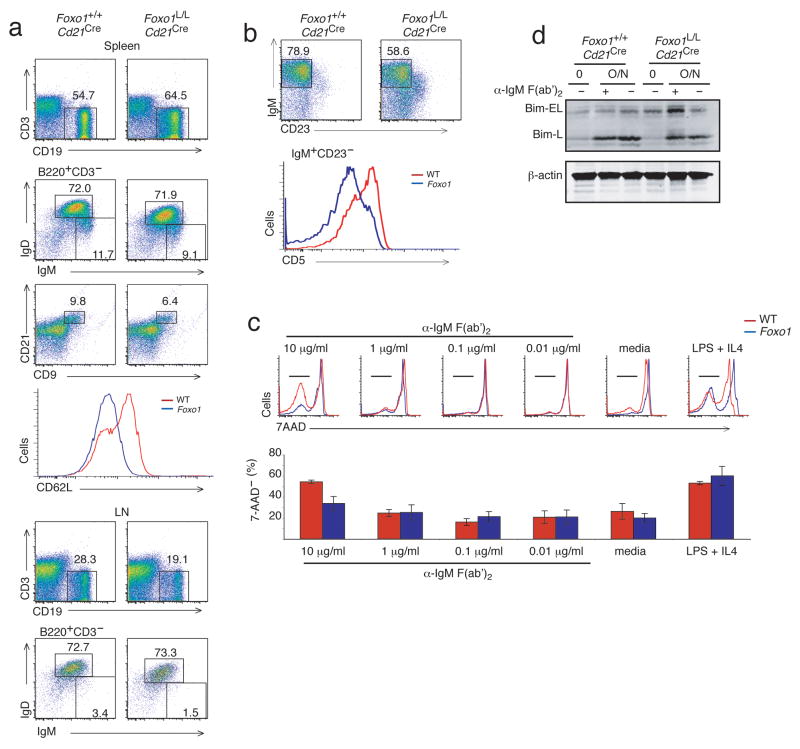
The pronounced defects in early B cell development presented in Foxo1L/Lmb1Cre and Foxo1L/LCd19Cre mice precluded analysis of peripheral B cell maturation and function. Therefore, Foxo1L/L mice were crossed to mice expressing Cre under control of the Cr2 promoter (Cd21Cre)30. These mice up-regulate Cre during the transitional stage of B cell development, after IL-7-dependent differentiation, Ig gene rearrangement and, presumably, RAG-mediated receptor editing has occurred. Flow cytometric analysis revealed that Foxo1L/LCd21Cre mice had a modest increase in the percentage of splenic B cells as compared to wild-type B cells. The percentage of Foxo1L/LCd21Cre B cells representing transitional (IgM+IgD− CD21−), marginal zone (CD9hiCD21hi) and mature (IgM+IgD+) B cells was normal relative to wild-type (Fig. 5a and data not shown). Although the expression of IgM and IgD was slightly reduced, no increase in IgM−IgD− B cells was observed, consistent with their postulated pre-B origin. Interestingly, there was a ~30% reduction in lymph node B cells, which is consistent with reduced expression of L-selectin (CD62L) and homing defects (Fig. 5a). Reduced expression of CD62L was also observed on peripheral B cells in Foxo1L/LCd19Cre mice (data not shown). In the peritoneal cavity, there was a reduction in B-1 cells that appeared to preferentially affect CD5+ B-1a cells (Fig. 5b). These findings suggest that Foxo1 regulates peripheral B cell homing via upregulation of CD62L.
Figure 5. Reduced lymph node and peritoneal B-1a cells in Foxo1L/LCd21Cre mice.
(a) Representative FACS profiles of spleen and lymph node from Foxo1+/+Cd21Cre and Foxo1L/LCd21Cre indicating the percentages of B cells of total cells and the percentage of B cells that are IgM+IgD+ or CD9+CD21+ marginal zone B cells. Representative FACS profile of the expression level of CD62L on B cells in the spleen. (b) FACS profiles indicating the percentage of IgM+CD23− B-1 cells and CD5+ B-1a vs CD5− B-1b subsets from the peritoneal cavity of Foxo1+/+Cd21Cre (red) and Foxo1L/LCd21Cre (blue) mice. (c) Stimulation of purified splenic B cells from Foxo1+/+Cd21Cre (red) and Foxo1L/LCd21Cre (blue) mice with titrated amounts of anti-IgM F(ab′)2 (48 h) and measurement of cell viability using 7AAD. Data is representative of triplicate stimulations done in 3 independent experiments (d) Immunoblot analysis for Bim expression in purified splenic B cells stimulated overnight with or without 10 μg/mL of anti-IgM F(ab′)2. FACS plots are representative of 3 mice/group.
Consistent with the reduced surface Ig expression, Foxo1L/LCd21Cre splenic B cells exhibited reduced survival in response to BCR crosslinking with anti-IgM F(ab′)2, as determined by flow cytometric analysis of cell death (Fig. 5c). The survival defect was dose-dependent and activation induced, since sub-mitogenic amounts of anti-IgM did not reveal differences between Foxo1L/LCd21Cre and wild-type B cells (Fig. 5c). These defects were associated with increased Bim expression, as was the case with Foxo1-deficient pro-B cells (Fig. 5d). The observed effects are BCR-specific as survival of Foxo1-deficent B cells was similar to wild-type when stimulated with lipopolysaccharide (LPS) and interleukin 4 (IL-4; Fig. 5c). Notwithstanding the effects of Foxo1 loss on surface BCR expression, Foxo1 does not appear to be required for B cell maintenance, but may contribute to survival post-activation.
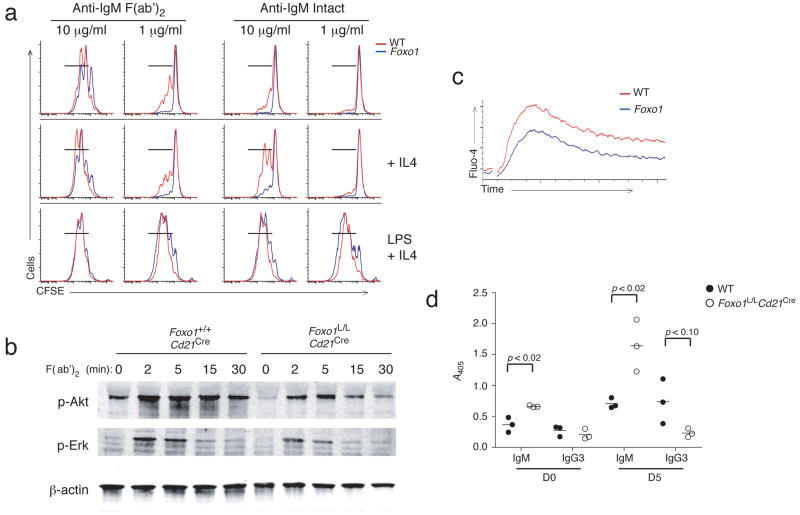
In accordance with decreased cell survival, Foxo1L/LCd21Cre B cells showed decreased proliferation in response to low concentrations of anti-IgM F(ab′)2 and intact anti-IgM stimulation, while proliferation of Foxo1-deficient B cells was similar to wild-type in response to LPS and IL-4 (Fig. 6a). Foxo1L/LCd21Cre B cells also showed decreased anti-IgM dependent phosphorylation of Akt and Erk (Fig. 6b) and had decreased calcium flux in response to anti-IgM F(ab)2 stimulation (Fig. 6c). Despite the BCR signaling defects observed in vitro, immunization of Foxo1L/LCd21Cre mice with the T-independent type 2 (TI-2) antigen, trinitrophenol (TNP)-Ficoll, elicited abundant production of TNP-specific serum IgM antibody (Fig. 6d). Naive Foxo1L/LCd21Cre mice also have slightly increased polyreactive natural antibody seen at day 0 (Fig. 6d). This observation correlates with the retained B-1b population in the peritoneal cavity, which is capable of responding to TI-2 antigens.
Figure 6. Foxo1L/LCd21Cre B cells have impaired responses to anti-IgM stimulation in vitro, but intact antibody responses to TI-2 antigens.
(a) Representative FACS plots of proliferation measurements of purified splenic B cells from Foxo1+/+Cd21Cre (red) and Foxo1L/LCd21Cre (blue) mice following CFSE labeling and stimulation (3 days) with 1 μg/mL or 10 μg/mL of either anti-IgM F(ab′)2 or intact anti-IgM in the presence or absence of IL-4; or stimulated with LPS and IL-4. CFSE partitioning was analyzed by FACS. (b) Immunoblot analysis of phospho-Akt and phospho-Erk from anti-IgM F(ab′)2 stimulated splenic B cells. (c) Calcium flux by purified splenic B cells from Foxo1+/+Cd21Cre (red) and Foxo1L/LCd21Cre (blue) mice loaded with Fluo-4 and stimulated with 10 μg/mL anti-IgM F(ab′)2. (d) ELISA to detect TNP-specific IgM and IgG3 present in the serum of Foxo1+/+Cd21Cre (open circles) and Foxo1L/LCd21Cre (filled circles) mice before and 5 days after IP immunization with 10 μg TNP-Ficoll in PBS. FACS profile and in vitro stimulation is representative of 3 independent experiments. ELISA data is representative of 2 independent experiments with 3 mice per group.
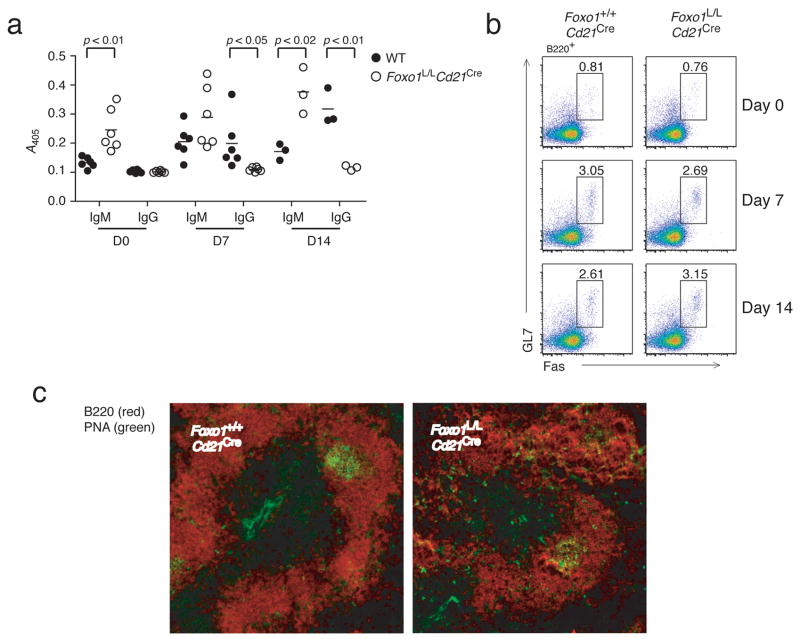
Strikingly, TI-2 immunization of Foxo1L/LCd21Cre mice produced very low titers of IgG3 antibody compared to wild-type (Fig. 6d). To determine if Foxo1L/LCd21Cre B cells can respond normally to T-dependent antigens, mice were immunized with nitrophenol-keyhole limpet hemacyanin (NP-KLH) and concentrations of serum immunoglobulin were analyzed 7 and 14 days later. Serum antibody from Foxo1L/LCd21Cre mice again showed an increase in NP-specific IgM antibody at days 7 and 14 but an absence of detectable IgG antibody at both timepoints (Fig. 7a). To discern whether the absence of IgG antibody is due to decreased germinal center formation, splenic cell suspensions and tissue sections were analyzed for the presence of germinal center B cells. In both analyses, Foxo1L/LCd21Cre mice showed germinal center responses to NP-KLH comparable to wild-type, suggesting that the lack of IgG antibody production is due to a general defect in class switch recombination (CSR) and not due to impaired T-B cell collaboration (Fig. 7b,c).
Figure 7. Foxo1 is required for class switch recombination, but not germinal center formation.
(a) ELISA to detect NP-specific IgM and IgG present in the serum of Foxo1+/+Cd21Cre (open circles) and Foxo1L/LCd21Cre (filled circles) mice before and 7 or 14 days after IP immunization with 100 μg of NP-KLH in alum. b) Splenic GL7+Fas+ germinal center B cells were quantified by FACS, and c) visualized by fluorescent imaging of frozen sections by PNA (green) and B220 (red) (D14 only). FACS plots and histology are representative of 3 mice/group.
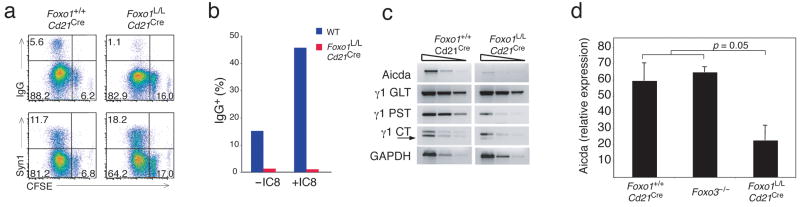
We have previously shown that in the absence of Pten, B cells do not undergo CSR in response to antigen or LPS and IL-4 stimulation both in vivo and in vitro, and that this CSR defect can be partially restored by the expression of a constitutively active Foxo143. To determine if Foxo1-deficient B cells can undergo CSR, cells were stimulated in vitro with LPS and IL-4 for 3 days and analyzed for proliferation, CSR and plasma cell formation. While wild-type B cells underwent both CSR and plasma cell formation efficiently upon stimulation, there was a striking decrease in CSR and enhanced plasmablast generation in the absence of Foxo1 (Fig. 8a). These cells also did not undergo CSR upon addition of the PI(3)Kδ inhibitor IC87114, which we have shown increases CSR in wild-type as well as in Pten-deficient B cells31, consistent with the role of Foxo1 as the downstream nuclear target of PI(3)K (Fig. 8b). To determine if expression of AID, the master regulator of both CSR and somatic hypermutation, was defective in Foxo1L/LCd21Cre B cells, RNA from in vitro stimulated cells was analyzed for Aicda expression. Foxo1-deficient B cells did not up-regulate expression of Aicda or generate post-switch IgG1 transcripts in response to LPS and IL-4 stimulation, but locus accessibility leading to germline transcript expression was unaffected (Fig. 8c). To corroborate these in vitro findings with in vivo expression of Aicda, RT-PCR was performed on cDNA prepared from sorted germinal center B cells from Foxo1L/LCd21Cre, Foxo3−/− and wild-type mice. As expected, Aicda expression was reduced in Foxo1L/LCd21Cre B cells, while Foxo3−/− and wild-type B cells showed similarly high expression (Fig. 8d). Thus, the Foxo1-associated defect in CSR is linked to Aicda transcription.
Figure 8. Foxo1 regulates AID expression.
(a) Flow cytometric analysis of LPS + IL-4 stimulated (3 days) B cells from Foxo1+/+Cd21Cre and Foxo1L/LCd21Cre mice (2 mice per group repeated in 3 independent experiments). Proliferation was measured by CFSE partitioning on plasmablasts (Syn1+) undergoing CSR (IgG+). (b) Percentage of class switched cells generated in the absence or presence of the PI(3)Kδ inhibitor IC87114 (+IC8). Data is representative of 3 independent experiments. (c) Representative semi-quantitative PCR from two independent experiments to measure Aicda expression and the generation of γ1 germline transcripts (GLT), γ1 post-switch transcripts (PST), and γ1 circular transcripts (CT-lower band). (d) QT-PCR of Aicda expression of splenic germinal center B cells from Foxo1+/+Cd21Cre, Foxo3a−/−, and Foxo1L/LCd21Cre mice (3 mice per group) 10 days after immunization with 200 μg of NP-KLH in alum.
Discussion
The generation and function of peripheral B cell subsets evolves from an elaborate series of cellular and molecular events that are often associated with developmental ‘checkpoints’. Transcriptional regulation of these events controls B cell commitment, survival, and proliferation, and B cell specific processes such as Ig gene rearrangement, CSR, and terminal differentiation into plasma cells. Here we demonstrate that Foxo1 exerts a nonredundant function in regulating many of these events.
Two recently reported microarray studies revealed that while Foxo3 expression does not vary through B cell development, there is a strong upregulation of Foxo1 seen at the early pro-B cell stage in humans and mice32, 33, suggesting that Foxo1 is the dominant family member expressed at this stage. We show that deletion of Foxo1 in nascent pro-B cells led to a block in B cell development, due to impaired expression of IL-7Rα. Expression of Il7ra is regulated by PU.1 in lymphoid progenitors34. Moreover, EBF, a downstream target of PU.1, can rescue IL-7Rα expression and B cell commitment in Pu.1-deficient fetal liver hematopoietic progenitors35. Since EBF controls mb1 expression and hence mb1-Cre expression, the requirement for Foxo1 in regulating sustained expression of Il7ra in early B cells occurs after the onset of EBF expression, suggesting a hierarchy of transcriptional control. Lack of IL-7Rα expression resulted in apoptosis of Foxo1-deficient pro-B cells. Interestingly, increased apoptosis of Foxo1-deficient pro-B cells correlated with increased Bim expression, despite earlier reports that Bim is a Foxo-induced gene product in several cell types36. The frequency of pro-B cells expressing Bcl-xL was reduced in the absence of Foxo1. This finding is consistent with reduced IL-7R expression, which induces Bcl-xL expression upon STAT5 activation37. IL-7-dependent pro-B cell survival was rescued upon ectopic Bcl-xL expression in vitro, but this manipulation was not sufficient to overcome IL-7R-dependent roles in early B cell proliferation and differentiation.
In addition to the lack of IL-7Rα expression, Foxo1-deficient B cells had reduced expression of Rag1 and Rag2, leading to impaired V(D)J rearrangement and cytoplasmic μ production in pro-B cells. Cd19Cre-mediated deletion resulted in a later impairment at the pre-B cell stage and reduced light chain gene rearrangement. These cells exited to the periphery and were present in the spleen but did not recirculate, perhaps due to reduced expression of L-selectin. It is also possible that Foxo1 regulates gene expression leading to the retention of developing B cells in the bone marrow. It has recently been observed that production of a functional BCR causes basal PI(3)K signaling and the downregulation of RAG38, 39. As Foxo proteins are negatively regulated by PI(3)K, this downregulation is likely attributable to the removal of Foxo proteins from the nucleus, equivalent to what is seen in Foxo1-deficient B cells. We show that Foxo1 can bind to the Erag enhancer region, although this may not be the only mechanism of Foxo1-dependent regulation of Rag. Interestingly, Foxp1, a Forkhead family member distantly related to the Foxo factors, has also been shown to regulate Rag expression in B cells via Erag binding28. By comparison, deletion of Foxo3 does not affect Ig gene rearrangement or assembly at the pro-B or pre-B cell stages. Thus, Foxo1 exerts a critical and nonredundant role in the rearrangement and editing of V(D)J segments.
During the preparation of this manuscript, two papers reported that Foxo factors contribute to RAG expression40, 41. In agreement with our findings, Amin and Schlissel showed that knockdown or forced expression of Foxo1, but not Foxo3, modulated Rag expression in early B cells40. In addition, Herzog et al. showed that overexpression of a constitutively nuclear Foxo3 molecule bearing mutated Akt phosphorylation sites was also able to up-regulate Rag expression41. Here we provide direct evidence that Foxo1 is required for Rag expression, and that Foxo3 is dispensable for Ig gene rearrangement. In vitro studies suggest that the Foxo factors bind similar if not identical DNA targets, consistent with the high degree of sequence similarity among their DNA-binding forkhead domains42. Thus, post-translational regulatory mechanisms, particularly those controlling protein stability, may contribute to the discrete functions of the individual Foxo factors. If this is the case, then expression of a constitutively active form of any Foxo factor may have similar effects, precluding the assignment of particular functions to individual Foxo factors.
Activation of mature B cells causes nuclear exclusion of Foxo1 and downregulation of Foxo1 transcription43–45. Here we show that Foxo1 plays a distinct role in peripheral B cell maturation and the antibody response. Deletion of Foxo1 in transitional B cells using Cd21Cre led to a reduction in lymph node B cells, consistent with reduced expression of L-selectin. Peritoneal B cells were also reduced and severely biased towards the CD5− B-1b subset, while the marginal zone B cell compartment was intact. These collective findings come as a surprise since our previous analyses of B cell-specific Pten-deficient mice revealed an expansion of the marginal zone and B-1a subsets in the presence of elevated PI(3,4,5)P3 and, presumably, inactive Foxo factors. Foxo1-deficient B cells also exhibited reduced BCR expression and consequent defects in BCR signaling, proliferation and survival. Similar to pro-B cells, Foxo1-deficient splenic B cells had increased expression of Bim upon stimulation with high concentrations of anti-IgM F(ab′)2. Despite reduced BCR responsiveness and an altered B-1 cell compartment, Foxo1-deficient B cells produced a robust T-independent immune response.
Foxo1L/LCd21Cre mice underwent normal germinal center formation and produced abundant antigen-specific IgM upon immunization with TD antigens; however class switched serum antibodies were not generated. This impairment was not due to impaired T cell help, as TI-2 responses show the same defect. Moreover, in vitro differentiation assays demonstrated normal or elevated plasma cell generation in the absence of accompanied CSR. These findings are consistent with our previous work demonstrating a negative regulatory role for PI(3)K in CSR31. In addition, we also showed that forced expression of nuclear Foxo1 increased the percent of class switched B cells and expression of Aicda in the presence or absence of a PI(3)K inhibitor31. We provide genetic evidence that Foxo1 exerts an essential and nonredundant role in CSR by up-regulating Aicda expression. Hence, PI(3,4,5)P3 appears to be necessary for the activation and cell cycle entry of resting B cells, but must be attenuated to allow proliferation-dependent CSR to proceed. This view is consistent with findings that strong BCR agonists suppress CSR46. Thus, while inactivation of Foxo1 does not strongly impact late B cell maturation and antigen-driven clonal expansion, it serves an essential role in CSR.
Together our findings establish pivotal functions for Foxo1 in early B cell maturation and peripheral immune function. These functions are attributed in large part to Foxo1-dependent transcription of Il7ra, Rag1, Rag2, L-selectin and Aicda, although other relevant targets certainly remain. Surprisingly, we found no evidence of enhanced survival or proliferation that would support previous suggestions that the Foxo proteins may be important for preventing B cell transformation. This observation does not appear to be due to redundant functions with Foxo3, as mice lacking both Foxo1 and Foxo3 in the B lineage presented a phenotype that is indistinguishable from that of Foxo1 alone. Future studies will address the specific role of Foxo1 In the context of other transcription factors regulating early and late B cell differentiation.
Methods
Mice
Mice with loxP sites flanking exon 2 of Foxo1 19 (Foxo1L/L) were crossed to Cd19-Cre 29 (Cd19Cre), mb1-Cre20 (mb1Cre), or Cd21-Cre30 (Cd21Cre) mice to delete Foxo1 at various stages of B cell development. Mice with a gene trap cassette inserted into the first intron of the Foxo3 gene were used for complete deletion of Foxo3 (Foxo3−/−)21. Mice were bred and housed at the Burnham Institute for Medical Research animal facility and all experiments received IACUC approval. Animals of 8–10 weeks of age were used for peripheral B cell analysis and 4–6 weeks of age for bone marrow cultures.
Flow Cytometry
Single cell suspensions (1 × 106) were first stained with biotinylated and/or FcBlock antibodies in FACS buffer (1% FBS, 0.1% azide in PBS) for 20 min at 4 °C, followed by incubation with a cocktail of FITC, PE, PerCPCy5.5, PeCy7, APC, and APC-Cy7 conjugated antibodies. For intracellular staining, cell surface staining was performed first, followed by buffer fixation/permeabilization (eBioscience) and intracellular staining. Antibodies used were: biotinylated- BP1 (6C3), CD9 (KM08), CD23 (B3B4), CD86 (GL1), IgD (11–26), IgG1 (A85-1), IgG2a/b (R2–40), IgG3 (R40–82); FITC-AnnV (#556419 BD-Pharmingen), CD2 (RM2–5), CD5 (53-7.3), CD21 (7G6), CD69 (HI.2F3), GL7 (GL7), H2Db (KH95), IgD (11–26), μ (II/41), κ (187.1), λ (R26–46), and anti-rabbit (#11-4839-81 eBiocience); PE-CD138 (281-2), CD23 (B3B4), IgD (11–26), CD43 (S7), CD3e (145-2C11), and CD127 (A7R34); PerCP-Cy5.5- B220 (RA3-6B2) and streptavidin (eBioscience); PeCy7- CD11b (M1/70), CD3 (145-2C11), and streptavidin (eBioscience); APC- CD127 (A7R34), B220 (RA3-6B2), CD19 (ID3), IgM (II/41), CD62L (MEL-14), and streptavidin (eBioscience); APC-Cy7- CD19 (ID3) and B220 (RA3-6B2) (BD-Pharmingen or eBioscience). Rabbit-anti-mouse Bim (#2819) and Bcl-xL (54H6) was purchased from Cell Signaling Technology. Data was collected on a FacCanto (BD) and analyzed using FlowJo software (TreeStar).
Semi-Quantitative RT-PCR and QT-PCR
Bone marrow pro-B cells were purified by initial depletion of CD25, CD2, IgM, CD11b+ cells using biotinylated antibodies followed by depletion using anti-biotin beads (Militenyi). B cells were further purified (85–90% pro-B) using B220+ positive selection (Militenyi). RNA was purified from cells using TRIZOL LS (Invitrogen) as according to the manufacturer. RT reactions were performed using Superscript II first strand synthesis kit (Invitrogen). PCRs were performed on serially diluted cDNA for 30 cycles using previously reported primer pairs47. Aicda QT-PCR was performed on RNA from purified B cells from immunized mice as described31. Aicda expression was normalized to Gapdh first and then to the percent of Fas+GL7+ germinal center B cells, as determined by FACS.
Immunizations and ELISAs
For T-dependent immunizations, animals were immunized ip with 100 μg of NP-KLH precipitated in alum (Imject, Pierce) and serum was collected on day 0, 7, and 14. For T-independent type 2 immunizations, mice were immunized with 10 μg of TNP-Ficoll and serum was collected on day 0 and 5. ELISAs for antigen-specific antibody was performed as described previously31. For Aicda expression, animals were immunized ip with 200 μg of NP-KLH in alum and analyzed 10 days later.
Cell Culture
Red blood cells from splenic single cell suspensions were lysed using ACK (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA) for 5 min at 4 oC. B cells were purified by negative selection using CD43-labeled magnetic beads (Miltenyi). For CFSE cultures, B cells were labeled with 2.5 μM Carboxyfluorescein succinimidyl ester (CFSE) for 10 min at 20 °C in PBS, washed and cultured in C/10 RPMI at 2 × 106 cells/ml in a 96-well flat bottom plate for 3 days with goat anti-mouse-IgM F(ab′)2 (Jackson) or rabbit anti-mouse intact-IgM (Jackson) at 10 μg/ml, 1 μg/ml or 0.1 μg/mlL; with or without 10 ng/ml IL-4, (RnD Systems) or in 20 μg/mL LPS (Sigma) with 10 ng/ml IL-4.
Calcium Flux
Purified splenic B cells were labeled with Fluo-4 (Invitrogen) for 60 min at 37 °C. Cells were washed and resuspended at 2 × 106 cells/ml and collected on the cytometer for 15 sec prior to stimulation. Cells were stimulated with 10 μg/ml anti-IgM F(ab′)2 and collected on the cytometer for 3 min.
Bone Marrow Infection
Viral supernatants were generated from transfected Phoenix-eco cells as previously described31. B cells were positively selected from bone marrow using B220-labeled magnetic beads (Miltenyi). Cells were cultured for 48 h in OMEM with 15% FBS (Hyclone), 100 U/ml Pen/Strep, 2 mM L-glut, and 55 μM 2-mercaptoethanol with the addition of 5 ng/ml IL-7 (RnD Systems). Cells were resuspended at 1 × 106 cells/ml in MSCV-Bcl-xL viral supernatants that was previously incubated with 8 μg/ml polybrene and diluted 2-fold with OMEM. Cells were spun at 1000 × g for 90 min at 30°C and then placed at 37 °C for 60 min. Cells were then collected and resuspended in C/15 OMEM with 5 ng/ml IL-7 and cultured for an additional 48 h, then superinfected with either empty virus (MSCV-IRES-Thy1.1) or MIT-Cre. Cells were returned to culture with IL-7 and analyzed 2 and 5 days later by FACS. For RT-PCR, PCR and ChIP experiments, cells were collected 3 days after MIT-Cre infection.
Southern Blot
DNA was purified from FACS-sorted pro-B cells or magnetic-bead sorted splenic B cells using the salt extraction method and resuspended at 25 ng/ml in TE buffer (10 mM Tris, pH 8, 1 mM EDTA). PCRs for DH to JH, distal VH (VHJ558) to (D)JH (JH4), proximal VH (VH7183) to (D)JH (JH4), Vκ (VκDeg) to Jκ (MAR35K), Vκ to Recombination Sequence (RS), or constant Cμ were performed on 100, 25, 6.25, and 1.56 ng/ml for 30 cycles as previously described48. Radiolabeled probes were synthesized using DNA polymerase Klenow fragment and PCR template generated from genomic DNA from wild-type splenic B cells. Southern blotting of heavy and light chain rearrangement were performed as described49, 50.
Chromatin immunoprecipitation
ChIP assay was performed essentially as described for the anti-acetylated histone H3 Ab-based ChIP assay kit (Upstate Biotechnology) with minor modifications. In brief, 37% formaldehyde solution was added directly to pro/pre-B cell cultures to a final concentration of 1% and cells were incubated for 10 min at 37 °C. Cells were harvested and washed twice with ice-cold PBS containing protease inhibitors (Roche) and lysed with SDS lysis buffer for 10′ on ice. The lysate was sonicated to an average length of 200–1000 bp and clarified by centrifugation. The samples were diluted 10-fold with ChIP dilution buffer then precleared with protein A/G beads (Calbiochem) for 1 h followed by incubation with either normal rabbit IgG (Jackson), anti-Foxo1 (Santa Cruz or Cell Signaling Technology), or anti-E47 (Becton-Dickinson) overnight at 4 °C. Protein A/G beads were added in the final hour of incubation prior to harvest. After washing and elution of beads per the manufacturer’s instructions, cross-linking were reversed by heating at 65 °C overnight followed by proteinase K treatment. DNA was purified using the Qiagen PCR purification kit, and PCR reactions were performed on eluted DNA for 30 cycles (94 °C, 15 s, 60 °C, 15 s, 72 °C, 1 min) using previously published primers28. For specificity control, PCR reactions were performed on eluted DNA for 35 cycles (94 °C, 30 s, 55 °C, 30s, 72 °C, 1 min) using primers 5′-CACGTTTATTGGTTCCTCTGTC-3′ and 5′-CCAGCTGCTGACTATGTCCC-3′ for a region in the conserved noncoding sequence X in the aicda gene.
Biochemistry
For short-term stimulations, purified B cells were stimulated with 10 μg/ml of anti-IgM F(ab′)2 for the indicated times in PBS at 2 × 107 cells/ml. For overnight stimulations, cells were cultured in C/10 RPMI at 2 × 107 cells/ml. Reactions were stopped by the addition of cold PBS and cells were immediately pelleted and lysed with RIPA (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Sodium deoxycholate and 0.1% SDS) including DNAse, protease, and phosphatase inhibitors. Westerns were probed with anti-Bim, anti-pAkt(S473), and p-ERK (Cell Signaling Technology), and primary antibodies detected with the Odyssey system (Licor) using anti-rabbit DyeLight800 (Pierce) or anti-rabbit-680 (Invitrogen).
Immunohistochemistry
Spleens were frozen in OCT (Tissue-Tek) and 8 micron sections were stained with anti-PNA-FITC and anti-B220-APC (pseudo-colored red with Slidebook).
Statistics
Groups of 3–8 mice were used in statistical analysis. p values were calculated using the student’s t-test.
Supplementary Material
Acknowledgments
We thank K. Rajewsky (Harvard) and M. Reth (MaxPlanck Institute) for providing Cd21Cre and mb1Cre mice, respectively; B. Finlay (BIMR) and K. Mowen (TSRI) for the Bcl-xL and Cre expressing retroviruses, respectively. S. Hedrick (UC-San Diego) and members of the Rickert lab for discussions and reading of the manuscript. Supported by the National Institutes of Health grant AI059447 to R.C.R. R.A.D. is an American Cancer Society Research Professor and supported by the Robert A. and Renee E. Befler Foundation Institute for Innovative Cancer Science.
References
- 1.Nutt SL, Kee BL. The Transcriptional Regulation of B Cell Lineage Commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 3.Wang JH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 4.Bain G, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 6.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 7.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Ma S, Turetsky A, Trinh L, Lu R. IFN Regulatory Factor 4 and 8 Promote Ig Light Chain {kappa} Locus Activation in Pre-B Cell Development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 10.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 12.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 13.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110delta isoform of PI3K. Trends Immunol. 2007;28:80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki A, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–667. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 18.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. PNAS. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Corcoran AE, et al. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. Embo J. 1996;15:1924–1932. [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh N, Yasunaga M, Hirashaima M, Yoshida O, Nishikawa SI. Role of IL-7 and KL in activating molecules controlling the G1/S transition of B precursor cells. Int Immunol. 1996;8:317–323. doi: 10.1093/intimm/8.3.317. [DOI] [PubMed] [Google Scholar]
- 25.Fang W, et al. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity. 1996;4:291–299. doi: 10.1016/s1074-7613(00)80437-9. [DOI] [PubMed] [Google Scholar]
- 26.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 27.Bertolino E, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 28.Hu H, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 29.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of Resting Mature B Lymphocytes Depends on BCR Signaling via the Ig[alpha]/[beta] Heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Omori SA, et al. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hystad ME, et al. Characterization of early stages of human B cell development by gene expression profiling. J Immunol. 2007;179:3662–3671. doi: 10.4049/jimmunol.179.6.3662. [DOI] [PubMed] [Google Scholar]
- 34.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 35.Medina KL, et al. Assembling a Gene Regulatory Network for Specification of the B Cell Fate. Developmental Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004;172:4770–4778. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 38.Verkoczy L, et al. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. 2007;178:6332–6341. doi: 10.4049/jimmunol.178.10.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110delta is required for down-regulation of RAG expression in immature B cells. J Immunol. 2007;178:1981–1985. doi: 10.4049/jimmunol.178.4.1981. [DOI] [PubMed] [Google Scholar]
- 40.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 42.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 43.Yusuf I, Zhu X, Kharas MG, Chen J, Fruman DA. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood. 2004;104:784–787. doi: 10.1182/blood-2003-09-3071. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, et al. Analysis of the Major Patterns of B Cell Gene Expression Changes in Response to Short-Term Stimulation with 33 Single Ligands. J Immunol. 2004;173:7141–7149. doi: 10.4049/jimmunol.173.12.7141. [DOI] [PubMed] [Google Scholar]
- 45.Hinman RM, Bushanam JN, Nichols WA, Satterthwaite AB. B cell receptor signaling down-regulates forkhead box transcription factor class O 1 mRNA expression via phosphatidylinositol 3-kinase and Bruton’s tyrosine kinase. J Immunol. 2007;178:740–747. doi: 10.4049/jimmunol.178.2.740. [DOI] [PubMed] [Google Scholar]
- 46.Rush JS, Hasbold J, Hodgkin PD. Cross-linking surface Ig delays CD40 ligand- and IL-4-induced B cell Ig class switching and reveals evidence for independent regulation of B cell proliferation and differentiation. J Immunol. 2002;168:2676–2682. doi: 10.4049/jimmunol.168.6.2676. [DOI] [PubMed] [Google Scholar]
- 47.Reynaud D, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ait-Azzouzene D, et al. An immunoglobulin C kappa-reactive single chain antibody fusion protein induces tolerance through receptor editing in a normal polyclonal immune system. J Exp Med. 2005;201:817–828. doi: 10.1084/jem.20041854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 50.Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.