Abstract
We reported recently that the human opportunistic pathogen Pseudomonas aeruginosa strain PA14 kills Caenorhabditis elegans and that many P. aeruginosa virulence factors (genes) required for maximum virulence in mouse pathogenicity are also required for maximum killing of C. elegans. Here we report that among eight P. aeruginosa PA14 TnphoA mutants isolated that exhibited reduced killing of C. elegans, at least five also exhibited reduced virulence in mice. Three of the TnphoA mutants corresponded to the known virulence-related genes lasR, gacA, and lemA. Three of the mutants corresponded to known genes (aefA from Escherichia coli, pstP from Azotobacter vinelandii, and mtrR from Neisseria gonorrhoeae) that had not been shown previously to play a role in pathogenesis, and two of the mutants contained TnphoA inserted into novel sequences. These data indicate that the killing of C. elegans by P. aeruginosa can be exploited to identify novel P. aeruginosa virulence factors important for mammalian pathogenesis.
Until recently, systematic genetic analysis of the pathogenic interaction between a eukaryotic host and a bacterial pathogen has been hampered by the lack of a genetically tractable system in which both the pathogen and host are amenable to low-cost, high-throughput genetic screens. Ideally, the eukaryotic host should be a model genetic organism such as Caenorhabditis elegans, Drosophila melanogaster, or Arabidopsis thaliana for which a complete genome sequence is available or will be available in the near future. Similarly, it would be advantageous if the genome sequence of the bacterial pathogen were also available. A final advantageous feature of a model pathogenesis system would be the use of a bacterial pathogen in the model that is also an important human pathogen.
We recently developed a C. elegans–Pseudomonas aeruginosa pathogenesis system that has all of the features described above (1, 2). The genome sequence of C. elegans is essentially complete (3), and the P. aeruginosa strain PA01 is available at http://www.pseudomonas.com. P. aeruginosa is a ubiquitous Gram-negative bacterium that is an important opportunistic human pathogen that is capable of causing disease in humans whose immune system has been compromised, who have sustained major trauma, or who are inflicted with cystic fibrosis (4). P. aeruginosa also has been reported to cause disease in plants, insects, and a variety of vertebrates (5–8). Interestingly, we showed that a clinical isolate of P. aeruginosa strain PA14 is not only a pathogen of mice but also kills C. elegans (1, 2), and we identified two distinct modes of PA14-mediated killing of C. elegans that appear to be mechanistically distinct. When PA14 is grown on low-nutrient media, C. elegans killing occurs over the course of several days and is referred to as “slow killing.” In contrast, when PA14 is grown on high-osmolarity media, C. elegans killing occurs over the course of several hours and is referred to as “fast killing.” Slow killing requires live bacteria and is correlated with the accumulation of PA14 in the C. elegans gut (2), whereas fast killing is mediated at least in part by low-molecular-weight toxins, including phenazines, and does not require live bacteria (1).
In previous publications, our laboratory showed that P. aeruginosa strain PA14 also causes disease in a variety of plants including lettuce and A. thaliana (8, 9). Using a plant–P. aeruginosa pathogenesis system, we identified a set of PA14 mutants that is less pathogenic in a plant leaf infiltration assay; remarkably, most of these mutants also were less pathogenic in a mouse burn model (9). Importantly, we showed that 6 of 10 genes identified as encoding important virulence factors for mouse and A. thaliana pathogenesis are also required for effective killing of C. elegans (2).
In this paper we demonstrate how the C. elegans–P. aeruginosa slow-killing model can be exploited to identify previously unknown P. aeruginosa virulence-associated factors and to identify virulence-associated systems that appear to be universal among Gram-negative animal and plant bacterial pathogens. These results validate our strategy of using a simple nonvertebrate host to scan the entire P. aeruginosa genome for pathogenesis-related genes.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Media, and Bacterial Genetics.
The P. aeruginosa strain PA14 (8), the E. coli strains OP50 (10) and DH5α (Bethesda Research Laboratories), and the PA14 TnphoA mutants 25F1 and pho15 (9) have been described. Complete media for bacterial culture and maintenance were Luria broth and King’s broth (11, 12). Minimal medium was M9 (12). The broad host-range cloning vector pUCP18 (13), the E. coli cloning vector pCR2.1 (Invitrogen), and the plasmid pKDT17, which contains the P. aeruginosa lasR gene (14), have been described. Plasmid pCH3, which contains the E. coli dsbA cloned into pBAD18, was a gift from C. Guilhot (Harvard Medical School, Boston). E. coli DH5α was used for plasmid constructions and plasmids were introduced into P. aeruginosa PA14 by electroporation (15).
Screening for PA14 TnphoA Mutants with C. elegans Slow-Killing Defects.
P. aeruginosa PA14 was mutagenized with TnphoA as described (9). Individual PA14∷TnphoA clones were inoculated into 200 μl King’s B medium in microtiter plates containing rifampicin and neomycin at 100 and 200 μg/ml, respectively. Ten microliters of an overnight culture was spread on nematode growth medium (NG; modified from NGM as described in ref. 16; 0.35% peptone instead of 0.25%) in 5.5 cm Petri plates and incubated at 37°C for 24 h. After 8–24 h at room temperature (23–25°C) each plate was seeded with two L4-stage hermaphrodite C. elegans strain N2 (Bristol) (10). Plates were incubated at 25°C and examined for live worms after 5 days. On plates seeded with a nonpathogenic mutant, thousands of progeny worms were present at day 5 and the bacterial lawn was completely consumed, whereas very few or no live worms were found on the plates seeded with the wild-type strain and the bacterial lawn remained intact. Putative nonpathogenic or attenuated mutants identified in the preliminary screen were retested and subjected to a virulence assay to determine the kinetics of C. elegans killing.
C. elegans Slow- and Fast-Killing Assays.
Slow-killing kinetics of C. elegans by PA14 and its derivatives were determined by using the same procedure as for the mutant screen except that each NG plate was seeded with 40–50 L4 C. elegans. Plates were scored every 4–6 h, and three to four replicates per trial were performed. A worm was considered dead when it no longer responded to touch. Worms that died as a result of getting stuck to the wall of the plate were excluded from the analysis. Fast-killing kinetics were determined in the same way as for slow killing except that high-osmolarity peptone/glucose/sorbitol (PGS; 1% Bacto-Peptone/1% NaCl/1% glucose/0.15 M sorbitol/1.7% Bacto-Agar) medium was used instead of NG medium and the plates were seeded with 30–40 worms.
Arabidopsis and Mouse Pathogenicity Assays.
An Arabidopsis leaf-infiltration assay using ecotype Llagostera and a mouse full-thickness skin burn assay using inbred mice (strain AKR/J; The Jackson Laboratory) were performed as described (8, 9).
Molecular Analysis of TnphoA Mutants.
Standard DNA manipulation protocols were performed as described in ref. 17. The generation and sequencing of inverse PCR (IPCR) sequence tags corresponding to TnphoA insertions were carried out as described (1, 9). IPCR sequence tags were used to identify corresponding sequences in the P. aeruginosa PA01 genome database (http://www.pseudomonas.com) by using the program blastn (18). PA01 sequence was used to design oligonucleotide primers corresponding to mutants 25F1, pho15, 50E12, and 35A9, which then were used to amplify a region from the wild-type PA14 genome corresponding to each mutant. Each of these amplified fragments from PA14 was sequenced directly (using the Sequenase PCR Product sequencing Kit, United States Chemical) or cloned into pCR2.1 and sequenced using standard dideoxy-sequencing methods. The sequences were deposited in the GenBank database and assigned the accession numbers AF116282 (pho15), AF116283 (35A9), AF116284 (25F1), and AF116285 (50E12). The IPCR sequence tags were also deposited in GenBank and assigned the following accession numbers: AF116277 (12A1), AF116278 (41C1), AF116279 (44B1), AF116280 (48D9), and AF116281 (41A5). The predicted translation products of the identified ORFs in each of the sequences were compared with the GenBank database by using blastx (19).
Genetic Complementation Analysis of Mutants pho15, 25F1, and 50E12.
pho15(dsbA). Primers TMW8 (5′-GCACTGATCGCTGCGTAGCACGGC-3′) and TMW9 (5′-TGACGTAGCCGGAACGCAGGCTGC-3′) were used to amplify a 1,126-bp fragment from PA14 genomic DNA containing the PA14 dsbA coding region plus 176 bp of sequence upstream of the translational start. This fragment was cloned into pCR2.1 by using the TA cloning kit (Invitrogen) to generate pCRdsbA. A 1.2-kb SacI/XbaI fragment containing the dsbA ORF was subcloned from pCRdsbA into SacI/XbaI-digested pUCP18 to construct pPAdsbA, placing the transcription of dsbA under the control of a constitutive E. coli lacZ promoter. A 700-bp KpnI/SphI fragment containing the E. coli dsbA was subcloned from pCH3 into KpnI/SphI-digested pUCP18 to generate pEcdsbA, placing the E. coli dsbA under the control of a constitutive E. coli lacZ promoter.
25F1.
Mutant 25F1 contained TnphoA inserted into the first ORF of a three-gene operon, orf338, orf224, and orf252. A 1.8-kb PCR fragment containing 482 bp of upstream promoter sequence, the entire orf338, and a truncated orf224 was amplified from PA14 genomic DNA by using primers F2327 (5′-CGAGGAATCCAGTCGAGGTG-3′) and R4180 (5′-GCAAGATGCAGCCGAGAGTAG-3′). The product was cloned directly into pCR2.1 to construct plasmid pMT403. A SacI/XbaI fragment from pMT403, which contained the orf338 PCR product and 482 bp of upstream sequences, was cloned into the SacI/XbaI site of pUCP18 to construct pORF338, placing orf338 under the control of its native promoter. A 3.6-kb genomic fragment containing orf338, orf224, orf252, and upstream sequences was amplified from genomic PA14 DNA using primers RIF3115 (5′-GTCAGAATTCTCAGCTTGACGTTGTTGCCC-3′) and RIR6757 (5′-GTCAGAATTCGACTTCTATTACCGCGACGCC-3′), each containing an EcoRI site (underlined). The EcoRI digest of the PCR product was cloned into the EcoRI site of pUCP18 to construct plasmid p3-ORFs, which contains orf338, orf224, and orf252 under the control of their native promoters.
50E12.
Mutant 50E12 contained TnphoA inserted into a gene homologous to ptsP. Promoter prediction by neural network analysis (http://www-hgc.lbl.gov/projects/promoter.html) suggests that the PA14 ptsP homologue is cotranscribed with the upstream ORF, orf159. orf159 encodes a putative 159 residue polypeptide that is closely related to proteins with a core MutT domain of unknown function found in Haemophilus influenzae (GenBank accession no. Q57045). A 4.3-kb PCR fragment, containing the PA14 ptsP homologue (ptsPPa), orf159, and 1.2-kb upstream sequences, with an EcoRI site introduced at each end, was amplified from genomic PA14 DNA by using the primers RIF1698 (5′-GTCAGAATTCGATGTTCCAGTCCCAGATCCC-3′) and RIR6002 (5′-GTCAGAATTCCAGTAGACCACCGCCGAGAG-3′). This fragment was cloned into the EcoRI site of pUCP18 to generate p206-lac and p206-nat. In p206-lac, the transcription of orf159 and ptsPPa is under the control of both a constitutive E. coli lacZ promoter and their native promoters. In p206-nat, orf159 and ptsPPa are only under the control of their native promoters.
Biochemical and Physiological Characterization of PA14 TnphoA Mutants.
Supernatants from log-phase Luria broth cultures were tested for proteolytic and elastolytic activities as described (20). Pyocyanin was assayed by absorbance at 520 nm in acidic solution after growth in King’s A broth (11) modified by the addition of 100 μM FeCl3 (21). Phospholipase C was assayed by using p-nitrophenylphosphorylcholine as a substrate, as described (22).
RESULTS
Identification of PA14 Mutants Defective in C. elegans Slow Killing.
A total of 2,400 prototrophic P. aeruginosa PA14 TnphoA insertion mutants were screened individually using the C. elegans slow-killing assay for mutants that failed to kill or exhibited attenuated killing (see Materials and Methods for details). Eight mutants (12A1, 35H7, 35A9, 44B1, 41A5, 41C1, 48D9, and 50E12), which consistently gave a lower rate of C. elegans killing relative to the parental PA14 strain, were chosen for further study. DNA blot analysis showed that each TnphoA mutant contained a single transposon insertion (data not shown). All of the mutants grew at the same rate as the wild-type strain in NG medium and in minimal M9 medium, indicating that the attenuated pathogenicity phenotypes observed were not simply a result of growth defects of the mutants.
To determine whether bacterial virulence genes (factors) mediating slow killing are relevant to pathogenesis in other hosts, the eight mutants obtained by screening in the nematode slow-killing assay were tested for their virulence in an Arabidopsis leaf-infiltration assay and in a mouse full-thickness burn model (8, 9). As summarized in Table 1, six (12A1, 35H7, 48D9, 50E12, 41C1, and 41A5) of the eight mutants were less pathogenic in Arabidopsis, and at least five (12A1, 48D9, 50E12, 35A9, and 44B1) were less pathogenic in mice. Mutant 35H7 (which contains a TnphoA insertion in gacA) was not tested in the mouse burn model because we showed previously that other PA14 gacA mutants exhibited reduced virulence in this model (8, 9). Mutant 41C1 also may be reduced in virulence in the mouse burn model, but a test at a lower-inoculation dose is required to reach a definitive conclusion. These data confirmed our previous conclusion that there is a large amount of overlap between the PA14 virulence factors required for pathogenesis in mice and plants and for slow killing of C. elegans (2).
Table 1.
Pathogenicity of P. aeruginosa PA14 mutants on various hosts
| Strain Isolation Number | Pathogenicity phenotypes
|
Gene in which TnphoA was inserted or closest published homologue (organism; GenBank accession no.) | ||
|---|---|---|---|---|
| C. elegans killing LT50, h* | Growth in Arabidopsis leaf† | % Mouse mortality (n) 5 × 105‡ | ||
| PA14 | 39.6 ± 2.8 | 2.9 × 107 | 100 (>16) | |
| 12A1 | >90 | 1.7 × 106 | 50 (8) | lasR (P. aeruginosa; P25084) |
| 35H7d | >90 | 1.2 × 104 | NT§ | gacA (P. aeruginosa; Q51373) |
| 48D9 | >90 | 1.0 × 104 | 50 (8) | lemA (P. syringae; P48027) |
| 50E12 | >90 | 2.0 × 105 | 0 (16) | ptsP (A. vinelandii; Y14681) |
| 35A9 | 61.2 ± 0.8 | 7.6 × 106 | 53 (17) | mtrR (N. gonorrhoeae; P39897) |
| 44B1 | 62.7 ± 2.3 | 1.0 × 107 | 56 (18) | No matches |
| 41C1 | 51.1 ± 0.3 | 2.4 × 105 | 81 (16) | aefA (E. coli; P77338) |
| 41A5 | 46.2 ± 0.4 | 1.3 × 104 | 100 (8) | No matches |
A mutant is considered attenuated in nematode pathogenicity if the mean time required to kill 50% of the worms feeding on it (LT50 from three to four replicates) is 2 SDs less than the LT50 of parental PA14 in the same experiment; for calculations of LT50 see ref. 2. A LT50 > 90 is given to mutant strains that kill less than 30% worms at 90 h.
Colony-forming unit per cm2 leaf area of bacterial counts at 4 days postinoculation of 103 bacteria; means of four to five samples. Mutants are defined as less pathogenic when the mean value of bacterial counts is 2 SDs lower relative to wild type within the same experimental set.
Six-week-old male inbred strain mice (from The Jackson Laboratories), weighing between 20 and 30 g, were injected with 5 × 105 bacterial cells. n, total number of mice tested. The number of animals that died of sepsis was monitored each day for 7 days.
Another gacA mutant 1D7 was isolated independently from a plant screen. Mutant 1D7 has been tested on mice and showed 50% mortality (9).
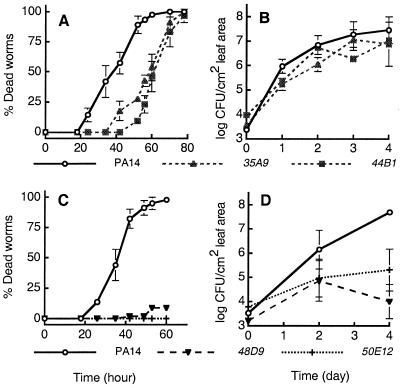
The killing of C. elegans by four representative PA14 TnphoA slow-killing mutants and the growth of these same mutants in Arabidopsis leaves are shown in Fig. 1. Mutants 35A9 and 44B1 exhibited reduced pathogenicity in nematodes but grew to wild-type levels in Arabidopsis (Fig. 1 A and B). Mutants 48D9 and 50E12 exhibited reduced pathogenicity in both plants and nematodes (Fig. 1 C and D). All four of these mutants are less pathogenic in mice.
Figure 1.
Pathogenicity phenotypes of representative P. aeruginosa PA14 TnphoA mutants on C. elegans and Arabidopsis relative to the wild-type strain. C. elegans mortality rates under slow-killing conditions mediated by P. aeruginosa PA14 TnphoA mutants are shown in A and C, and growth of the same mutants in Arabidopsis leaves are shown in B and D. (A and B) Mutants 35A9 (▴) and 44B1 (■) exhibited reduced pathogenicity in nematodes but grew the same as the parental strain, PA14 (○) in Arabidopsis. (C and D) Mutants 48D9 (▾) and 50E12 (+) exhibited reduced pathogenicity in both plants and nematodes compared with PA14 (○).
In a separate publication, we described the isolation of six additional TnphoA PA14 mutants (1G2, 3E8, 6A6, 8C12, 23A2, and 36A4) that exhibit reduced killing in the C. elegans fast-killing assay (1). Interestingly, although at least four of these mutants also exhibited reduced pathogenicity in the mouse burn model, only one [36A4, a hrpM homologue (1, 23)] exhibited a phenotype in the C. elegans slow-killing assay (data not shown). All eight mutants described in this paper that were isolated in the slow-killing assay killed C. elegans as fast as wild type under the high-osmolarity fast-killing conditions (see Materials and Methods, data not shown). These observations confirmed the conclusion reported elsewhere that slow and fast killing of C. elegans are mechanistically distinct (2).
Genetic Analysis of TnphoA Target Sites and Biochemical and Physiological Characterization of PA14 TnphoA Mutants.
To determine which genes were disrupted in the eight PA14∷TnphoA slow-killing mutants, IPCR was used to amplify DNA sequences adjacent to the sites of the TnphoA insertions in each of the mutants. DNA fragments corresponding to the IPCR products were identified, sequenced, and analyzed as described in Materials and Methods.
The DNA sequence analysis, summarized in Table 1, showed that both novel and known genes were identified. In addition to the eight mutants identified in the nematode screen, two PA14 mutants isolated previously in the plant infection model as less pathogenic, pho15 and 25F1 (9), were subjected to additional analysis because they are also attenuated in C. elegans slow killing. We reported previously that pho15 contains a TnphoA insertion in a homologue of the E. coli dsbA gene (9) and that the insertion in 25F1 did not correspond to a known gene. With further sequence analysis, we confirm the assignment of pho15 as dsbA and report that 25F1 has a TnphoA insertion in an ORF with homology to orfT from Chlorobium tepidum (see below).
A detailed description of the DNA sequence analysis and biochemical analysis of each of the eight mutants that correspond to previously described genes [12A1 (lasR), 35H7 (gacA), 48D9 (lemA), pho15 (dsbA), 25F1 (orfT), 50E12 (pstP), 35A9 (mtrR), and 41C1 (aefA)] is presented in the following sections.
Mutant 12A1.
The TnphoA insertion in 12A1 is inserted into codon 154 of the previously described lasR gene of P. aeruginosa PA01 (27). The phenotype of 12A1, as with other known lasR mutants, is pleiotropic and includes decreased elastase and protease production (27, 28). In contrast to known lasR phenotypes, however, 12A1 produces two to three times more pyocyanin than the parent PA14 strain does at stationary phase. Previously, we showed that PA14 accumulated in the lumen of C. elegans during the slow-killing assay in contrast to E. coli or a PA14 gacA mutant, which failed to accumulate (2). The lasR mutant expressing green fluorescent protein from plasmid pRR54GFP19–1 (2) also failed to accumulate in the worm gut (data not shown).
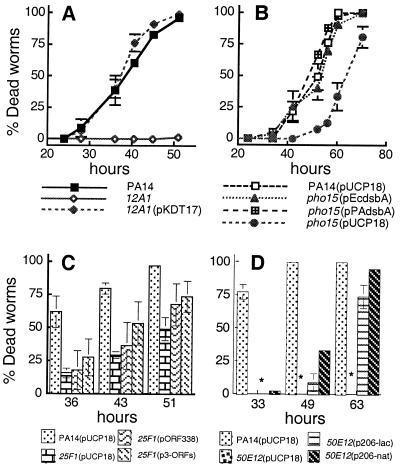
Fig. 2A shows that the defective slow-killing phenotype of 12A1 was restored completely to wild-type killing levels when the P. aeruginosa lasR gene under the control of the constitutive lacZ promoter was expressed in trans in strain 12A1 (pKDT17). Elastase production, but not the overproduction of pyocyanin (data not shown), also was restored to wild-type levels in 12A1 (pKDT17), suggesting that 12A1 may harbor a second mutation that results in the up-regulation of pyocyanin production that is not related to the attenuated pathogenicity phenotype.
Figure 2.
Complementation analysis of the ability of P. aeruginosa PA14 mutants to mediate slow killing of C. elegans. (A) Slow killing of 1-day-old adult C. elegans by wild-type PA14, 12A1, and 12A1 (pKDT17; expressing PAO1 lasR under the control of a constitutive E. coli lacZ promoter). (B) Slow killing of L4 larval-stage C. elegans by PA14 (pUCP18; vector control), pho15 (pUCP18; vector control), pho15 (pEcdsbA; expressing E. coli dsbA under the control of the constitutive lacZ promoter), and pho15 (pPAdsbA; expressing P. aeruginosa dsbA under the control of a constitutive E. coli lacZ promoter). (C) Slow killing of L4 larval-stage C. elegans by PA14 (pUCP18; vector control), 25F1 (pUCP18; vector control), 25F1 (pORF338; expressing orf338 from its native promoter), and 25F1 (p3-ORFs; expressing orf338-orf224-orf252 from their native promoters). (D) Slow killing of L4 larval-stage C. elegans by 50E12 (pUCP18; vector control), PA14 (pUCP18; vector control), 50E12 (p206-lac; expressing the putative orf159-ptsPPa operon under the control of a constitutive E. coli lacZ promoter), and 50E12 (p206-nat; expressing the putative orf159-ptsPPa operon under the control of its native promoter). ∗, Zero mortality was observed for worms feeding on 50E12(pUCP18). For all experiments shown, each data point represents means ± SD of three to four replicates. At least two independent experiments were performed for each analysis.
Mutant 35H7.
There is a TnphoA insertion within codon 188 of the previously described GacA protein in P. aeruginosa strain PA14 (8) in mutant 35H7. Mutant 35H7 was severely defective in killing C. elegans under slow-killing conditions, similar to two other gacA mutants that we had tested previously (2). We also tested a derivative of one of these latter gacA mutants, described in ref. 8, in which the mutated gacA gene had been replaced with a 2.4-kb DNA fragment containing the wild-type gacA gene. The restored wild-type strain killed C. elegans as efficiently as PA14 (data not shown).
Mutant 48D9.
TnphoA is inserted between codon 491 and 492 of a gene encoding a 925-aa homologue of the P. syringae LemA protein, a sensor kinase belonging to a family of bacterial two-component regulators (26). The cognate response regulator of LemA in P. syringae is GacA (29), and GacA + LemA have been shown to affect the expression of a variety of virulence factors in animal and plant pathogens (8, 30, 31).
Mutant pho15.
Fig. 2B shows that the pathogenicity-defective phenotype of pho15 in C. elegans was fully restored by constitutive expression of the E. coli or the PA14 dsbA gene in trans.
Mutant 25F1.
In 25F1, TnphoA is inserted within codon 100 of a putative gene (orf338) that encodes a 338-aa protein, the first gene of a putative 3-gene operon. The predicted downstream genes (orf224 and orf252) encode 224- and 252-aa proteins, respectively. orf338 is 28.5% identical (37.7% similar) to orfT of C. tepidum (GenBank accession no. U58313), the function of which is unknown. orf224 has significant homology to mannose-1-phosphate guanylyltransferases (MPGs) from eukaryotes, archaeobacteria, cyanobacteria, and mycobacteria, but it is not clear whether orf224 encodes a functional MPG because all known MPGs consist of 359–388 aa residues, whereas orf224 encodes a 224-aa polypeptide. orf252 encodes a homologue of E. coli DjlA (ref. 32; GenBank accession no. P31680), which may play a role in the correct assembly, activity, and/or maintenance of a number of membrane proteins, including the two-component histidine kinase signal-transduction systems (33).
As shown in Fig. 2C, pORF338 and p3-ORFs, which express orf338 and all three ORFs (orf338, orf224, and orf252) from their native promoters, respectively, both partially complemented the slow-killing phenotype of 25F1. These results indicate that the TnphoA insertion in orf338 likely is responsible for the pathogenicity phenotype but do not rule out the possibility of a polar effect of TnphoA insertion on downstream genes or that the downstream genes orf224 and orf252 also may play a role in PA14 virulence.
Mutant 50E12.
The TnphoA insertion in 50E12 is inserted within codon 39 of a predicted 759-aa protein that is 86.7% identical (90.4% similar) to the Azotobacter vinelandii PtsP protein (25), which is predicted to encode Enzyme INtr, a presumptive transcriptional regulator of RpoN-dependent operons (34). The A. vinelandii PtsP protein is required for accumulation of poly-β-hydroxybutyrate (25).
As shown in Fig. 2D, plasmid p206-lac, which expresses orf159 and ptsPPa under the control of the constitutive lacZ promoter, and plasmid p206-nat, in which transcription of orf159 and ptsPPa is controlled only by their native promoter (see Materials and Methods), both partially complemented 50E12. In both cases, wild-type killing levels were restored, although it took longer for the complemented strains to kill 100% of the worms than PA14. Importantly, partial complementation also was observed in the burned-mouse assay using p206-nat. Mouse mortality was 39% for the complemented strain compared with 100% and 0% mortality when infected by the wild-type strain and 50E12, respectively (data not shown).
Mutant 35A9.
The TnphoA in 35A9 is inserted in the first codon of a putative 210-aa protein (encoded by orf210) that is most closely related (31.5% identity) to the N. gonorrhoeae MtrR protein (24) that belongs to the TetR family of helix–turn–helix-containing bacterial transcriptional regulators (Prosite: PS01081). orf210 is adjacent to and divergently transcribed from three genes that are homologous to components of the energy-dependent efflux system in P. aeruginosa. Analyses of sequences from P. aeruginosa PA01 show that together, these four genes define a new energy-dependent efflux system in P. aeruginosa (M.-W.T. and F.M.A., unpublished data). The other energy-dependent efflux systems in P. aeruginosa described previously are the mexR, mexA-mexB-oprK system, the nfxB, mexC-mexD-oprJ system, and the nfxC, mexE-mexF-oprN system, which function as multidrug efflux pumps (35–37).
Mutant 41C1.
TnphoA is inserted in a homologue of the putative E. coli integral membrane protein AefA, which is a member of the UPF0003 protein family (Prosite: PS01246). The function of these proteins is not known.
DISCUSSION
By screening 2,400 TnphoA mutants of P. aeruginosa strain PA14, we have identified 8 P. aeruginosa mutants that exhibit reduced “slow” killing of C. elegans. Importantly, six of these mutants are less pathogenic in an Arabidopsis–leaf infiltration model, and at least five are less virulent in a mouse full-thickness skin burn model. These data demonstrate that the C. elegans slow-killing model can be used to efficiently identify P. aeruginosa virulence genes (factors) required for mammalian and plant pathogenesis.
An important feature of the C. elegans slow-killing screen is its ability to identify P. aeruginosa mutants that are only slightly impaired in their ability to kill C. elegans. This high degree of sensitivity is a consequence of the following features of slow killing. First, the longer it takes for hermaphrodite worms to be killed, the more progeny are produced. Second, early larval stages are apparently more resistant to killing by P. aeruginosa. Thus, attenuated bacterial mutants that delay killing only by a few hours result in the production of partially resistant progeny by the survivors, which effectively “amplifies” a weak defect into a readily observable phenotype. That is, on plates containing attenuated PA14∷TnphoA mutants, two seeded L4-stage hermaphrodites will produce hundreds of progeny worms. Severely debilitated PA14 mutants allow the production of thousands of progeny worms that completely consume the bacterial lawn, whereas no live worms or very few worms are found after seeding onto plates containing wild-type PA14. In contrast to the C. elegans screen, it would be extremely difficult to identify mildly attenuated P. aeruginosa mutants using a vertebrate model because of the large number of animals that would have to be tested to observe a statistically significant reduction in pathogenicity. Interestingly, some of the PA14 mutants that were only moderately attenuated in C. elegans killing, such as 35A9 and 44B1, were markedly less pathogenic in mice.
A variety of biochemical assays revealed that a majority of the PA14 mutants that were obtained in the C. elegans slow-killing screen were indistinguishable from the wild-type parent and would not have been identified by screening solely on the basis of a defect in a known pathogenicity-related factor. Consistent with this observation, many of the genes identified by the mutant screen are either completely novel or have not been defined previously as virulence factors in P. aeruginosa or other pathogenic bacteria.
The analysis of eight TnphoA mutants identified in this paper, nine TnphoA mutants isolated in a plant screen (9), six TnphoA mutants isolated in a C. elegans fast-killing screen (1), and three PA14 mutants that had been constructed by marker exchange in the toxA, plcS, and gacA genes (8) indicated that the mutants fall into several categories. In creating these categories, we have combined mutants that affect either fast or slow C. elegans killing for the sake of simplicity. As summarized in Table 2, 15 class I mutants corresponding to 12 different genes are less pathogenic in all 3 hosts, 2 class II mutants are less pathogenic in nematodes and mice, 5 class III mutants are less pathogenic in plants and mice, 3 class IV mutants are less pathogenic in plants and nematodes, and 1 class V mutant is less pathogenic on nematodes. Remarkably, among 23 different genes that, when mutated, are less pathogenic in nematodes or plants, 19 were identified as playing a significant role in pathogenesis in a mouse burn model. Moreover, some of the mutants that did not show a significant decease in pathogenicity in the mouse burn model, such as 41C1 (see Table 1), might have shown a defect if they had been tested at a lower-inoculation dose. Importantly, mutations in none of these genes affect the growth of PA14 in vitro in either minimal or rich medium.
Table 2.
Phenotypes of PA14∷TnphoA mutants
| Class | Phenotype | Mutant: |
|---|---|---|
| I | Less pathogenic in nematodes, plants, and mice | toxA, gacA, ID7 (gacA), 35H7 (gacA), 25F1 (orfT), 34H4, pho15 (dsbA), pho34B12, 12A1 (lasR), 48D9 (lemA), 50E12 (pstP), 3E8 (phzB), 6A6 (phzB) 8C12, 36A4 (hrpM) |
| II | Less pathogenic in nematodes and mice | 35A9 (mtrR), 44B1 |
| III | Less pathogenic in plants and mice | plcS, 33A9, 33C7, 25A12, 16G12 |
| IV | Less pathogenic in plants and nematodes; | 41A5, 23A2, 41C1 (aefA) |
| V | Less pathogenic in nematodes only | IG2 |
For most of the mutants, virulence in mice was determined at an inoculum of 5 × 105 cells, a relatively high dose that results in 100% mortality with PA14 wild type. It is conceivable that more of the mutants would be classified as being attenuated in mouse pathogenesis if a lower-inoculation dose had been used.
The summary of mutant phenotypes shown in Table 2 highlights the advantages of using several nonvertebrate hosts in screens for mutants involved in mammalian pathogenesis. As exemplified by class II mutants that are less pathogenic in nematodes and mice but not in plants and by class III mutants that are less pathogenic in plants and mice but not in nematodes, a significant proportion of the mutants that exhibited attenuated virulence in the mouse model would have been missed if only one screen in a single nonvertebrate host had been used.
To date, we have screened a total of 2,500 TnphoA-generated mutants of PA14 in a plant pathogenesis assay (9), 2,400 in the C. elegans slow-killing pathogenesis assay (this paper), and 3,300 in the C. elegans fast-killing pathogenesis assay (1). This represents approximately 25% of the total number that needs to be tested to give a 95% probability of testing each gene in each of the assays (9). Among the genes isolated from the plant and nematode screens, 37H7 and 1D7 are both alleles of gacA and 3E8 and 6A6 are both alleles of phzB. The high proportion of previously uncharacterized virulence-related genes isolated from these multihost screens and the low number of multiple alleles isolated indicate that many P. aeruginosa genes that are functionally important in pathogenicity remained to be identified.
Given the relatively large number of P. aeruginosa PA14 mutants involved in this study, it was not feasible to demonstrate for each of the mutants that the TnphoA insertion is the cause of the pathogenicity-related phenotype(s) and that it is the disruption of the ORF interrupted by TnphoA that causes the mutant phenotype(s) rather than disruption of downstream gene expression due to polar effects of the transposon insertion. Nevertheless, five mutants corresponding to three known virulence factors (gacA, lasR, and dsbA) and two novel virulence factors [orf338 (25F1) and ptsP (50E12)] were chosen for additional genetic analysis. The results, some of which are presented in Fig. 2, show that at least in the context of the C. elegans slow-killing model, the mutant phenotypes associated with insertions in these five genes could be correlated directly with the impaired killing phenotype. In two cases (dsbA and lasR), the insertions in these genes could be complemented in trans by plasmids that expressed only the mutated gene. In the case of gacA, a nonpolar insertion in gacA was shown to cause the same phenotype as a polar insertion (2) and a reconstructed wild-type gacA gene was shown to have the same level of C. elegans killing as wild-type PA14 (data not shown). In two cases, an operon containing the mutated gene (the orf159, ptsPPA operon corresponding to 50E12 and the orf338, orf224, and orf252 operon corresponding to 25F1) was able to partially complement the impaired killing phenotype. Moreover, in the case of 50E12, partial complementation also was obtained in the mouse burn model.
Among known virulence factors, the C. elegans screen identified LasR, LemA, and GacA as important for pathogenicity in nematodes, plant, and mice (Table 2, class I). Interestingly, these proteins form a hierarchical cascade of interacting proteins, regulating the transcription and export of virulence factors, including ToxA (another class I factor) (28), via the type II secretion machinery (38, 39). LasR has been identified previously in P. aeruginosa strain and, together with LasI, is one of two quorum-sensing systems found in P. aeruginosa. The lasR-lasI system is a global regulator of many virulence-associated genes (40). GacA, together with its cognate sensor, LemA, are members of a two-component signaling pathway. GacA functions upstream of the lasR-lasI modulon in a complex, cell-density-dependent signal-transduction pathway regulating several exoproducts and virulence factors (41). Thus, we show that the C. elegans screen is efficient in identifying several interacting virulence-associated systems that appear to be universal among Gram-negative animal and plant bacterial pathogens. In addition, we show that gacA and lasR mutants fail to accumulate in the gut of C. elegans, suggesting that the establishment and/or proliferation of bacteria within the host also may be dependent on this regulatory cascade. Further screening using plants and nematodes as host likely will identify other components of this complicated virulence-related pathway as well as the effector molecules that are under the regulation of these genes.
From an evolutionary perspective, the requirement for functional lasR, lemA, and gacA gene products as pathogenicity factors in plants, nematodes, and mice suggests that quorum sensing and regulated export of proteins are general features of pathogenesis in all hosts. The lasR, lemA, and gacA genes are present in many plant and animal bacterial pathogens as well as in saprophytes. We speculate it is likely that LasR, LemA, and GacA initially served as master regulators, enabling ancestral prokaryotic organisms to adapt to their environment. Later, with the appearance of eukaryotes, LasR, LemA, and GacA evolved to regulate a variety of genes that allowed prokaryotes to invade and establish residency inside eukaryotes.
In conclusion, the work presented here in combination with two other recent publications from our laboratory (1, 2) show that the C. elegans–P. aeruginosa pathogenesis model that we have developed has a variety of useful features currently not found in any other pathogenesis models, including those utilizing A. thaliana (42) and D. melanogaster (43) as model genetic hosts, in which genetic dissection is limited largely to the host because genome sequencing of their pathogens have not been initiated. These features include the ability to carry out high-throughput genetic identification of both pathogen and host genes involved in the pathogenic interaction between a pathogen and a host for which complete genome sequences are now available.
Acknowledgments
We thank B. H. Iglewski for kindly providing pKDT17, C. Guilhot for pCH3, and J. Morris for help with bioinformatics. This work was supported by a grant from Shriner’s Hospitals for Children and by a grant from Hoechst AG to Massachusetts General Hospital.
ABBREVIATIONS
- NG
nematode growth
- IPCR
inverse PCR
Footnotes
References
- 1.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 2.Tan M-W, Mahajan-Miklos S, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Wood R E. Hosp Pract. 1976;11:91–100. doi: 10.1080/21548331.1976.11706983. [DOI] [PubMed] [Google Scholar]
- 5.Elrod R P, Braun A C. Science. 1941;94:520–521. doi: 10.1126/science.94.2448.520. [DOI] [PubMed] [Google Scholar]
- 6.Lysenko O. J Insect Pathol. 1963;5:78–82. [Google Scholar]
- 7.Schroth M N, Cho J J, Green S K, Kominos S D. In: Pseudomonas aeruginosa: Ecological Aspects and Patient Colonization. Young V M, editor. New York: Raven; 1977. [Google Scholar]
- 8.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 9.Rahme L G, Tan M-W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King E O, Ward M K, Raney D E. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 13.Schweizer H P. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 14.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith A W, Iglewski B H. Nucleic Acids Res. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 18.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gish W, States D J. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 20.Toder D S, Ferrell S J, Nezezon J L, Rust L, Iglewski B H. Infect Immun. 1994;62:1320–1327. doi: 10.1128/iai.62.4.1320-1327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essar D W, Eberly L, Hadero A, Crawford I P. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacherer P, Défago G, Haas D. FEMS Microbiol Lett. 1994;116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay P, Williams J, Mills D. J Bacteriol. 1988;170:5479–5488. doi: 10.1128/jb.170.12.5479-5488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan W, Spratt B G. Mol Microbiol. 1994;11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 25.Segura D, Espin G. J Bacteriol. 1998;180:4790–4798. doi: 10.1128/jb.180.18.4790-4798.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hrabak E M, Willis D K. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambello M J, Iglewski B H. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gambello M J, Kaye S, Iglewski B H. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rich J J, Kinscherf T G, Kitten T, Willis D K. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J P, Normark S. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]
- 31.Frederick R D, Chiu J, Bennetzen J L, Handa A K. Mol Plant–Microbe Interact. 1997;10:407–415. doi: 10.1094/MPMI.1997.10.3.407. [DOI] [PubMed] [Google Scholar]
- 32.Clarke D J, Jacq A, Holland I B. Mol Microbiol. 1996;20:1273–1286. doi: 10.1111/j.1365-2958.1996.tb02646.x. [DOI] [PubMed] [Google Scholar]
- 33.Kelley W L, Georgopoulus C. Mol Microbiol. 1997;25:913–931. doi: 10.1111/j.1365-2958.1997.mmi527.x. [DOI] [PubMed] [Google Scholar]
- 34.Reizer J, Reizer A, Merrick M J, Plunkett G R, Rose D J, Saier M H., Jr Gene. 1996;181:103–108. doi: 10.1016/s0378-1119(96)00481-7. [DOI] [PubMed] [Google Scholar]
- 35.Li X-Z, Nikaido H, Poole K. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 37.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 38.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunski A. FEMS Microbiol Rev. 1992;103:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 39.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 40.Pesci E C, Iglewski B H. Trends Microbiol. 1997;5:132–134. doi: 10.1016/S0966-842X(97)01008-1. , and discussion, 134–135. [DOI] [PubMed] [Google Scholar]
- 41.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 42.Glazebrook J, Rogers E E, Ausubel F M. Annu Rev Genet. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann J A, Reichhart J-M. Trends Cell Biol. 1997;7:309–316. doi: 10.1016/S0962-8924(97)01087-8. [DOI] [PubMed] [Google Scholar]