Abstract
Protein synthesis is catalyzed in the peptidyl transferase center (PTC), located in the large (50S) subunit of the ribosome. No high-resolution structure of the intact ribosome has contained a complete active site including both A- and P-site tRNAs. Additionally, though structures of the 50S subunit found no ordered proteins at the PTC, biochemical evidence suggests specific proteins are capable of interacting with the 3′ ends of the tRNA ligands. Here we present structures at 3.5 Å and 3.55 Å resolution of the 70S ribosome in complex with A- and P-site tRNAs that mimic pre- and post-peptidyl transfer states. These structures demonstrate that the PTC is very similar between the 50S subunit and the intact ribosome. Additionally they reveal interactions between ribosomal proteins L16 and L27 and the tRNA substrates, helping to elucidate the role of these proteins in peptidyl transfer.
Introduction
The ribosome is responsible for the synthesis of proteins in all organisms. It is composed of two subunits (designated 50S and 30S in bacteria), contains three RNAs, and approximately 50 proteins. The structure of the 50S ribosomal subunit from the archaeon Haloarcula marismortui found no ordered proteins within 18 Å of the active site of the enzyme 1. Subsequent structural studies on the 50S subunit using substrate analogs 2, as well as biochemical studies on the 70S ribosome 3, all suggest that the ribosome is an RNA-based enzyme. However, in the absence of a structure of the intact ribosome containing ordered A- and P-site substrates within the peptidyl transferase center (PTC), two questions important to our understanding of catalysis by the ribosome remain: Are there any structural differences in the PTC when studied in the context of the intact 70S ribosome compared to the 50S subunit alone? What is the role of proteins located within the PTC in catalyzing peptidyl transfer?
Structurally, there has been disagreement as to the validity of studies performed on the 50S subunit alone. Differences between the conformation of the PTC in the intact 70S ribosome compared to that in the 50S subunit have been reported 4. Additionally the relevance of small oligonucleotide mimics of tRNA used in structural studies of the PTC in the 50S subunit has been questioned 5.
Biochemically, several previous studies have suggested that in E. coli, specific ribosomal proteins are located at or near the active site. In bacteria, the protein L27 can be cross-linked to the 3′ end of both the A- and P-site tRNAs in the peptidyl transferase center 6,7. Deletion of L27, or even its first three N-terminal residues, reduces the rate of peptidyl transfer 8. Additionally, deletion of L16, a protein predicted to interact with the A-site substrate, causes defects in tRNA binding and the rate of peptidyl transfer 9,10. These data suggest that, at least in bacteria, proteins play a supporting role in catalysis by the ribosome, and are closer to the active site than previously reported. However no structural data have yet elucidated the possible role of these proteins in peptidyl transfer.
A recent high-resolution structure of the 70S ribosome revealed weak density for the N-terminal tail of L27 (ref. 11), which localizes between the 3′ ends of the A- and P-site tRNAs. However no density was observed for the side chains of the first nine amino acids, precluding any conclusions about detailed interactions of L27 with the tRNAs in the PTC. Furthermore, the A-site tRNA was almost entirely disordered, with the exception of the anticodon stem loop in the 30S subunit. The absence of an ordered A-site tRNA in the PTC prevented study of the complete 70S ribosomal active site, as well as any interactions between the aminoacyl tRNA and protein L16.
Here we report two structures of the 70S ribosome from Thermus Thermophilus with a complete peptidyl transferase center including an A-site tRNA for which the CCA and amino acid at its 3′ end are ordered within the PTC. Together, these structures provide a more complete description of the ligand-bound ribosome. Furthermore, density for the N-terminal tail of L27 as well as that for protein L16 were clearly resolved, thus revealing their interactions with the ribosomal substrates. These interactions provide insights into the role of these proteins in facilitating peptidyl transfer by the ribosome. Finally, the structures should dispel concerns about the validity of previous structural work on the PTC using the 50S subunit.
Results
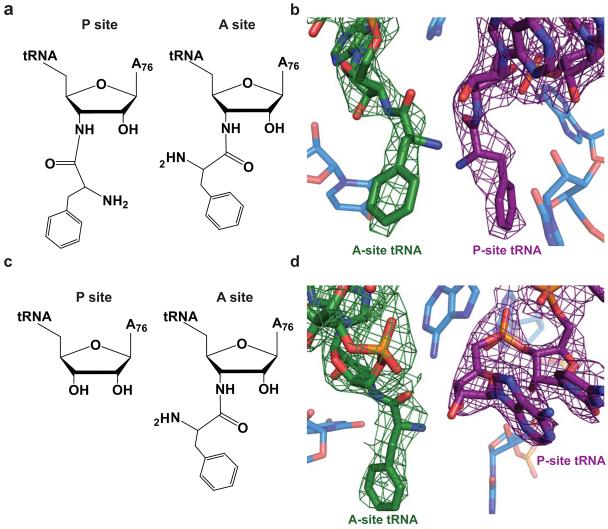
The crystal structures reported here were determined using modified tRNAs in which the ester linkage between the amino acid and A76 was replaced with an amide. Due to its greater stability, these substrates allow the crystallization of aminoacylated tRNAs without appreciable deacylation. Using these modified tRNAs, we have successfully determined two structures of the 70S ribosome: in the first, both A- and P-site tRNAs are aminoacylated as they would be just prior to peptidyl transfer (Figure 1a, b); the second contains a deacylated P-site tRNA, mimicking the state just following peptidyl transfer but prior to translocation (Figure 1c,d). Density for both the A- and P-site tRNAs and mRNA was observed.
Figure 1.
Ribosomal substrates in the peptidyl transferase center. a) Chemical diagram of the pre-peptidyl transfer state of the ribosomal active site. In this structure both the A- and P-site tRNAs contain an amide linkage between residue A76 and the phenylalanine amino acid. b) Model of the ribosomal active site in the pre-peptidyl transfer state including representative 3Fo-2Fc density for the A- and P-site tRNAs in green and purple, respectively. c) Chemical diagram of the post-peptidyl transfer state in which the A site contains an amide-linked Phe-tRNAPhe and the P site contains tRNAfMet. d) Model of the post-peptidyl transfer in the peptidyl transferase center including 3Fo-2Fc density for the A- and P-site tRNAs in green and purple, respectively.
The P-site tRNA, as in earlier studies, is well ordered in both structures. Interactions between the P-site tRNA and its nearby proteins such as S9, S13 and L5, as well as Helix 69 of the 23S RNA appear unchanged as a result of A-site binding. Additionally, the previously reported 11 distortion of the major groove of the P-site tRNA that occurs upon binding to the ribosome is observed in both structures reported here. In both the pre- and post-peptidyl transfer structures, though regions of the A-site tRNA appear to be dynamic, clear density was observed for the A-site anticodon stem loop (ASL), CCA-tail and amino acid, and acceptor arm (Supplementary Fig. 1 online). Only weak density for the E-site tRNA was observed in either structure. Comparison of the pre- and post-peptidyl transfer structures reveals no specific changes within the PTC between the two states.
A-site binding interactions
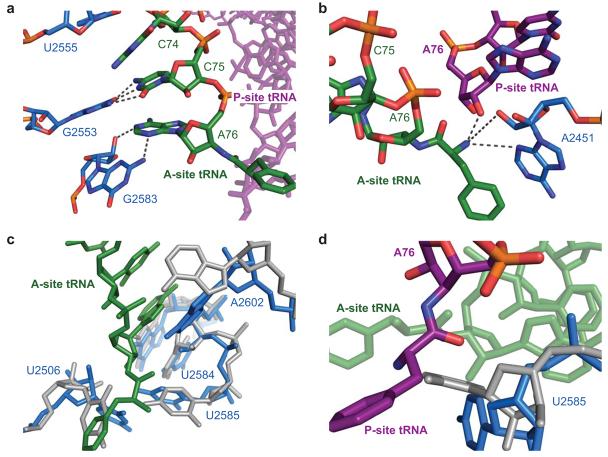
In both structures, stabilizing interactions between the 3′ end of the A-site tRNA and the ribosome were observed. C74 stacks with U2555 (E. coli numbering is used throughout) of the 23S RNA. C75 forms a Watson-Crick base pair with residue G2553 12 and A76 forms a Class I A-minor interaction with the G2583 (ref. 13) (Figure 2a). These interactions stabilize the CCA tail of the A-site tRNA, orienting the amino acid for nucleophilic attack on the peptidyl-ester. Additionally, the α-amine is within hydrogen bonding distance of both N3 and the 2′OH of residue A2451, as well as the 2′OH of A76 of the peptidyl-tRNA (Figure 2b).
Figure 2.
Interactions of the ribosomal substrates with the 23S RNA in the peptidyl transferase center. a) Model of the ribosome and the stabilizing interactions between the CCA tail of A-site tRNA, shown in green, and the 23S RNA, shown in light blue as determined in the pre-peptidyl transfer structure. b) Interactions of the A-site amino acid backbone with the 23S RNA, shown in blue, and the peptidyl-tRNA, shown in purple. For ease of illustration, the structure of the post-peptidyl transfer state of the ribosome is displayed. c) Superposition of the 23S RNA from Selmer et al. 11 (displayed in gray), which contains an empty 50S A site, with that of the pre-peptidyl transfer structure presented here, containing an occupied A site (displayed in blue). Shifts in residues U2584, U2585, U2506, and A2602 are characteristic of A-site accommodation. d) Model of the conformational change of residue U2585 that, upon binding of A-site tRNA, exposes the peptidyl-tRNA ester for nucleophilic attack.
Induced changes upon A-site binding
In both structures a series of conformational changes in the 23S RNA, characteristic of A-site accommodation, are observed (Figure 2c,d) (Supplementary Video 1 online). Prior to binding of an A-site substrate U2585 is positioned to protect the peptidyl ester from hydrolysis (Figure 2d). Following binding of an A-site substrate, U2506 shifts in order to avoid a steric clash with the A-site amino acid. Residue U2584 shifts slightly in order to maintain stacking and to allow G2583 to form an A-minor interaction with A76 (Figure 2c). Residue A2602 has also shifted as a result of A site binding, and residue U2585 swings 90° away from the P site, exposing the peptidyl-tRNA ester for nucleophilic attack (Figure 2d) 14.
Structure and Interactions of the N-terminal tail of protein L27
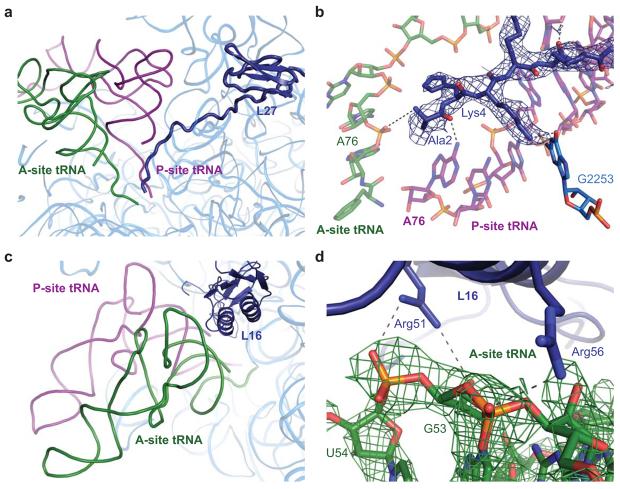
The N-terminal tail of L27 was known from previous high resolution structures 11 to localize between the 3′ ends of the A- and P-site tRNAs (Figure 3a). In both structures reported here, clear density was observed for the amino acid side chains of the protein, making it possible to model the entire N-terminal tail (Figure 3b). No density was detected for the N-terminal methionine of L27, suggesting it is not present in the reported structures. Stabilizing interactions between L27 and the 23S RNA and peptidyl-tRNA were observed (Figure 3b). Lys4 is within hydrogen bonding distance of the O6 of G2253 of the 23S RNA. Ala2 and Gly6 of L27 are also positioned to interact with the peptidyl-tRNA through the O2 of residue C1 and a non-bridging phosphate oxygen of G65. Additionally, there is a possibility that the N-terminal amine of residue Ala2 and the non-bridging phosphate oxygen of A76 of the A-site tRNA could form an interaction, though it is not directly visible in this structure (Figure 3b).
Figure 3.
Interactions of the ribosomal proteins L27 and L16 with the ribosomal substrates. a) Overview of protein L27 in relation to the A- and P-site tRNAs, shown in green and purple, respectively. The protein, shown in dark blue, contains a globular domain and an N-terminal extension that localizes between the 3′ ends of the ribosomal tRNAs. b) Predicted interactions of protein L27 with the ribosomal substrates and 23S RNA (shown in light blue). The modeled interactions were observed in both structures containing occupied A sites, though the post-peptidyl transfer structure is displayed here as it contained moderately better electron density for L27. A representative 3Fo-2Fc electron density map is displayed in blue. c) Overview of protein L16 in relation to the ribosomal substrates. The protein is located adjacent to the elbow of the A-site tRNA. d) Interactions between the conserved residues Arg51 and Arg56 of protein L16, shown in dark blue, with the backbone of the A-site tRNA, shown in green. Representative 3Fo-2Fc density, as determined in the pre-peptidyl transfer structure, is displayed in green for the region of the A-site tRNA predicted to interact with L16.
Interactions between protein L16 and the aminoacyl-tRNA
The protein L16 is positioned directly above the elbow of the A-site tRNA (Figure 3c). In both structures, though most clearly in the post-peptidyl transfer structure, the acceptor stem of the A-site tRNA is well ordered and clear density was observed for the region predicted to contact L16 (Figure 3d). Two well-conserved arginine residues, Arg56 and Arg51, appear to interact with the backbone of the A-site tRNA. An interaction between Arg56 and the non-bridging phosphate oxygen of G53 was observed. Additional interactions between the N2 of Arg51 and the non-bridging phosphate oxygen of U54 and the 2′OH of G53 were also observed.
Discussion
This study attempts to answer some important remaining questions about the nature of the PTC of the ribosome. What is the structure of the PTC when bound by full-length A- and P-site tRNA substrates within the intact 70S ribosome, and how similar is this to the PTC of the 50S subunit? Are small oligonucleotide mimics of tRNA appropriate models for the tRNA substrates at the PTC? What, if any, is the role of proteins in peptidyl transfer?
In order to address these questions, it was necessary to obtain a structure of the 70S ribosome in complex with ordered A-site tRNA substrates. In the previous 70S structure from our laboratory11, all but the anticodon stem loop of the A-site tRNA was disordered, which was likely due in part to its spontaneous deacylation during crystallization. Additionally, the low concentration of Mg2+ that remained after cryoprotection of these crystals likely also destabilized the deacylated tRNA further. Using a chemically synthesized adenosine analog, acylated tRNAs were produced containing an amide linkage between the 3′ terminal A76 and the amino acid. This modification ensured that the amino acid linkage would be stable throughout crystallization, while also trapping complexes in a pre- or post-peptidyl transfer state. Further, this allowed for crystallization of these structures with the native α-amino group of the A-site tRNA intact, while previous 50S structures used a hydroxyl mutation to slow the rate of reaction 14.
Similarity of the PTC in the ribosome and the isolated 50S subunit
The interactions in the 70S ribosome of the A- and P-site tRNA substrates with the PTC are very similar to those that have been observed in the context of the 50S subunit 14 (1VQN, Supplementary Fig. 3 online). Additionally, while previous structures have used a hydroxyl group in place of the native α-amine, the structures presented here were crystallized with an unaltered amino acid. However, all of the interactions described in previous 50S structures between the backbone of the A-site amino acid were also observed in the context of the 70S ribosome (Figure 2b). Thus the positioning of the α-amino group in relation to the 2′OH of A76 of the peptidyl-tRNA is consistent with a substrate-assisted mechanism for catalysis 15. These structures are also consistent with the proposed proton-shuttle mechanism 16 in which the 2′OH of A76 simultaneously accepts and donates a proton from the α-amine to the 3′O leaving group.
Thus, we do not observe any fundamental differences between the PTC in the 50S subunit when compared to the 70S ribosome. This conclusion is further supported by kinetic studies that showed the rate of peptidyl transfer by the 50S subunit was comparable to that of the intact ribosome provided that full-length tRNA substrates were used rather than puromycin or C-puromycin 17. The validity of using small oligonucleotides to mimic tRNAs for structural study of the 50S subunit has also been questioned 5. While it appears that full-length tRNAs form additional interactions with the ribosome that enhance the rate of peptidyl transfer, their conformation within the PTC is very similar to that of the oligonucleotide mimics previously studied in the context of the 50S subunit 14.
Interactions of the 3′ ends of tRNAs with the PTC
The interactions described here between the 70S ribosome and its A-site substrate are independent of tRNA identity, as they rely only on the conserved CCA tail and amino acid backbone. Given that the ribosome uses more than 20 different aminoacyl-tRNA substrates, it seems reasonable that the primary interactions with both A- and P-site tRNAs do not require the amino acid side chain, as it varies substantially across the library of amino acids. However there is biochemical evidence that the presence, and type, of amino acid side chain can substantially affect tRNA binding, particularly in the A site of the ribosome 18,19. It was initially demonstrated that Phe-tRNAPhe had a substantially higher affinity for the A site 20 than that of deacylated tRNAPhe 21,22. This observation cannot be attributed to interactions with the amino acid backbone alone, as it was later shown that the specific amino acid side chain had substantially different effects on the stability of binding of the full tRNA 18. On the other hand, some studies have suggested that there is no difference in binding affinity for acylated vs. deacylated tRNA in the P site of the ribosome 18,19. In a previous 50S structure, it was noted that an esterified tyrosine in the A site could stack on residue A2451 and possibly form a hydrogen bond with residue G2061 1. In the structures reported here, no specific stacking interactions with the phenyl ring in the A site could be determined. However, the A-site binding pocket is notably composed of hydrophobic and aromatic residues. Further, while the phenylalanine side chain was clearly resolved in the A site even in initial unbiased difference maps, density for the phenyl ring was much less pronounced in the P site. This may arise from an inherent flexibility of the side chain in the P site making it less likely to contribute to binding, consistent with the existing biochemical data.
Accommodation of A-site tRNA
In addition to the expected interactions of the aminoacyl-tRNA with the ribosome, conformational changes characteristic of A-site accommodation were also observed. Upon binding of an A-site substrate, a series of conformational changes in the 23S RNA expose the peptidyl-tRNA ester for nucleophilic attack by the α-amine. Accommodation is induced by the binding of any A-site substrate containing at least residue C74 23,24 consistent with observations that binding of even a deacylated tRNA in the A site increases the rate of hydrolysis of the peptidyl-tRNA. The structures presented here confirm that similar changes occur upon binding of an A-site tRNA on both the 50S subunit as well as the intact ribosome. Recent crystal structures of termination complexes containing release factors RF1 25 and RF2 26 in complex with the 70S ribosome, observed similar changes upon their binding. In particular residue U2585, which protects the peptidyl ester when the A site is unoccupied, shifts to the induced conformation upon binding of both RF1 and RF2. Thus, as suggested previously 14, induced conformational changes in 23S RNA that expose the peptidyl-tRNA ester for hydrolysis are a feature of both peptidyl transfer and peptide release.
In both structures containing accommodated full-length A-site substrates, only weak density was observed for the E-site tRNA. In previous structures determined using the same crystallization conditions, strong E-site density was observed for a 70S ribosome lacking an ordered 50S A-site tRNA 11, or containing RF2 in the A site 26. Previous structures showing ligands in both the E and A sites have generally contained a sub-stoichiometric amount of tRNA or a protein factor in the A site. More studies need to be performed to determine whether the lower stoichiometry observed in the E site here is related to previous suggestions of a general allostery between A and E sites 27.
The role of proteins in peptidyl transfer
The structures reported here confirm that catalysis by the ribosome is essentially carried out by its RNA components. However both structures suggest that at least two proteins, L27 and L16, play a role in facilitating peptidyl transfer. No interactions between proteins and substrates at or near the PTC were observed in structures of the 50S subunit from the archaeon Haloarcula marismortui 1,28. However archaea do not contain a homolog of L27, which is known to cross-link to the 3′ ends of the tRNA substrates in bacteria 7,29. Further, the relevant regions of protein L10e, an archaeal homolog of L16, were disordered in these 50S structures, possibly because they lacked a full-length tRNA substrate. In particular, structural alignment of L10e with protein L16 shows that L10e contains an extended loop that is not present in bacteria (Supplementary Fig. 2a online). This loop is well conserved in both sequence and length across diverse sub-kingdoms of archaea (Supplementary Fig 2b. online) and is predicted to localize between the acceptor-arms of the A- and P-site tRNAs. However no structural data for this region of L10e is available, as the loop region has not been modeled in any studies of the archaeal 50S subunit 28.
The N-terminal tail of L27 stabilizes the tRNA substrates in the PTC
The reported structures suggest that the N-terminal tail of L27 may be involved in stabilizing the 3′ ends of the ribosomal substrates. Use of an amide-linked tRNA appears to have stabilized the CCA conjugated amino acid in the A site, possibly helping to order L27. The improved density for L27 in both structures suggests that it lacks the N-terminal methionine, unlike a previous ribosome structure from our laboratory 11. The retention of the N-terminal methionine of L27 varies widely across species 30-32 and the existing mass spectrometry data from T. Thermophilus were also inconclusive 33.
The improved density for this region of the protein also made it possible to elucidate interactions between the protein and the peptidyl-tRNA. As the observed interactions required only the first 6 amino acids of L27, the structures are consistent with cross-linking experiments that localized the contact between L27 and the P-site tRNA to the amino-terminus of the protein 8. It was shown that removal of the first 3-6 residues of L27 substantially reduced the cross-linking efficiency between the protein and P-site tRNA, and deletion of the first 9 residues abolished cross-linking entirely.
An interaction may also be possible between the N-terminal amino acid of L27 and A76 of the A-site tRNA. In these two ground-state structures, the N-terminal amine of residue Ala2 appears to be ∼4 Å from the phosphate of A76, just outside hydrogen-bonding distance. This interaction between L27 and the A-site tRNA was predicted by a recent molecular dynamics study 34, and would also explain observations that deletion mutants of L27 are defective in A-site tRNA binding 6. If indeed this interaction were possible, L27 could not only stabilize each ribosomal substrate individually, but could also help to position their 3′ ends relative to each other to facilitate peptidyl transfer, explaining the reduction in peptidyl transfer when the protein or even its first three N-terminal residues are removed 8. In archaea, the extended loop of L10e that is disordered in structures of the 50S subunit is expected to localize near the acceptor arms of the A- and P-site tRNAs. It is tempting to speculate that this loop plays the role of the N-terminal tail of L27, thus providing an explanation for the nearly universal conservation of specific residues within this region of L10e (Supplementary Fig. 2a,b).
The role of L16 in facilitating aminoacyl-tRNA binding
Along with elucidating the role of protein L27, the structures suggest that ribosomal protein L16 may also stabilize the binding of A-site tRNA. It has been known for many years that L16 is required for peptidyl transferase activity 9,35 and is important for binding of A-site substrate 10. From previous lower resolution structures, interactions between L16, located above the elbow of the aminoacyl-tRNA, and the A-site substrate have been predicted 36 (Figure 3c). With the high-resolution structure of the ribosome containing a full A-site substrate presented here, we can describe the details of these interactions. Two conserved arginine residues, Arg51 and Arg56, were observed to interact with the elbow region of the A-site tRNA. Interactions between the A-site tRNA and Arg56 had additionally been predicted through a recent NMR study 37. Interestingly all contacts were non-sequence specific, involving only backbone phosphates and ribose 2′OHs. Thus these interactions, which can form with any tRNA that binds to the ribosomal A site, rationalize the longstanding biochemical data in which the mutation of L16 specifically decreases the affinity of A-site tRNA for the ribosome 10.
Interactions between protein and the elbow region of the A-site tRNA may be a conserved mechanism for stabilizing tRNA binding across the various kingdoms. For example, regions of the archaeal protein L10e share a similar fold to L16. Moreover, residues 51 and 56 of L10e are highly conserved arginines across all archaea 38 and therefore may interact with the A-site tRNA in a similar manner to that seen in these structures.
The data reported here represent structures of the 70S ribosome with ordered, full-length A- and P-site tRNAs in the peptidyl transferase center. They show that the structure of the peptidyl transferase center and the conformation of the tRNA substrates in the intact ribosome are very similar to those seen previously in the context of the 50S subunit using small oligonucleotide mimics of the aminoacyl ends of tRNA. Furthermore they elucidate interactions between the proteins L16 and L27 and the ribosomal substrates. This study provides direct structural evidence for the role of these two proteins in stabilizing A- and P-site tRNA binding, thereby facilitating peptidyl transfer by the ribosome.
Methods
Purification of 70S ribosomes, tRNA, and mRNA
Thermus Thermophilus 70S ribosomes and deacylated E. coli tRNAfMet were purified as previously described 11. mRNA was obtained from Dharmacon of sequence
5′ GGC AAG GAG GUA AAA AUG UUC AAA 3′ for complexes containing Phe-tRNAPhe in the A site and tRNAfMet in the P site and of sequence
5′ GGC AAG GAG GUA AAA UUC UUC AAA 3′ for complexes containing Phe-tRNAPhe in the A and P sites.
Synthesis of modified 3′-Amino-3′-deoxy tRNAs
All modified Phe-tRNAPhe containing an amide linkage between A76 and the amino acid, were obtained as previously reported with minor modifications 39. The 3′-amino-3′-deoxy adenosine nucleoside was synthesized as previously described 40, using the published synthetic scheme through step 6. The nucleoside triphosphate was then prepared as described by Ludwig 41, and deprotected in H2O using H2/10%Pd/C overnight. tRNAPhe was purified from E. coli and the 3′ terminal AMP was removed through incubation with snake venom phosphodiesterase (10 ug mL−1 of reaction) at 37° C. 3′-Amino-3′-deoxy adenosine was incorporated by incubation of the modified adenosine triphosphate with CCA adding enzyme in a buffer of 50 mM glycine pH9 and 10 mM MgCl2 as described 39. The full length tRNA was then acylated enzymatically 11, resulting in a final yield of ∼35-50%.
Complex formation and crystallization
Complexes were formed as previously described in buffer G (5 mM HEPES pH 7.5, 10 mM MgoAc, 50 mM KCl, 10 mM NH4Cl, and 6 mM β-mercaptoethanol) 11. Ribosomes were incubated with a 2-fold excess of mRNA for 6 min and a 4-fold excess of the P and A-site tRNAs for 30 min each at 55° C. Paromomycin was added to a final concentration of 100 μM and complexes were incubated for 20 min at room temperature. Following addition of Deoxy Big Chap (Hampton Research) to a concentration of 2.8 mM, crystals were grown via vapor diffusion in sitting drop trays through addition of 2 μL reservoir solution (0.1 M Tris-HAc pH 7, 0.2 M KSCN, 3.5-5.5% (w/v) PEG20K and 3.5-5.5% (w/v) PEG550MME) with 2.4 μL of the 70S complex. Crystals were grown at 20° C over 3 weeks. Crystals of the pre-peptidyl transfer state were cryo-protected stepwise until reaching a final solution of 0.1 M Tris-HAc pH 7, 0.2 M KSCN, 5% (w/v) PEG20K, 10 mM MgoAc, and 30% (w/v) PEG550MME in which they were incubated overnight. Crystals of the post-peptidyl transfer state were cryo-protected stepwise until reaching a final solution of 0.1 M Tris-HAc pH 7, 0.2 M KSCN, 5% (w/v) PEG20K, and 30% (w/v) PEG400 in which they were incubated for 1 hour. Crystals were then harvested and frozen by plunging into liquid nitrogen, and data were collected at 100 K.
Data collection and refinement
X-ray diffraction data were collected at X06SA at the Swiss Light Source, Villigen, Switzerland. Data were collected on several regions of any given crystal and the exposure time was adjusted to ensure that a complete data set could be obtained from a single crystal with minimal radiation damage. All data were integrated and scaled using XDS 42. The previous high resolution 70S structure 11 without its tRNA and mRNA ligands was used as a starting model for refinement. Refinement was conducted using CNS first through a rigid body refinement of each of the two 70S molecules in the asymmetric unit; an additional rigid body refinement where each domain of the ribosome, the tRNAs and ribosomal proteins were defined as separate rigid-body groups, followed by two rounds of energy minimization and B-factor refinement. The tRNA and mRNA ligands were built into the unbiased difference density from the initial round of refinement and the described refinement scheme was performed after the addition of each ligand. The amino acids attached to the tRNA were omitted until the remainder of the active site had been correctly built, and were then placed into the unbiased difference density using previous 50S structures as a guide 43. The N-terminal tail of L27 was initially refined with the polyalanine model published in our previous structure 11. A registry error was corrected and side chains were placed based on difference Fourier maps, which showed clear positive difference density for the amino acid side chains for the first 9 residues. The side chain orientation was confirmed by difference Fourier density obtained from a parallel refinement in which the first 15 residues of L27 were completely omitted from the initial model. Data and refinement statistics are reported in Table 1.
Table 1.
Summary of crystallographic data and refinement
| Pre-peptidyl transfer | Post-peptidyl transfer | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell Dimensions | ||
| a, b, c (Å) | 210.864, 450.456, 628.880 | 212.13, 450.80, 629.62 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0-3.4 (3.4-3.5) | 50.0-3.3 (3.4-3.3) |
| Rsym (%) | 20.2 (97.2) | 18.2 (188.6) |
| I/σI | 7.42 (1.04)* | 9.08 (0.97)** |
| Completeness (%) | 97.1 (75.0) | 99.4 (99.2) |
| Redundancy | 5.60 (2.48) | 5.30 (5.22) |
| Refinement | ||
| Resolution (Å) | 50.0-3.5 | 50.0-3.3 |
| No. reflections | 789,129 | 890,245 |
| Rwork/Rfree | 22.12/27.05 | 23.44/28.16 |
| No. atoms | ||
| Protein | 45311 | 45311 |
| RNA | 98088 | 98077 |
| Ions | 743 | 743 |
| B-factors | ||
| Protein | 110 | 119 |
| RNA | 97 | 105 |
| Ions | 75 | 77 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.007 |
| Bond angles (°) | 1.5 | 1.2 |
I/Iσ=2 at 3.55 Å resolution
I/Iσ=2 at 3.5 Å resolution
Supplementary Material
Acknowledgements
We thank Martin Schmeing for guidance with refinement and interpretation of the data, Elaine Stephens for mass spectrometry work, Jesse C. Cochrane for critical review of the manuscript, and Martin Fuchs and Clemens Schulze-Briese for their advice and help with data collection at the Swiss Light Source. This work was supported by the Medical Research Council UK, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. RV is the recipient of a Gates-Cambridge scholarship.
Footnotes
Accession codes
The atomic coordinates and structure factors of the pre-peptidyl transfer and post-peptidyl transfer structures have been deposited in the PDB with accession codes XXXX and YYYY, respectively. They will also be made available on http://www.mrc-lmb.cam.ac.uk/ribo/
References
- 1.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 2.Steitz TA. Structural insights into the functions of the large ribosomal subunit, a major antibiotic target. Keio J Med. 2008;57:1–14. doi: 10.2302/kjm.57.1. [DOI] [PubMed] [Google Scholar]
- 3.Rodnina MV, Beringer M, Wintermeyer W. Mechanism of peptide bond formation on the ribosome. Q Rev Biophys. 2006;39:203–25. doi: 10.1017/S003358350600429X. [DOI] [PubMed] [Google Scholar]
- 4.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 5.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 6.Wower IK, Wower J, Zimmermann RA. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J Biol Chem. 1998;273:19847–52. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 7.Wower J, Hixson SS, Zimmermann RA. Labeling the peptidyl transferase center of the Escherichia coli ribosome with photoreactive tRNA(Phe) derivatives containing azidoadenosine at the 3′ end of the acceptor arm: a model of the tRNA-ribosome complex. Proc Natl Acad Sci U S A. 1989;86:5232–6. doi: 10.1073/pnas.86.14.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005;20:427–35. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Moore VG, Atchison RE, Thomas G, Moran M, Noller HF. Identification of a ribosomal protein essential for peptidyl transferase activity. Proc Natl Acad Sci U S A. 1975;72:844–8. doi: 10.1073/pnas.72.3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazemie M. Binding of aminoacyl-tRNA to reconstituted subparticles of Escherichia coli large ribosomal subunits. Eur J Biochem. 1976;67:373–8. doi: 10.1111/j.1432-1033.1976.tb10701.x. [DOI] [PubMed] [Google Scholar]
- 11.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 12.Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–64. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 13.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–4. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 15.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–6. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 16.Dorner S, Panuschka C, Schmid W, Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31:6536–42. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wohlgemuth I, Beringer M, Rodnina MV. Rapid peptide bond formation on isolated 50S ribosomal subunits. EMBO Rep. 2006;7:699–703. doi: 10.1038/sj.embor.7400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–83. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- 19.Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 20.Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–31. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 21.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–55. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 22.Schilling-Bartetzko S, Franceschi F, Sternbach H, Nierhaus KH. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J Biol Chem. 1992;267:4693–702. [PubMed] [Google Scholar]
- 23.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–98. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 24.Caskey CT, Beaudet AL, Scolnick EM, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci U S A. 1971;68:3163–7. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–7. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 26.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008 doi: 10.1126/science.1164840. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 28.Ban N, Nissen P, Hansen J, Moore PB, Steitz TS. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 29.Maguire BA, Manuilov AV, Zimmermann RA. Differential effects of replacing Escherichia coli ribosomal protein L27 with its homologue from Aquifex aeolicus. J Bacteriol. 2001;183:6565–72. doi: 10.1128/JB.183.22.6565-6572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen R, Mende L, Arfsten U. The primary structure of protein L27 from the peptidyl-tRNA binding side of Escherichia coli ribosomes. FEBS Lett. 1975;59:96–99. doi: 10.1016/0014-5793(75)80349-8. [DOI] [PubMed] [Google Scholar]
- 31.Odintsova TI, et al. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003;22:249–58. doi: 10.1023/a:1025068419698. [DOI] [PubMed] [Google Scholar]
- 32.Strader MB, VerBerkmoes NC, Tabb DL, Connelly HM, Barton JW, Bruce BD, Pelletier DA, Davison BH, Hettich RL, Larimer FW, Hurst GB. Characterization of the 70S ribosome from Rhodopseudomonas palustris using an integrated “top-down” and “bottom-up” mass spectrometric approach. J. Proteome Res. 2004;3:965–978. doi: 10.1021/pr049940z. [DOI] [PubMed] [Google Scholar]
- 33.Suh MJ, Hamburg DM, Gregory ST, Dahlberg AE, Limbach PA. Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics. 2005;5:4818–31. doi: 10.1002/pmic.200402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trobro S, Aqvist J. Role of ribosomal protein L27 in peptidyl transfer. Biochemistry. 2008;47:4898–906. doi: 10.1021/bi8001874. [DOI] [PubMed] [Google Scholar]
- 35.Hampl H, Schulze H, Nierhause KH. Ribosomal components from Escherichia coli 50S subunits involved in the reconstitution of peptidyl transferase activity. J. Biol. Chem. 1981;258:12810–12815. [PubMed] [Google Scholar]
- 36.Noller HF, et al. Structure of the ribosome at 5.5 A resolution and its interactions with functional ligands. Cold Spring Harb Symp Quant Biol. 2001;66:57–66. doi: 10.1101/sqb.2001.66.57. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura M, et al. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J Mol Biol. 2004;344:1369–83. doi: 10.1016/j.jmb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Belova L, Tenson T, Xiong L, McNicholas PM, Mankin AS. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc Natl Acad Sci U S A. 2001;98:3726–3731. doi: 10.1073/pnas.071527498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraser TH, Rich A. Synthesis and aminoacylation of 3′-amino-3′-deoxy transfer RNA and its activity in ribosomal protein synthesis. Proc Natl Acad Sci U S A. 1973;70:2671–5. doi: 10.1073/pnas.70.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robins MJ, Miles RW, Samano MC, Kaspar RL. Syntheses of puromycin from adenosine and 7-deazapuromycin from tubercidin, and biological comparisons of the 7-aza/deaza pair. J Org Chem. 2001;66:8204–10. doi: 10.1021/jo010935d. [DOI] [PubMed] [Google Scholar]
- 41.Ludwig J. A new route to nucleoside 5′-triphosphates. Acta Biochim Biophys Acad Sci Hung. 1981;16:131–3. [PubMed] [Google Scholar]
- 42.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 43.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–48. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.