Abstract
Objective
The cause of preeclampsia remains unknown and the diagnosis can be uncertain. We used proteomic-based analysis of urine to improve disease classification and extend the pathophysiological understanding of preeclampsia.
Study design
Urine samples from 284 women were analyzed by mass spectrometry-based proteomics (SELDI). In the exploratory phase, 59 samples were used to extract the proteomic fingerprint characteristic of severe preeclampsia requiring mandated delivery and develop a diagnostic algorithm. In the challenge phase we sought to prospectively validate the algorithm in 225 women screened for a variety of high and low-risk conditions, including preeclampsia. Of these, 19 women were followed longitudinally throughout pregnancy. Presence of biomarkers was interpreted relative to clinical classification, need for delivery and other urine laboratory measures (ratios of protein-to-creatinine and soluble fms-like tyrosine kinase-1-to-placental growth factor). In the translational phase biomarker identification by tandem mass spectrometry and validation experiments in urine, serum and placenta were employed to identify, quantify and localize the biomarkers or related proteins.
Results
We report that women with preeclampsia appear to present a unique urine proteomic fingerprint which predicts preeclampsia in need for mandated delivery with highest accuracy. This characteristic proteomic profile also has the ability to distinguish preeclampsia from other hypertensive or proteinuric disorders in pregnancy. Pregnant women followed longitudinally who developed preeclampsia displayed abnormal urinary profiles >10 weeks prior to clinical manifestation. Tandem mass spectrometry followed by de-novo sequencing identified the biomarkers as non-random cleavage products of SERPINA-1 and albumin. Of these, the 21-aminoacid C-terminus fragment of SERPINA-1 was highly associated with severe forms of preeclampsia requiring early delivery. In preeclampsia, increased and aberrant SERPINA-1 immunoreactivity was found in urine, serum and placenta where it localized predominantly to placental villi and placental vascular spaces adherent to the endothelium. In addition, significant perivascular deposits of misfolded SERPINA-1 aggregates were exclusively identified in preeclamptic placentas.
Conclusion
Proteomics-based characterization of urine in preeclampsia identified a proteomic fingerprint composed of SERPINA-1 and albumin fragments which can accurately diagnose preeclampsia and shows promise to discriminate it from other hypertensive proteinuric diseases. These findings provide insight into a novel pathophysiological mechanism of preeclampsia related to SERPINA-1 misfolding which may offer new therapeutic opportunities in the future.
Keywords: proteomics, prematurity, preeclampsia, birth, mandated delivery, proteinuria
INTRODUCTION
Preeclampsia complicates 5–8% of pregnancies in the United States.1 Geographic, social, economic and racial differences are thought to be responsible for incidence rates up to 3 times higher in some populations.2,3 Preeclampsia has been implicated in 20% of pregnancy-related maternal deaths and continues to be the leading cause of a mandated preterm delivery.4,5,6 As a pregnancy-specific hypertensive disorder, preeclampsia is defined as new-onset elevated blood pressure accompanied by proteinuria after 20 weeks of gestation.7 Compelling evidence suggests that key disturbances at the fetal-maternal interface (endothelial cell dysfunction, oxidative stress, circulatory and metabolic alterations) play a central role in the etiology and pathogenesis of the clinical manifestations of this syndrome leading to progressive deterioration of maternal and fetal condition, with early delivery as the only definitive treatment.8
Because delivery of the fetus and the placenta is the only intervention proven to prevent high maternal morbidity and mortality, management involves balancing the risks of prematurity against severity of preeclampsia and response to treatment.9 This decision is frequently complicated by the difficulty in discriminating preeclampsia from other conditions that are also characterized by hypertension and proteinuria, such as chronic hypertension or glomerulonephropathies. Therefore, discovery of relevant biomarkers to aid with the accurate prediction, rapid confirmation of the diagnosis, and treatment monitoring of preeclampsia remains crucial.
Proteomics seems to have all the necessary attributes to provide the needed breakthrough in understanding the pathophysiology of preeclampsia by discovery of biomarkers that may also point to novel inroads into prevention or treatment of this syndrome.10 Since renal pathology is a hallmark of preeclampsia~11 and the degree of proteinuria frequently reflects disease severity,12 we turned to proteomic profiling of urine aiming to develop a rapid and non-invasive diagnostic and prognostic modality for preeclampsia based on qualitative rather then quantitative changes in urinary proteins. Furthermore, by identifying the aberrantly excreted proteins (biomarkers) we hoped to gain new insights into the mechanisms responsible for this condition.
METHODS
Patients and samples
We studied urine samples from 284 women. Pregnant subjects were enrolled at Yale-New Haven Hospital from March 2004 to December 2006. The non-pregnant women were enrolled at Fletcher Allen Health Care Hospital. The research protocols were approved by both Yale University and University of Vermont Human Investigational Committees. All women provided written informed consent.
In all instances gestational age was established based on last menstrual period and/or ultrasonographic examination prior to 20 weeks of gestation. At enrollment, women were clustered into clinical categories based on well recognized clinical and laboratory criteria.7 Severe preeclampsia (sPE) was defined as systolic blood pressure of >160 mm Hg or diastolic >110 mm Hg on at least 2 occasions 6 hours apart, >5 grams protein excretion / 24 hour urine collection, and/or persistent +3 proteinuria on dipstick testing. Other elements of the sPE definition included: in utero growth restriction (IUGR) <10th percentile, persistent neurological symptoms (headache, visual disturbances), epigastric pain, oliguria (less than 500 mL/24 h), serum creatinine >1.0 mg/dL or any elements of HELLP syndrome: hemolysis, elevated liver enzymes (>2 times the normal), low platelet count (<100,000 cells/µL). Chronic hypertension (crHTN) was defined as a sustained elevation in blood pressure prior to pregnancy or before 20 weeks gestational age.7 Superimposed preeclampsia (spPE) was diagnosed in the context of crHTN accompanied by new onset proteinuria or a sudden increase in proteinuria (if present in early pregnancy), or a sudden increase in blood pressure that met severe criteria and did not respond to medical therapy, or the presence of other sPE criteria, with the exclusion of isolated IUGR.7 Mild PE (mPE) was defined as a blood pressure of at least 140/90 mmHg and urinary protein excretion of at least 300 mg/24 hours urine collection (or at least 1+ or greater on dipstick testing), each on two occasions 4–6 hours apart and no diagnosis of either sPE or spPE. A cut-off of 150 mg/24 hours urine collection was used to judge proteinuria for non-pregnant women.13
Serum and urine samples were collected contemporaneously as previously described. 14 In select cases, placental tissue was collected at delivery for histology and mRNA extraction. Placental tissues from gestational age-matched women with spontaneous preterm birth in the absence of histological evidence of infection or inflammation were used as the appropriate control for preeclamptic placentas.
Study design
The study was undertaken in 3 phases: exploratory, challenge and translational.15 The purpose of the exploratory phase was to extract the urinary proteomic profile characteristic of preeclampsia requiring mandated delivery which was defined as an intentional intervention because of a deteriorating maternal or fetal condition in the context of preeclampsia. To minimize inaccuracies at this stage we selected the exploratory phase samples based on stringent clinical criteria. In point, we abstracted for each patient the clinical information into a semi-quantitative variable named the “objective clinical score of preeclampsia severity (OCS-sPE)” (see Appendix Section I.a. for details). Women with sPE (n=38) were elected based on a minimum of 3 criteria of severity7 which mandated a clinically indicated delivery. Twenty-one healthy asymptomatic women at similar gestational ages who did not develop preeclampsia during the remainder of their pregnancy and delivered at term were chosen as controls (CRL). SELDI profiles were generated from urine aliquots stored at −80°C. The characteristics of the women that provided urine samples for the exploratory phase are presented in Appendix Section I.b.
The purpose of the challenge phase was to validate the proteomic profile in a population different than that used for its development by determining the ability to identify preeclampsia requiring mandated delivery and differentiate this condition from other clinical contexts such as mPE or crHTN. The ability of the proteomic profile to predict disease severity based on need for delivery was further compared to that of other analytes measured in a random urine specimen and proposed to have diagnostic and prognostic value for preeclampsia, specifically the urinary soluble fms-like tyrosine kinase-1 (sFlt-1)-to-placental growth factor (PlGF) and protein-to-creatinine ratios.14,16,17,18
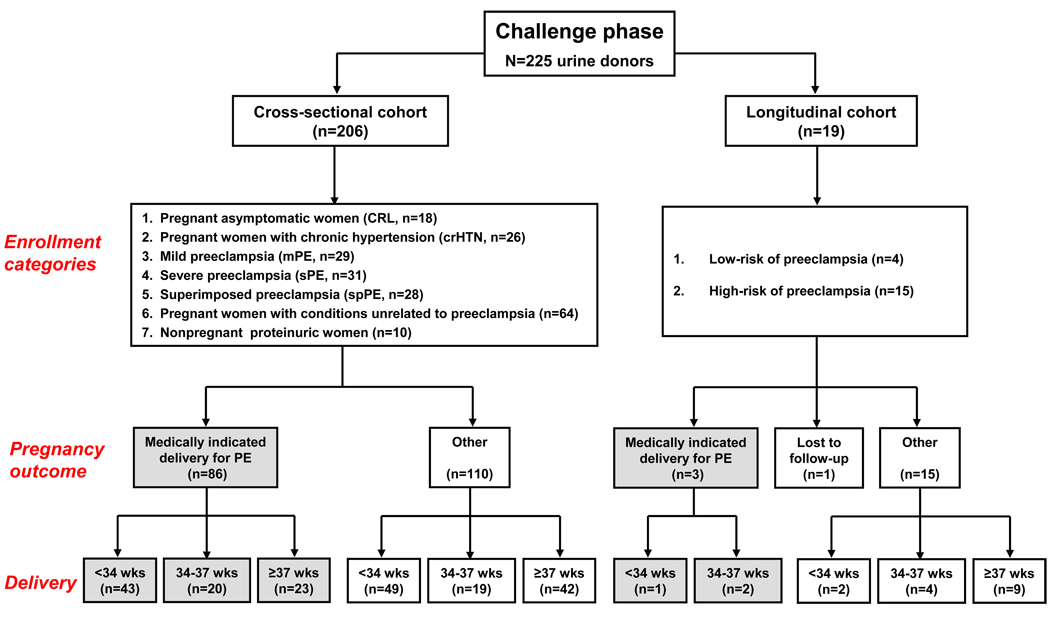
The design of the challenge phase is illustrated in Figure 1 and was based on the rational that test accuracy can vary with the extent, stage of the disease and associated morbidities.19 Additionally, since preeclampsia is a progressing disease and by definition a clinical diagnosis for which no acceptable gold standard is yet available,20 we chose as outcome measure the need for delivery rather than the clinical classification at enrollment. We reasoned that an indication for mandated delivery belongs to a team, is the last management resort when all other strategies have failed, its resulting outcome is final, cannot be revoked and thus less subject to bias.
Figure 1. Flow-chart and distribution of urine samples for the challenge phase.
Pregnancy-associated conditions unrelated to preeclampsia were preterm labor: n=47; preterm premature rupture of the membranes: n=11, placenta praevia: n=4, sepsis: n=1 and isolated IUGR: n=1. Of the women enrolled in the cross-sectional cohort, 86 had a medically-indicated delivery for preeclampsia. The remaining 110 pregnant women delivered at different gestational ages. Deliveries at <37 weeks in this group occurred either spontaneous (n=50) or were medically-indicated for clinical conditions such as: placenta previa (n=4), non-reassuring fetal status (n=10) or idiopathic intra-uterine fetal demise (n=2). Out of the 19 women enrolled in the longitudinal cohort of the challenge phase, 3 developed sPE or spPE requiring mandated delivery at <37 weeks. Abbreviations: PE: preeclampsia; sPE: severe preeclampsia; mPE: mild preeclampsia; crHTN: chronic hypertension; CRL: healthy pregnant control; wks: weeks.
A total of 225 consecutive women were enrolled prospectively. Proteomic spectra were acquired using fresh samples of urine and all samples were scored by investigators unaware of the clinical classification or outcome. The cross-sectional challenge cohort consisted of 206 women who had a single random sample of urine analyzed (at entry into the study). At the end of enrollment and for the purpose of data analysis, several subsets of women were identified based on the clinical classification at the time of their urine sampling: 1) asymptomatic pregnant controls (CRL: n=18, gestational age: 25 [7–41] weeks); 2) pregnant women with chronic hypertension (crHTN: n=26, gestational age: 33 [20–39] weeks); 3) mild preeclampsia (mPE: n=29, gestational age: 36 [24–40] weeks); 4) severe PE (sPE: n=31, gestational age: 36 [24–40] weeks); 5) superimposed preeclampsia (spPE: n=28, gestational age: 34 [16–39] weeks); 6) pregnancy-associated conditions unrelated to preeclampsia (n=64, gestational age: 28 [21–34] weeks); 7) nonpregnant proteinuric women (n=10).
The longitudinal cohort included 19 randomly selected asymptomatic women at low (n=4) or high-risk (n=15) for developing preeclampsia. In all, we performed serial proteomic profiling of their fresh urine samples from the first trimester until 6 weeks postpartum. Pre-pregnancy conditions defining the high-risk population included: crHTN, history of sPE in a prior pregnancy, diabetes, diabetic nephropathy, nephrolitiasis, membranous glomerulonephritis and sickle cell disease with history of crises.
In the 3rd phase (translational) we identified the discriminatory biomarkers and conducted validation experiments in urine, serum and placental samples.
SELDI profiling and development of the urinary proteomic scores (UPS) of preeclampsia
During the exploratory phase, SELDI conditions were first optimized by systematically screening combinations of ProteinChip® array surfaces (Ciphergen Biosystems, Fremont CA) and various binding/washing protocols as previously described.21 We found that an on-spot application of 1.5-µg of total unfractionated urine protein provided optimal signal-to-noise (S/N) ratios. After screening >200 experimental conditions, two (reverse phase hydrophobic surfaces H4 and H50) were considered optimal based on several peak clusters in the 2.3–17.5~kDa mass region unique to the sPE samples. Arrays were incubated for 1-hour with the samples (6-µL/spot) diluted to 0.25 mg/mL total protein. Following incubation, unbound proteins were removed by washing each spot with the respective buffer. After drying, 1-µL 20% saturated α-cyano-4-hydroxycinnamic acid solution (CHCA) was added and the arrays read in the SELDI-TOF mass spectrometer.
To extract the discriminatory peaks (biomarkers) and define which combination of biomarkers is optimal, we applied the principles of Mass Restricted (MR) scoring as described earlier 21,22 with minor modifications (see Appendix Section I.c. for details). The first goal of the MR scoring method is to identify the minimal combination of SELDI peaks (identified by their molecular mass) with discriminatory value and second, to reduce all proteomic information into a numeric variable that characterizes each sample and can be further compared to other diagnostic modalities or tested prospectively against outcome using standard statistical methods. 23 Two objective urinary proteomic scores were designed: a Boolean score (UPSb) representing the sum of Boolean indicators (1:~present; 0:~absent) assigned to each biomarker and a ranked score (UPSr) with merged semi-quantitative information of biomarkers present calculated as UPSr~=~∑x(S/N)/10+1, where “x” includes the biomarkers with Boolean indicators of 1 (i.e. present).
To determine the intra - and inter-observer variability of the proteomic scores and their stability with varying conditions and times of urine storage, SELDI analyses and calculations of UPSb and UPSr were performed on 8 samples of fresh urine (3 CRL and 3 sPE) by three independent investigators each performing the analysis in triplicate. In addition, the investigators performed the same SELDI analysis after aliquots of the 8 samples were maintained at either −80°C, +4°C or +20°C for 24h, 3 days and 7 days, respectively.
Identification of discriminatory proteomic biomarkers
Tandem mass spectrometric (MS/MS) peptide sequencing was accomplished using a quadrupole time of flight instrument (Q-TOF™II, Micromass Ltd, UK) equipped with a PCI1000 interface (Ciphergen) which allows for peptides to be sequenced directly from the arrays without any offline purification necessary.24 Protein identification was achieved by database searching using Mascot software (Matrix Science, London, UK). Methodological details on identification of the discriminatory proteomic biomarkers components of the UPS scores are provided in Appendix Sections I.d. Following identification by MS/MS of some of the peaks of the UPS score, we conducted in vitro experiments by spiking urine samples devoid of biomarkers (CRL samples with UPS scores of 0, n=3) with purified precursors (Sigma, St. Louis, MO) in the attempt to identify whether the biomarkers of the UPS profile can be re-created by the interaction of the precursor with the urine milieu.
Other experimental procedures are described in Appendix Sections I.e.
Statistical analysis
All data sets were subjected to normality testing using the Kolmogorov-Smirnov method. Data is reported as either mean with standard deviation (SD) or 95% confidence intervals (for normally distributed data) or as median with range (for non-normally distributed data). Data sets were compared with either Student’s t test or Mann-Whitney test as appropriate. Correlation analysis was performed using Sperman’s rank order correlation. Longitudinal data was compared by 2-way ANOVA. Proportions were compared with Fisher’s exact tests. Test accuracy (cases correctly classified / total number of cases), sensitivity, specificity, and likelihood ratios were measured on receiver operator characteristic (ROC) plots using MedCalc (Broekstraat, Belgium) statistical software. Intra- and inter-rater variability of the proteomic scores (ordinal scale) was estimated by Cronbach's alpha coefficient for scale reliability 25 and agreement in diagnosis (nominal scale) with either Cohen’s kappa method (two raters) or by the method of Fleiss-Cuzick, an extension of the Cohen’s kappa method for 3 raters per subject.26 Agreement level calculations were performed using StatsDirect software (v.2.5.7, StatsDirect Ltd, Cheshire, UK) and interpreted as “very good” (kappa = 0.81–1.00), good (kappa = 0.61–0.80) moderate (kappa = 0.41–0.60), fair (kappa = 0.21–0.40) or poor (kappa <0.20). Multiple stepwise logistic regression analysis was used to adjust p-values and odds ratios (ORs), respectively, for potential and combined influences of other parameters. Variables are entered into the model if p<0.05 and removed for p>0.1. A p-value of less than 0.05 was used to indicate significance.
RESULTS
Characterization of the urinary proteomic signature of severe preeclampsia
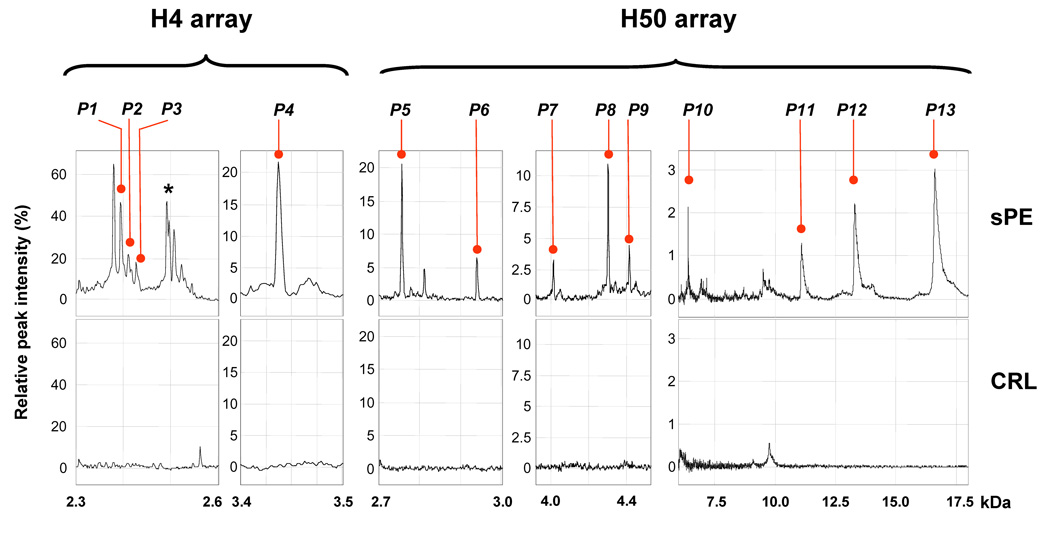
At the end of the exploratory phase through analysis of the SELDI-TOF spectra, we determined that the proteomic profile characteristic of preeclampsia requiring delivery includes 13 biomarkers peaks (P1 to P13, Figure 2). This dictates that UPSb can range from 0 (all biomarkers absent) to 13 (all 13 biomarkers present) while UPSr can range from 0 to infinity depending on the sum of the S/N ratios of the biomarkers present. In Appendix Section II.a. we list the observed molecular masses and the frequency with which each of the 13 biomarkers was detected in the exploratory phase. sPE women had significantly elevated UPS scores compared to controls (sPE: UPSb: 9 [8–13] and UPSr 29 [13–49] vs CRL: UPSb: 1 [0–6] and UPSr 1 [0–8], P<0.001). In ROC analysis we determined that the combination of an UPSb >6 and an UPSr >8 distinguished with 100% sensitivity and 100% specificity a diagnosis of sPE from CRLs at the time of initial clinical screening.
Figure 2. The urinary proteomic fingerprint of severe preeclampsia requiring delivery.
Representative SELDI profile from a patient with severe preeclampsia (sPE) at 30 weeks gestational age illustrating the 13 biomarker components (P1–P13, marked with red bullets) of the urinary proteomic scores (UPSb=13 and UPSr=47) presented in comparison with a SELDI profile of urine from a healthy pregnant control patient (both UPSb and UPSr of 0) at the same gestational age (CRL). kDa: kilodaltons. The asterisk marks a complex of peaks not part of the UPS profile that accompanies P1–P3 complex.
Challenge phase - prospective validation of the proteomic profiling of urine to predict preeclampsia requiring mandated delivery
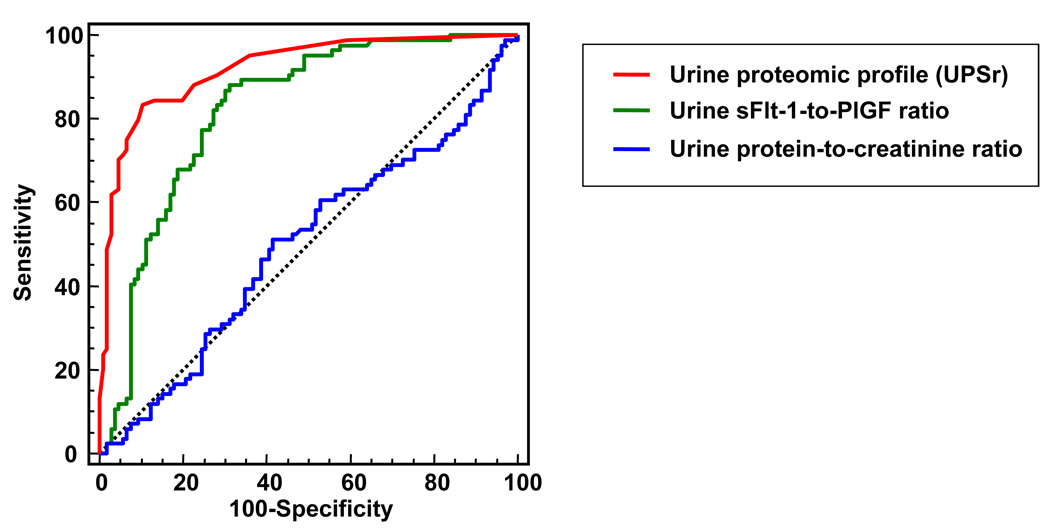
Of the women enrolled in the cross-sectional cohort, 86 had a mandated delivery for preeclampsia which in 73% occurred preterm and in 50% of cases at <34 weeks (Figure 1). Test performances of the urine proteomic scores were compared with those of other tests from a random urine sample such as protein-to-creatinine ratio, and the sFlt-1-to-PlGF ratio (Figure 3). The urine sFlt-1-to-PlGF ratio predicted a medically-indicated delivery for preeclampsia better than the protein-to-creatinine ratio (difference in ROC areas: 0.32 [95%CI: 0.22–0.42], P<0.001) and also better than either urinary analyte alone (P<0.001). However, the performance of the urine proteomic scores was superior to sFlt-1-to-PlGF ratio and to all the other tests (difference in ROC areas between uFP and UPSr: 0.10 [95%CI: 0.04–0.17], P=0.002) (Figure 3). The accuracies of the urine analytes and proteomic scores to predict a preeclampsia-related indicated delivery are listed in Table 1. A subanalysis of the women enrolled in the non-hypertensive groups that delivered either at term or preterm in the absence of preeclampsia revealed that gestational age per se did nor correlate with the proteomic scores (for UPSb: Sperman’s r=0.08, P=0.427 and for UPSr: r=0.07, P=0.468).
Figure 3. Diagnostic performances of the proteomic profile relative to other analytes assessed in the same random urine sample.
Receiver operating characteristic (ROC) curves of the proteomic score UPSr (red line), protein-to-creatinine ratio (blue line) and soluble fms-like tyrosine kinase 1 [sFlt-1]-to-placental growth factor [PlGF] ratio (green line) for predicting a preeclampsia-related indicated delivery. Analysis included all consecutive pregnant women who contributed with urine samples to the cross-sectional challenge phase (n=196).
Table 1.
Diagnostic accuracies of urine analytes and proteomic scores for predicting need for delivery in the cross-sectional challenge phase.
| Analytes | ROC area [95%CI] |
P value |
Cut-off* | Sens. [95%CI] |
Spec. [95%CI] |
+LR | −LR | Accuracy |
|---|---|---|---|---|---|---|---|---|
| Protein-to-creatinine ratio (mg/mgc) | 0.51 [0.43–0.58] |
0.901 | > 8.4 | 52 [41–63] |
59 [49–68] |
1.3 | 0.8 | 55.9 |
| sFlt-1-to-creatinine ratio (pg/mgc) | 0.71 [0.64–0.78] |
<0.001 | >28.0 | 63 [52–73] |
72 [62–80] |
2.2 | 0.5 | 67.7 |
| PlGF-to-creatinine ratio (pg/mgc) | 0.77 [0.71–0.83] |
<0.001 | < 57.6 | 85 [75–92] |
67 [57–76] |
2.6 | 0.2 | 74.5 |
| sFlt-1-to-PlGF ratio | 0.82 [0.75–0.87] |
<0.001 | > 1.3 | 88 [79–94] |
69 [59–77] |
2.8 | 0.2 | 77.1 |
| UPSb | 0.92 [0.87–0.95] |
<0.001 | ≥ 5 | 85 [76–92] |
86 [79–92] |
6.2 | 0.2 | 85.7 |
| UPSr | 0.92 [0.87–0.96] |
<0.001 | ≥ 7 | 84 [74–91] |
89 [82–94] |
7.7 | 0.2 | 86.7 |
| Non-reassuring urine profile: UPSb ≥5 and UPSr ≥7 | 85 [77–93] |
87 [81–96] |
6.7 | 0.2 | 86.2 | |||
represents the cut-off with highest accuracy.
Abbreviations: Sens: sensitivity; Spec: specificity; +LR: positive likelihood ratio; −LR: negative likelihood ratio; mgc: milligrams creatinine; sFlt-1: soluble fms-like tyrosine kinase-1; PlGF: placental growth factor
We next tested whether addition of the “non-reassuring” proteomic status to the clinical assessment would have additive value in predicting outcome. In a stepwise multivariate logistic regression a clinical diagnosis of sPE at enrolment and the “non-reassuring” proteomic urinary status were independent predictors of the need for delivery. The best model included the clinician’s assessment (OR: 45.8 [95%CI:9.2–228.5], P<0.001), a“non-reassuring” urine pattern (OR: 12.4 [95%CI:4.7–33.2], P<0.001) and gestational age at assessment (OR: 1.2 [95%CI:1.0–1.3], P=0.004). Variables excluded from the model (P>0.1) were maternal age, gravidity, parity, sFlt-1-to-PlGF and protein-to-creatinine ratios. When we restricted our analysis to women screened for sPE at less than 34 weeks (n=139) of gestation a “non-reassuring” urinary profile was the only variable with additive value (P<0.001) to the clinical assessment to predict the severity of sPE requiring a mandated preterm delivery at <34 weeks, independent of gestational age. Moreover, the additive predictive value was maintained at a level of P<0.001 when we restricted the analysis to the women classified initially as mPE, crHTN and spPE. In the clinical context where at the time of initial clinical assessment differentiation of mPE, uncontrolled crHTN and presumed spPE from sPE is most difficult based on clinical criteria alone, a non-reassuring SELDI profile was the only predictor of preeclampsia requiring delivery (OR: 9.1 [95%CI:2.8–29.6.0] improving disease classification to 81% from 67% based on clinical signs and symptoms alone (P<0.001). Variables excluded from the model were gestational age, parity, urine sFlt-to-PlGF or protein-to-creatinine ratios.
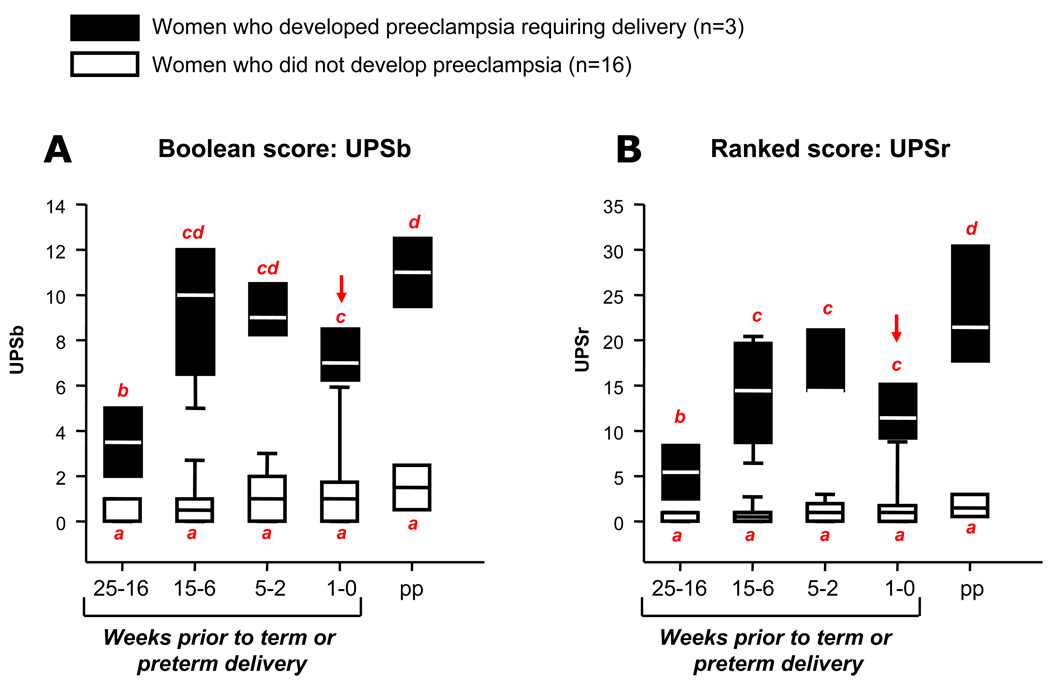
Out of the 19 women enrolled in the longitudinal cohort of the challenge phase, 3 developed sPE requiring delivery (Figure 2). We found that the subgroup of patients that ultimately developed sPE had significantly higher median UPS scores at all times even up to 25 weeks prior to delivery (Figure 4). While the UPS scores in patients who did not develop sPE did not change throughout pregnancy and postpartum, we found a progressive increase in UPS scores in women who ultimately developed sPE. Evolution of urinary proteomic scores of women who developed sPE was characterized by the increase both in the number of biomarkers present in the urine (2-way ANOVA: UPSb P<0.001) and their relative abundance (UPSr P<0.001) at least 10 weeks prior to onset of clinical manifestations (Figure 4). We further determined that none of the non-pregnant women had the proteomic fingerprint of preeclampsia present despite significant proteinuria (median [range]: 332 [162–543] mg/24 hours). This allowed us to conclude that our biomarkers are specific of preeclampsia and not of proteinuric conditions unrelated to pregnancy.
Figure 4. Evolution of urinary proteomics scores during pregnancy in the longitudinal cohort of the challenge phase.
19 asymptomatic pregnant women at low or high risk for developing preeclampsia were followed longitudinally from the first trimester until 6 weeks postpartum. UPSb (Panel A) and UPSr (Panel B) scores were generated prospectively. Black boxes represent the women who developed preeclampsia requiring an mandated preterm delivery (n=3). Open boxes represent women who had a normal course of their pregnancy (n=16). On the x axis are the time periods in weeks prior to each patient’s delivery date (time 0). pp = postpartum. On the y axis are the values of the urinary proteomic scores (UPSb and UPSr, respectively). The arrow indicates the time-point where women manifested clinical signs and symptoms. The data is presented as percentiles with median. The ends of the boxes define the 25th and 75th percentiles, the line inside the box defines the median and the whiskers show the largest and smallest values. 2-way ANOVA: p<0.01 for both time periods and outcome. Bars with at least one common red letter are not statistically different at a value of p>0.05.
Challenge phase – ability of the proteomic profile to distinguish preeclampsia from other hypertensive disorders
The results of the challenge phase revealed that in preeclampsia the urinary proteomic profile is a dynamic entity with an increase in the number of biomarkers and their abundance which parallels disease progression. Based on an ROC analysis we established that pregnant women with proteomic scores UPSb ≥5 and UPSr ≥7 at the time of initial assessment have either sPE or spPE requiring at some point in pregnancy a medically-indicated delivery. Therefore such scores should be deemed “non-reassuring”.
To determine whether proteomic profiling can aid clinicians to distinguish preeclamptic women that would require delivery from those with other hypertensive disorders which can potentially be managed expectantly, we first investigated the frequency of a “non-reassuring” urine profile in all pregnant women with known outcomes enrolled in the cross-sectional cohort of the challenge phase (n=196). Of all women categorized clinically as sPE, 94% (29/31) had a “non-reassuring” profile. Of the spPE women, 89% (25/28) had the “non-reassuring pattern” and thus correctly classified based on clinical criteria. Conversely, 66% of the (19/29) of the cases classified as mPE also had a “non-reassuring” pattern, while crHTN and asymptomatic non-hypertensive women exhibited the proteomic profile in only 27% (7/26) and 9% (7/82) of cases, respectively. This suggests that clustering hypertensive disorders in pregnancy on clinical criteria alone can be subject to bias and supports the need of an additional test such as proteomic profiling of the urine for correct classification.
Intra, inter-observer variability and stability of the proteomic scores
Using agreement measures we report “very good” Cronbach alpha values of 0.990 (intra-observer) and 0.975 (inter-observer) for the nominal classification of urinary profiles as reassuring/non-reassuring and an average kappa (Fleiss and Cuzick extension) of 0.943 (intra-observer, “very good”) and 0.720 (inter-observer, “good”) for the UPS scores as ordinal variables. Storage of the urine in various conditions did not alter the proteomic scores (kappa ranging from 1: “perfect agreement” to 0.75: “good”).
Biomarker identification
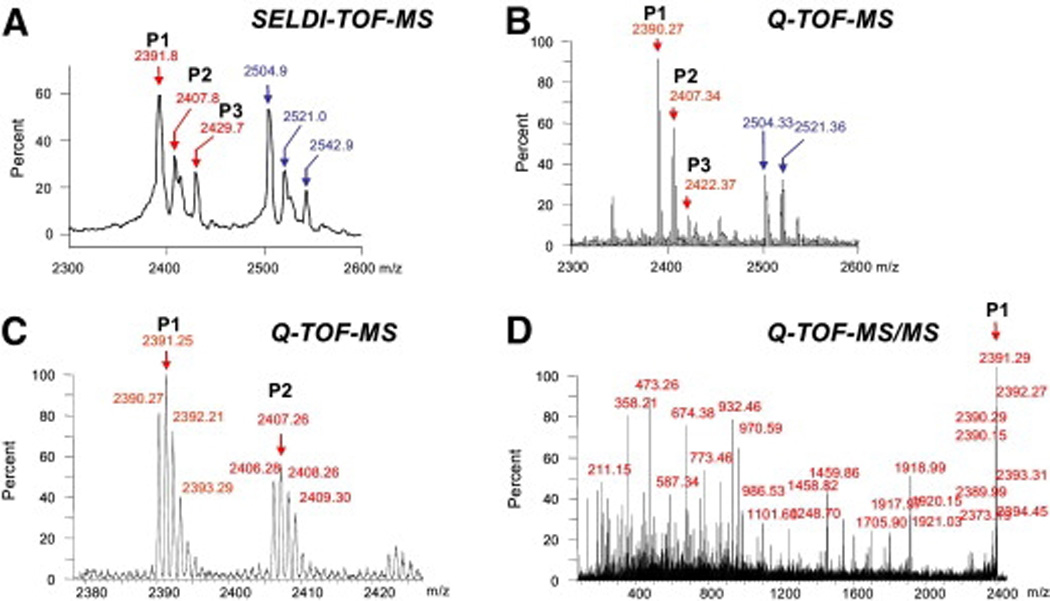
Using tandem mass spectrometry we identified corresponding sequences for 5 of the biomarker components of the profile: P1, P2, P3, P5 and P7 (Figure 2) which matched peptide fragments of human SERPINA-1 (SwissProt P01009) and albumin (SwissProt P02768). The results obtained from the MS/MS analysis on selected peptides are presented in Table 2. Additional results illustrating the correspondence of biomarkers between detection in the SELDI-TOF and the Q-TOF mass spectrometer are displayed in Appendix Section II.b. The complex P1–P3 at 2390–2430 Da corresponded to the 21-aa C-terminus fragment of SERPINA-1 [aa~398–418] in either non-oxidized form (P1) or with either one (P2) or two (P3) methionine residues (positions 398 and/or 409) in oxidized form. Although not part of the final score, the complex of peaks that accompanies P1–P3 (Figure 2, asterisk) was identified as the 22-aa C-terminus peptide fragment of SERPINA-1 [aa 397–418]. The biomarker peak P7 corresponded to the 24-aa N-terminus fragment of SERPINA-1 after cleavage of the signal peptide.27 We attempted a similar identification strategy for the remaining peaks of the UPS profile but the amount of corresponding protein was below the current technical capability for identification. Using a strategy detailed in Appendix Section II.c. we determined that P6, P8, P10, P11, P12 and P13 are originating from human albumin.
Table 2.
Results obtained from the MS/MS analysis on selected peptides on the ProteinChip Tandem Interface
| Peak | Experimental mass (Daltons) |
Mowse score; expected value |
Sequence | Calculated mass (Daltons) |
SwissProt accession & residues |
|
|---|---|---|---|---|---|---|
| SELDI TOF-MS |
Q-TOF- MS |
|||||
| P1 | 2391.88 | 2390.27 | 72; 0.00097 | MIEQNTKSPLFMGKVVNPTQK | 2390.84 |
P01009 aa 398–418 |
| P2 | 2407.81 | 2406.28 | 63; 0.0068 | MoxIEQNTKSPLFMGKVVNPTQK | 2406.83 |
P01009 aa 398–418 |
| P3 | 2429.72 | 2422.37 | 58; 0.039 | MoxIEQNTKSPLFMoxGKVVNPTQK | 2422.82 |
P01009 aa 398–418 |
| P5 | 2755.18 | 2753.50 | 52; 0.086 | DAHKSEVAHRFKDLGEENFKALVL | 2754.10 |
P02768 aa 25–48 |
| P7 | 4011.59 | 4010.28 | 34; 10 | EDPQGDAAQKTDTSHHDQDHPT FNKITPNLAEFAFS |
4011.20 |
P01009 aa 25–60 |
Mox: oxidized methionine
Translational implications of presence of SERPINA-1 fragments in urine of patients with sPE
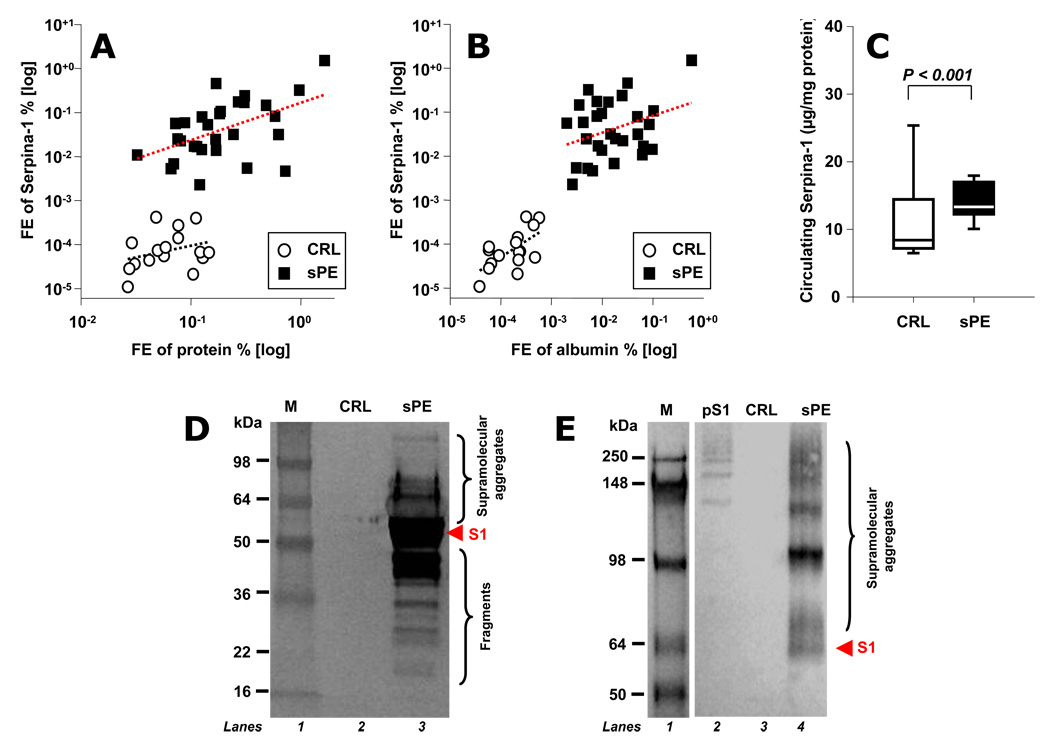
By studying the combinations in which the biomarker peaks occurred in patients with sPE, we found that the urinary appearance of the 21-aa C-terminus fragment of SERPINA-1 in the urine (P1) and at least one of its oxidized forms (P2 and/or P3) is a specific hallmark of preeclampsia severity. We thus became interested on how SERPINA-1 homeostasis is disturbed in urine, serum and placenta of women with sPE from a quantitative and qualitative perspective. Examination of fractional excretion of SERPINA-1 revealed that sPE women had significantly elevated urinary excretions of SERPINA-1 immunoreactivity (450-fold, P<0.001) even after correcting for the degree of non-specific proteinuria (275-fold, P<0.001, Figure 5A) or albuminuria (3.2-fold, P<0.001, Figure 5B). Despite this excessive excretion, serum levels of immunoreactive SERPINA-1 remained significantly elevated (P<0.001, Figure 5C). We identified by western blotting that SERPINA-1 immunoreactivity excreted in urine by sPE women was heterogeneous and included multiple molecular weight forms both below (fragments) and above (supramolecular aggregates) the 52kDa mass of monomeric SERPINA-1 while urine samples of CRL women were devoid of SERPINA-1 immunoreactivity (Figure 5D and E).
Figure 5. Quantitative and qualitative changes in SERPINA-1 immunoreactivity in urine and serum of women with severe preeclampsia.
Fractional excretion (FE) of SERPINA-1 (proportion of SERPINA-1 immunoreactivity excreted in the urine compared with that filtered by the glomeruli) relative to that of total proteins (Panel A) and albumin (Panel B), in logarithmic format. Despite the significant urinary loss, circulating SERPINA-1 levels are increased in women with severe preeclampsia (sPE) (n=29) compared to controls (CRL, n=16) at similar gestational ages (sPE: 32.7±4.4 weeks vs. CRL: 30.4±1.5 weeks, p=0.151) (Panel C). Western blot of SERPINA-1 immunoreactivity in urine from representative CRL and sPE women under denaturing (Panel D) or native conditions (Panel E). The red arrow points to monomeric SERPINA-1 (S1: 52 kDa). The bands above 52 kDa represent supramolecular aggregates while those below represent fragments. These results suggest that women with sPE excrete a complex pattern of SERPINA-1 fragments and aggregates in addition to P1–P3 biomarkers identified by us using SELDI. Lane 2 in panel E was loaded with SERPINA-1 polymerized in vitro (pS1) to demonstrate antibody reactivity against oligomeric SERPINA-1. Molecular weight markers (M) in kilodaltons (kDa) are loaded in Lane 1.
By immunohistochemistry we found that placentas from pregnancies complicated by sPE (n=10) had increased stromal (P<0.001), endothelial (P<0.001) and intra-vascular (P<0.001) SERPINA-1 staining compared to gestational age-matched controls (n=10) (Figure 6). The most conspicuous finding was the acellular positive material sequestered within the fetal vascular spaces and adhering to the vascular endothelium of the villi, virtually absent in non-preeclamptic placentas. In preeclamptic placentas SERPINA-1 positive accelular aggregates were also encountered more often in the intervillous space occupied by maternal blood. By using the monoclonal ATZ11 antibody~30,28 known to recognize oligomeric misfolded SERPINA-1 complexes but not the native or latent forms of monomeric SERPINA-1~29 we found an intense staining pattern in sPE placentas with predominant endothelial and perivascular localization while the villi of the non-preeclamptic placentas remained entirely free of staining.30
Figure 6. Immunohistochemistry of SERPINA-1 and of its supramolecular aggregates in human placenta.
Representative placental sections from a woman that delivered after a medically-indicated preterm delivery for severe preeclampsia (Panels a–;d & g) and a gestational age matched patient with spontaneous preterm birth (panels e–;g) were stained immunohistochemically with a polyclonal antibody recognizing the multiple immunoreactive forms (Panels a–;c & e–;f) and with a monoclonal antibody (ATZ11) recognizing only the supramolecular aggregates of SERPINA-1 (oligomers, misfolded forms and SERPINA-1-elastase complexes). Positive staining is shown by Ni-DAB (purple-black) deposits. PVS: placental villous stroma; PTR: placental trophoblast; PBV: placental blood vessels; Endo: endothelium. Panel g: sPE: omitted primary antibody. Magnification scales are presented for each panel in microns. Panels b, c, f & g: light methyl-green counterstain.
DISCUSSION
We used multi-dimensional proteomic technology to determine that women with preeclampsia requiring mandated delivery exhibit a urinary proteomic signature characterized by non-random fragments of SERPINA-1 and albumin. We provided evidence that appearance of the proteomic fingerprint in the urine precedes the onset of clinical symptoms and maintains diagnostic significance in the context of associated morbidities, thereby improving disease classification. Proteomic profiling of the urine was significantly better than clinical evaluation alone, or angiogenic factors to predict severity of preeclampsia and need for delivery. This could be a very important especially for the developing world where severe disease is associated with high mortality.31 We further provided evidence that proteomic profiling of the urine carries the potential to differentiate preeclampsia from uncontrolled hypertensive disorders when clinical criteria alone seem powerless. Lastly, our findings that a significant proportion of women with crHTN or mPE display the biomarkers characteristic for sPE supports the view that classification of hypertensive disorders based on clinical criteria is deficient. Proteomics may allow in the future identification of "new" molecular diseases relevant for reclassification of the preeclamptic syndrome, treatment selection and prognosis. In addition, knowledge of the identity of the biomarkers provided us with a new mechanistic view of preeclampsia.
Proteinuria is a fundamental criterion for diagnosis of preeclampsia and the result of endothelial impairment with acute atherosis at the level of renal glomeruli.11 The degree of proteinuria has been associated with the severity of preeclampsia32 and in direct relationship with perinatal mortality. 33 Current analysis of proteinuria for diagnosis and monitoring of severity of preeclampsia in clinical practice relays on quantitative determination of total protein in a 2-step approach. The urinary dipstick analysis has been shown to be inaccurate do to high false positive rate 34 and is used as screening. If positive, it leads to the measurement of total protein eliminated in a 24-hour period. However, 24-hour collections are cumbersome, often requiring hospitalization, subject to error and compliance issues and result in delay in diagnosis. Several studies attempting to provide evidence in support of random proteinuria after normalization for creatinine (protein-to creatinine ratio) as a quick surrogate measure of proteinuria of preeclampsia have yielded mixed results.35, 36 Despite this, this test is now accepted by the International Society for the Study of Hypertension in Pregnancy (ISSHP) as reliable measure of proteinuria of preeclampsia. 37
The current study aimed to provide a comprehensive qualitative and quantitative description of proteinuria of preeclampsia to create a customized combination of biomarkers that differentiate it from proteins present in urine of normal subjects and most importantly from proteinuria of other conditions unrelated to preeclampsia. As demonstrated by our current study the accuracy of diagnostic indices based on proteomic profiling to discriminate cases with significant morbidity was higher than that of protein-to-creatinine ratio and even that of sFlt-1-to-PlGF ratio in a challenge cohort of over 100 cases with various pathologies as applicable to a high-risk obstetrical service.
Aside from the practicality of producing a non-invasive diagnostic and prognostic test, knowledge on the identity of the biomarkers has leaded us to interesting and novel observations related to SERPINA-1 and albumin fragmentation. SERPINA-1 is an abundant plasma protein and the main blood-borne serine protease inhibitor. Although its primary function is the inhibition of neutrophil elastase 38 it also has activity against catepsin G, proteinase 3, pancreatic elastase, trypsin, chymotrypsin and collageanses and kallikrein. 39 The antiproteolytic activity is explained by formation of an equimolar enzyme-Serpina-1 complex. This results in the proteolytic cleavage of the reactive center peptide bond between Met358 and Ser359 of the secreted form of SERPINA-1.40 SERPINA-1 is synthesized by the liver, macrophages neutrophils and also by trophoblast.41 Increases in serum SERPINA-1 occur in diseases such as rheumatoid arthristis, vasculitis, infections and other diseases associated with an inflammatory component.42 Interestingly, studies have shown that even minor increases in levels of serum SERPINA-1 are associated with development of arterial hypertension and an increased risk of cardiovascular disease.43, 44 One explanation is that by inhibiting the activity of the kallikrein-kinin system 45, an upregulation of plasmatic SERPINA-1 favours the renin-angiotensin system leading to systemic vasoconstriction and hypertension.
Our experiments confirmed that women with sPE have higher serum, urine and placental SERPINA-1 immunoreactivity despite a tremendous urinary loss. Also significant may be that the C-terminus fragment of SERPINA-1 corresponding to P1-P2-P3 biomarkers is part of a sequence which has been shown to activate monocytes to a heightened pro-inflammatory state,46 an observation consistent with the increased cytokine levels reported in preeclampsia.47
Protein misfolding diseases are a growing body of pathological entities such as systemic amyloidosis, Alzheimer’s and prion diseases. These conditions are characterized by aberrations in tertiary and quaternary structures of specific proteins which result in deposits of aggregated oligomers and alterations in organ function.48 SERPINA-1 stands out because of its susceptibility to oxidative stress-induced fragmentation, misfolding, polymerization and aggregation.49 Supramolecular aggregates of misfolded SERPINA-1 have been recently implicated in pathophysiology of an emerging class of diseases known as serpinopathies characterized by accumulation of aggregates within hepatocytes and neurons leading to liver damage and encephalopathy.50 The high frequency of abnormal liver and neurological findings in women with preeclampsia supports the view that fragments or aberrant forms of SERPINA-1 may be direct players in the pathophysiological manifestations of this syndrome. Our finding of ATZ11-positive staining limited to sPE villi is indicative that placenta is also a target organ for SERPINA-1 aggregates and thus may explain its altered function in this condition.
It is known that normal urine contains small amounts of intact albumin and much larger quantities of low molecular weight albumin fragments. In this study we demonstrated that, in addition to the biomarkers generated by fragmentation of SERPINA-1, the urine of sPE patients also contains a consistent pattern of albumin fragments different from that of normal subjects. Though most dye-binding or antibody-based assays detect the intact form of albumin, they would not detect the fragments that we identified as specific biomarkers for preeclampsia.51 Because mass spectrometry technology can recognize these precise biomarkers, it may prove to be the ideal screening tool for women at low and high risk of preeclampsia. In support of the value of mass spectrometry is the observation that none of the albumin or SEPRINA-breakdown products are present in the urine of non-pregnant proteinuric women. Thus, in the future mass spectrometry may represent the solution to the frequent clinical dilemma of differentiating PE from other pre-existing hypertensive proteinuric disorders during gestation.
We do not know at this time if any of these specific SERPINA-1 or albumin cleavage products have a direct or facilitating role in the endothelial and/or glomerular damage characteristic of preeclampsia. However, our proteomics approach determined that two abundant serum proteins serve as a reservoir of novel peptides generated through a non-genomic posttranslational and non-random cleavage. For example, we have confirmed that despite elevated SERPINA-1 immunoreactivity in serum and placentas by immunoblot and immunohistochemistry, the mRNA levels of SERPINA-1 in sPE placentas by real time PCR remain unchanged compared to controls (data not shown). This suggests that placenta is not the source of the aberrantly elevated SERPINA-1, but rather its deposit. Furthermore, it is well-known that circulating SERPINA-1 levels are elevated not only in preeclampsia but also in acute inflammatory conditions. 52 Therefore it is not the elevation in SERPINA-1 level which is characteristic for preeclampsia but rather its pattern of fragmentation present in urine. Further studies are required to elucidate if besides being biomarkers, the fragments or misfolded forms of SERPINA-1 are also direct players in the pathophysiological manifestations of preeclampsia. Given the increasing interest in molecular and pharmacological chaperones which have been found to reduce the severity of several neurodegenerative disorders and other protein-misfolding diseases,53 our findings set the preliminary stage to test whether drugs that stabilize thermodynamically native protein structures can also prevent or halt the progress of preeclampsia.
Supplementary Material
ACKNOWLEDGEMENT
We are indebted to the fellows, residents and nurses in the Departments of Obstetrics and Gynaecology at Yale New Haven Hospital and University of Vermont College of Medicine who assisted with patient enrolment.
ROLE OF THE FUNDING SOURCE
The study has been funded by the Department of Obstetrics, Gynaecology and Reproductive Sciences, Yale University School of Medicine and by the Department of Health and Human Services / NIH RO1 HD 047321-01 (IAB). None of the funding sources had any involvement in study design, interpretation of data, writing of the report or decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FINANCIAL DISCLOSURE STATEMENT
Dr. Irina and Catalin Buhimschi are joint inventors on patent applications filed by Yale University regarding the use of urinary angiogenic factors and proteomic profiling for diagnosis or preeclampsia. Dr. Nathan Harris is an employee of Ciphergen Biosystems. Dr. Irina Buhimschi or Catalin Buhimschi have no financial relationship with Ciphergen Biosystems and have collaborated with Mr. Harris solely on scientific basis. None of the other authors have any financial relationships regarding the content of this manuscript.
REFERENCES
- 1.Sibai BM, Ewell M, Levine RJ, et al. Risk factors associated with preeclampsia in healthy nulliparous women: the Calcium for Preeclampsia Prevention (CPEP) study group. Am J Obstet Gynecol. 1997;177:1003–1010. doi: 10.1016/s0002-9378(97)70004-8. [DOI] [PubMed] [Google Scholar]
- 2.Samadi AR, Mayberry RM, Zaidi AA, Pleasant JC, McGhee N, Jr, Rice RJ. Maternal hypertension and associated pregnancy complications among African-American and other women in the United States. Obstet Gynecol. 1996;87:557–563. doi: 10.1016/0029-7844(95)00480-7. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization International Collaborative Study of Hypertensive Disorders of Pregnancy. Geographic variation in the incidence of hypertension in pregnancy. Am J Obstet Gynecol. 1988;158:80–83. [PubMed] [Google Scholar]
- 4.MacKay AP, Berg CJ, Duran C, Chang J, Rosenberg H. An assessment of pregnancy-related mortality in the United States. Paediatr Perinat Epidemiol. 2005;19:206–214. doi: 10.1111/j.1365-3016.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 5.Fronterhouse W, Christensen FC, Rayburn LA, Gilson GJ, Curet LB, Rayburn WF. Mandated preterm delivery: its prevalence and impact at a tertiary referral center. J Matern Fetal Med. 2001;10:162–165. doi: 10.1080/714904318. [DOI] [PubMed] [Google Scholar]
- 6.Ananth CV, Vintzileos AM. Epidemiology of preterm birth and its clinical subtypes. J Matern Fetal Neonatal Med. 2006;19:773–782. doi: 10.1080/14767050600965882. [DOI] [PubMed] [Google Scholar]
- 7.ACOG Committee on Practice Bulletins- Obstetrics. ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. Obstet Gynecol. 2002;99:159–167. doi: 10.1016/s0029-7844(01)01747-1. [DOI] [PubMed] [Google Scholar]
- 8.Buhimschi IA, Saade GR, Chwalisz K, Garfield RE. The nitric oxide pathway in pre-eclampsia: pathophysiological implications. Hum Reprod Update. 1998;4:25–42. doi: 10.1093/humupd/4.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Zamorski MA, Green LA. NHBPEP report on high blood pressure in pregnancy: a summary for family physicians. Am Fam Physician. 2001;64:263–270. 216. [PubMed] [Google Scholar]
- 10.Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19–50. doi: 10.1080/02648725.1996.10647923. [DOI] [PubMed] [Google Scholar]
- 11.Lindheimer MD, Katz AI. The normal and diseased kidney in pregnancy. In: Schrier RW, editor. Diseases of the Kidney and Urinary Tract. 7th edition. Philadelphia: Lippincott, Williams and Wilkins; 2001. pp. 2129–2165. [Google Scholar]
- 12.Ferrazzani S, Caruso A, De Carolis S, Martino IV, Mancuso S. Proteinuria and outcome of 444 pregnancies complicated by hypertension. Am J Obstet Gynecol. 1990;162:366–371. doi: 10.1016/0002-9378(90)90387-m. [DOI] [PubMed] [Google Scholar]
- 13.Silkensen JR, Kasiske BL. Laboratory assessment of kidney disease: clearance, urinalysis, and kidney biopsy. In: Brenner BM, editor. The Kidney. 7th edn. Philadelphia: WB Saunders; 2004. p. 1107. [Google Scholar]
- 14.Buhimschi CS, Norwitz ER, Funai E, Richman S, Guller S, Lockwood CJ, Buhimschi IA. Urinary angiogenic factors cluster hypertensive disorders and identify women with severe preeclampsia. Am J Obstet Gynecol. 2005;192:734–741. doi: 10.1016/j.ajog.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 15.Zhou XH, Obuchowski NA, McClish DK. Statistical Methods in Diagnostic Medicine. New York: Wiley & Sons; 2002. [Google Scholar]
- 16.Price CP, Newall RG, Boyd JC. Use of protein: creatinine ratio measurements on random urine samples for prediction of significant proteinuria: a systematic review. Clin Chem. 2005;51:1577–1586. doi: 10.1373/clinchem.2005.049742. [DOI] [PubMed] [Google Scholar]
- 17.Levine RJ, Thadhani R, Qian C, Lam C, Lim KH, Yu KF, Blink AL, Sachs BP, Epstein FH, Sibai BM, Sukhatme VP, Karumanchi SA. Urinary placental growth factor and risk of preeclampsia. JAMA. 2005;293:77–85. doi: 10.1001/jama.293.1.77. [DOI] [PubMed] [Google Scholar]
- 18.Buhimschi CS, Magloire L, Funai E, Norwitz ER, Kuczynski E, Martin R, Richman S, Guller S, Lockwood CJ, Buhimschi IA. Fractional excretion of angiogenic factors in women with severe preeclampsia. Obstet Gynecol. 2006;107:1103–1113. doi: 10.1097/01.AOG.0000207698.74104.4f. [DOI] [PubMed] [Google Scholar]
- 19.Weinstein S, Obuchowski NA, Lieber ML. Clinical evaluation of diagnostic tests. Am J Roentgenol. 184:14–19. doi: 10.2214/ajr.184.1.01840014. Review. [DOI] [PubMed] [Google Scholar]
- 20.Lindheimer MD, Davison JM. Renal biopsy during pregnancy: 'to b … or not to b …?'. Br J Obstet Gynaecol. 1987;94:932–934. doi: 10.1111/j.1471-0528.1987.tb02265.x. [DOI] [PubMed] [Google Scholar]
- 21.Buhimschi IA, Christner R, Buhimschi CS. Proteomic biomarker analysis of amniotic fluid for identification of intra-amniotic inflammation. BJOG. 2005;112:173–181. doi: 10.1111/j.1471-0528.2004.00340.x. [DOI] [PubMed] [Google Scholar]
- 22.Buhimschi CS, Weiner CP, Buhimschi IA. Clinical proteomics Part II. The emerging role of proteomics over genomics in spontaneous preterm labor/birth. Obstet Gynecol Survey. 2006;61:543–553. doi: 10.1097/01.ogx.0000228779.39568.59. [DOI] [PubMed] [Google Scholar]
- 23.Buhimschi IA, Buhimschi C. Proteomics of the amniotic fluid in assessment of the placenta. Relevance for preterm birth. Placenta 2008; 29, Suppl A. Trophoblast Res. 2008;22:S95–S101. doi: 10.1016/j.placenta.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merchant M, Weinberger S. Recent advancements in surface enhanced laser desorption/ionization time of flight mass spectrometry. Electrophoresis. 2000;21:1164–1167. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 25.Cronbach L. Coefficient alpha and the internal structure of tests. Psychometrika. 1951;16:297–333. [Google Scholar]
- 26.Fleiss JL, Cuzick J. The reliability of dichotomous judgements: unequal numbers of judges per subject. Applied Psychological Measurement. 1979;3:537–542. [Google Scholar]
- 27.Carrell RW, Jeppsson JO, Laurell CB, Brennan SO, Owen MC, Vaughan L, Boswell DR. Structure and variation of human alpha 1-antitrypsin. Nature. 1982;298:329–334. doi: 10.1038/298329a0. [DOI] [PubMed] [Google Scholar]
- 28.Callea F, Brisigotti M, Faa G, Lucini L, Eriksson S. Identification of PiZ gene products in liver tissue by a monoclonal antibody specific for the Z mutant of alpha 1-antitrypsin. J Hepatol. 1991;12:372–376. doi: 10.1016/0168-8278(91)90842-y. [DOI] [PubMed] [Google Scholar]
- 29.Janciauskiene S, Dominaitiene R, Sternby NH, Piitulainen E, Eriksson S. Detection of circulating and endothelial cell polymers of Z and wild type alpha 1-antitrypsin by a monoclonal antibody. J Biol Chem. 2002;277:26540–26546. doi: 10.1074/jbc.M203832200. [DOI] [PubMed] [Google Scholar]
- 30.James EL, Whisstock JC, Gore MG, Bottomley SP. Probing the unfolding pathway of alpha1-antitrypsin. J Biol Chem. 1999;274:9482–9488. doi: 10.1074/jbc.274.14.9482. [DOI] [PubMed] [Google Scholar]
- 31.Duley L. Maternal mortality associated with hypertensive disorders of pregnancy in Africa, Asia, Latin America and the Caribbean. Br J Obstet Gynaecol. 1992;99:547–553. doi: 10.1111/j.1471-0528.1992.tb13818.x. [DOI] [PubMed] [Google Scholar]
- 32.Chan P, Brown M, Simpson JM, Davis G. Proteinuria in pre-eclampsia: how much matters? BJOG. 2005;112:280–285. doi: 10.1111/j.1471-0528.2004.00395.x. [DOI] [PubMed] [Google Scholar]
- 33.Ferrazzani S, Caruso A, De Carolis S, Martino IV, Mancuso S. Proteinuria and outcome of 444 pregnancies complicated by hypertension. Am J Obstet Gynecol. 1990;162:366–371. doi: 10.1016/0002-9378(90)90387-m. [DOI] [PubMed] [Google Scholar]
- 34.Brown MA, Buddle ML. Inadequacy of dipstick proteinuria in hypertensive pregnancy. Aust N Z J Obstet Gynaecol. 1995;35:366–369. doi: 10.1111/j.1479-828x.1995.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Thompson D, Lieberman Use of a random urinary protein to creatinine ratio for the diagnosis of significant proteinuria during pregnancy. Am J Obstet Gynecol. 2001;185:808–811. doi: 10.1067/mob.2001.117349. [DOI] [PubMed] [Google Scholar]
- 36.Meyer NL, Mercer BM, Friedman SA, Sibai BM. Urinary dipstick protein: a poor predictor of absent or severe proteinuria. Am J Obstet Gynecol. 1994;170:137–141. doi: 10.1016/s0002-9378(94)70398-1. [DOI] [PubMed] [Google Scholar]
- 37.Brown MA, Lindheimer MD, de Swiet M, Van Assche A, Moutquin JM. The classification and diagnosis of the hypertensive disorders of pregnancy: statement from the International Society for the Study of Hypertension in Pregnancy (ISSHP) Hypertens Pregnancy. 2001;20:IX–XIV. doi: 10.1081/PRG-100104165. [DOI] [PubMed] [Google Scholar]
- 38.Coakley RJ, Taggart C, O'Neill S, McElvaney NG. Alpha1-antitrypsin deficiency: biological answers to clinical questions. Am J Med Sci. 2001;321:33–41. doi: 10.1097/00000441-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 39.Lisowska-Myjak B. AAT as a diagnostic tool. Clin Chim Acta. 2005;352(1–2):1–13. doi: 10.1016/j.cccn.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Carrell RW. alpha 1-Antitrypsin: molecular pathology, leukocytes, and tissue damage. J Clin Invest. 1986;78:1427–1431. doi: 10.1172/JCI112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergman D, Kadner SS, Cruz MR, Esterman AL, Tahery MM, Young BK, Finlay TH. Synthesis of alpha 1-antichymotrypsin and alpha 1-antitrypsin by human trophoblast. Pediatr Res. 1993;34:312–317. doi: 10.1203/00006450-199309000-00015. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O. glycoprotein (orosomucoid), alpha1-antitrypsin, and haptoglobin: a comparison of a large cohort to the world's literature. J Clin Lab Anal. 2000;14:265–270. doi: 10.1002/1098-2825(20001212)14:6<265::AID-JCLA3>3.0.CO;2-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engstrom G, Janzon L, Berglund G, Lind P, Stavenow L, Hedblad B, Lindgarde F. Blood pressure increase and incidence of hypertension in relation to inflammation-sensitive plasma proteins. Arterioscler Thromb Vasc Biol. 2002 Dec 1;22:2054–2058. doi: 10.1161/01.atv.0000041842.43905.f3. [DOI] [PubMed] [Google Scholar]
- 44.Engstrom G, Lind P, Hedblad B, Stavenow L, Janzon L, Lindgarde F. Long-term effects of inflammation-sensitive plasma proteins and systolic blood pressure on incidence of stroke. Stroke. 2002;33:2744–2749. doi: 10.1161/01.str.0000034787.02925.1f. [DOI] [PubMed] [Google Scholar]
- 45.Schmaier AH. The plasma kallikrein-kinin system counterbalances the renin-angiotensin system. J Clin Invest. 2002;109:1007–1009. doi: 10.1172/JCI15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janciauskiene S, Moraga F, Lindgren S. C-terminal fragment of alpha1-antitrypsin activates human monocytes to a pro-inflammatory state through interactions with the CD36 scavenger receptor and LDL receptor. Atherosclerosis. 2001;158:41–51. doi: 10.1016/s0021-9150(00)00767-x. [DOI] [PubMed] [Google Scholar]
- 47.Kupferminc MJ, Peaceman AM, Wigton TR, Rehnberg KA, Socol ML. Tumor necrosis factor-alpha is elevated in plasma and amniotic fluid of patients with severe preeclampsia. Am J Obstet Gynecol. 1994;170:1752–1757. [PubMed] [Google Scholar]
- 48.Aguzzi A, Haass C. Games played by rogue proteins in prion disorders and Alzheimer's disease. Science. 2003;302:814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 49.Matheson NR, Wong PS, Travis J. Enzymatic inactivation of human alpha-1-proteinase inhibitor by neutrophil myeloperoxidase. Biochem Biophys Res Commun. 1979;88:402–409. doi: 10.1016/0006-291x(79)92062-x. [DOI] [PubMed] [Google Scholar]
- 50.Lomas DA, Belorgey D, Mallya M, Onda M, Kinghorn KJ, Sharp LK, Phillips RL, Page R, Crowther DC, Miranda E. Polymerisation underlies alpha1-antitrypsin deficiency, dementia and other serpinopathies. Front Biosci. 2004;9:2873–2891. doi: 10.2741/1444. [DOI] [PubMed] [Google Scholar]
- 51.Gudehithlu KP, Pegoraro AA, Dunea G, Arruda JA, Singh AK. Degradation of albumin by the renal proximal tubule cells and the subsequent fate of its fragments. Kidney Int. 2004;65:2113–2122. doi: 10.1111/j.1523-1755.2004.00633.x. [DOI] [PubMed] [Google Scholar]
- 52.Travis J, Shieh BH, Potempa J. The functional role of acute phase plasma proteinase inhibitors. Tokai J Exp Clin Med. 1988;13:313–320. [PubMed] [Google Scholar]
- 53.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.