Abstract
The cell adhesion molecule L1 regulates axonal guidance and fasciculation during development. We previously identified the regulatory region of the L1 gene and showed that it was sufficient for establishing the neural pattern of L1 expression in transgenic mice. In the present study, we characterize a DNA element within this region called the HPD that contains binding motifs for both homeodomain and Pax proteins and responds to signals from bone morphogenetic proteins (BMPs). An ATTA sequence within the core of the HPD was required for binding to the homeodomain protein Barx2 while a separate paired domain recognition motif was necessary for binding to Pax-6. In cellular transfection experiments, L1-luciferase reporter constructs containing the HPD were activated an average of 4-fold by Pax-6 in N2A cells and 5-fold by BMP-2 and BMP-4 in Ng108 cells. Both of these responses were eliminated on deletion of the HPD from L1 constructs. In transgenic mice, deletion of the HPD from an L1-lacZ reporter resulted in a loss of β-galactosidase expression in the telencephalon and mesencephalon. Collectively, our experiments indicate that the HPD regulates L1 expression in neural tissues via homeodomain and Pax proteins and is likely to be a target of BMP signaling during development.
Keywords: gene regulation, neural development, promoters
Cell adhesion molecules (CAMs) are cell surface glycoproteins that regulate developmental processes such as axonal guidance, fasciculation, and synapse formation—all of which are essential for wiring of the embryonic nervous system (1). A particular CAM of the Ig superfamily called L1 is an important modulator of neuron-neuron and neuron-glia interactions during development of both the central and peripheral nervous systems (2, 3). L1 is an integral membrane protein composed of six Ig domains, five fibronectin type three repeats, and a cytoplasmic domain (4). Mutations in the human L1 gene are responsible for a variety of congenital neural disorders, including hydrocephalus, mental retardation, and agenesis of the corpus callosum (5).
The expression of L1 is particularly dynamic during neuronal migration and path finding, changing in granule cells during their migration in the cerebellum (6), and in commissural neurons after their axons cross the floor plate into the contralateral spinal cord (7). These observations suggest that understanding the regulation of L1 expression would be a significant step in relating the place-dependent appearance of this CAM to its role in morphogenesis. We therefore have sought to identify cis-regulatory elements and associated trans-factors that either activate or repress the expression of the L1 gene.
In a previous study (8), we identified the L1 promoter and found that a 15-kb segment of the mouse L1 gene extending from the promoter to the fourth exon (see Fig. 1A) was required to produce a neural pattern of expression in transgenic mice. However, deletion of a single neuron restrictive silencer element (NRSE) located within the second intron of the L1 gene resulted in extraneural expression of a β-galactosidase-containing reporter gene L1lacZΔNRSE. This study established a role for the NRSE in silencing L1 expression outside of the nervous system, but did not identify positive regulatory elements that induce L1 gene expression in either neural or non-neural cells.
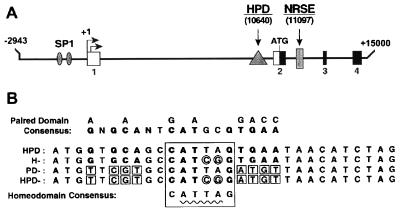
Figure 1.
(A) Location of DNA control elements in the mouse L1 gene. The first four exons are indicated by boxes labeled 1–4. The transcription initiation sites are demarcated by rightward pointing arrows. Upstream of this position is the L1 promoter containing two SP1 motifs. An NRSE that binds to the factor REST/NRSF is located within the second intron, and the HPD is located in the first intron. (B) Sequences of HPD, H−, PD−, and HPD− probes. Drawn above the HPD is the consensus paired domain binding sequence. Nucleotide similarities between this consensus sequence and the HPD are indicated in boldface type. Mutations in the PD− probe are indicated by small squares. Drawn below the PD− probe is the consensus homeodomain binding sequence CATTAG. The location of this motif in all four probes motif is indicated by a large box. Mutations made within the ATTA core sequence in the HD− variant are indicated by small circles. Both sets of mutations were included in the HPD− probe.
To identify positive elements that control L1 gene expression, we pursued our observations that, in the mice carrying the L1lacZΔNRSE transgene, extraneural L1 expression appeared in tissues that are known to receive signals from differentiation factors of the bone morphogenetic protein (BMP) family. Recent studies have shown that BMPs modulate the activity of several homeodomain and Pax proteins (9–14) and can increase the synthesis of L1 mRNA in the neural cell line, Ng108–15 (15). We therefore searched for DNA regulatory elements within the L1 gene that respond to BMPs, focusing on elements that also respond to cues from homeodomain and Pax transcription factors.
In particular, we investigated the role in the regulation of L1 gene expression of a DNA element called the HPD that contains binding sites for both homeodomain and paired domain (Pax) proteins. This element previously had been identified as a binding site for HoxA1 and Pax-6 (16), but was not demonstrated to regulate L1 gene expression because the L1 gene promoter had not been isolated. Our subsequent identification of an additional 10 kb of the L1 gene, which included the true promoter and a large segment of the first intron (8), prompted us to re-examine the function of the HPD in the context of these additional regulatory sequences.
MATERIALS AND METHODS
The cDNAs encoding murine Pax-6, Pax-3, and Pax-1 were provided by Peter Gruss (Max-Planck Institute, Stuttgart, Germany). The cDNAs for HoxA1, Otx-1, and Otx-2 were generated by reverse transcriptase–PCR. Barx2 cDNAs were isolated in a previous study (17). All cDNAs were cloned into a pcDNA3 vector (Invitrogen) containing an amino terminal myc-tag, and the corresponding proteins were synthesized in COS7 cells after transfection as described (18).
Double-stranded HPD, H−, PD−, and HPD− oligonucleotides were labeled with the Klenow fragment of DNA polymerase I and α-32P-dCTP (6,000 Ci/mmol). Binding reactions and gel mobility-shift experiments were performed as described (18). Competitor DNAs (in 200-fold molar excess) were added simultaneously with probe. For supershift analyses, a monoclonal Pax-6 antibody (Babco) or a myc tag antibody (Sigma) were preincubated with extracts before addition of probe.
L1-luciferase reporter constructs were constructed as described (8). Three additional constructs also were prepared: 4.4E2, L1–11ΔHPD, and L1lacZΔHPD (see Fig. 3). For 4.4E2, a 4.4-kb XbaI fragment containing the 3′ end of the first intron and the second exon was inserted into the pGL2basic vector (Promega). L1–11ΔHPD was constructed by PCR-mediated deletion of the HPD sequence from the L1–10 plasmid (generating the plasmid L1–10ΔHPD) and subsequent insertion of a fragment containing exons 2–4 into the Sse8387I site of L1–10ΔHPD to yield L1–11ΔHPD. These same manipulations also were performed on the L1lacZ plasmid to generate L1lacZΔHPD.
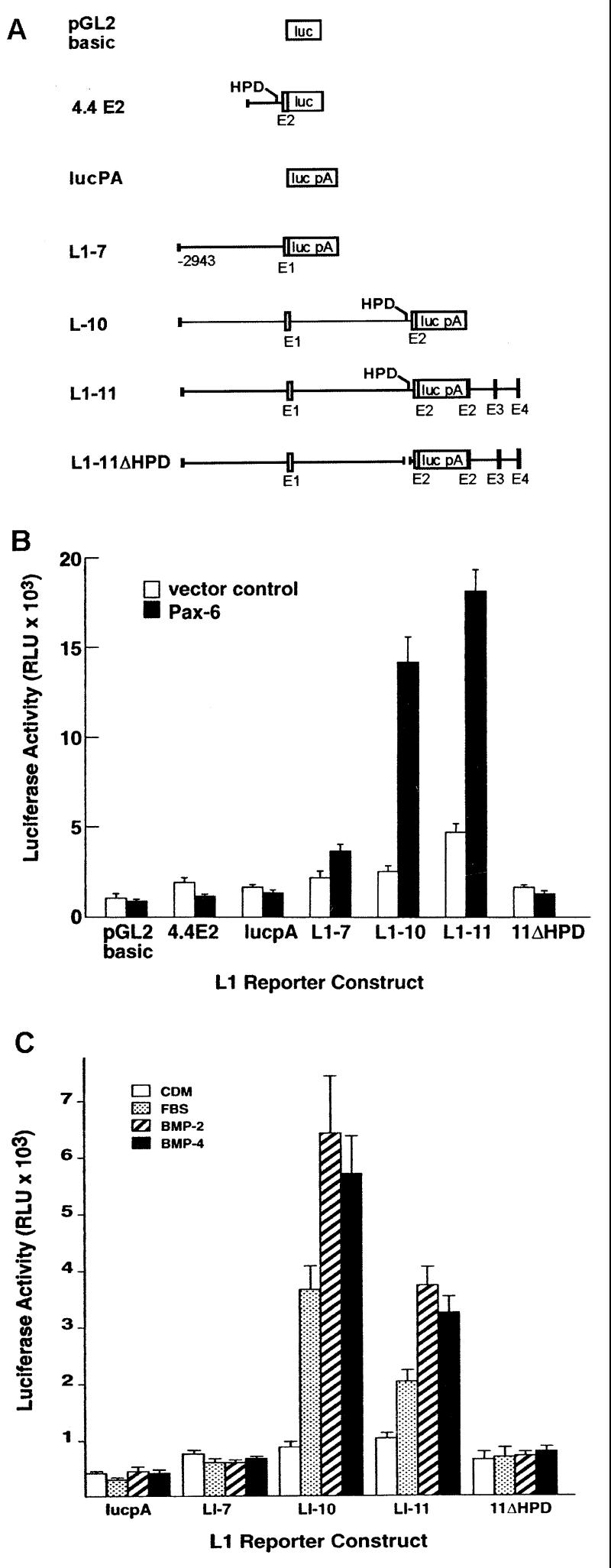
Figure 3.
(A) L1-luciferase reporter constructs used to examine induction of L1 expression by Pax-6 and BMPs. The construct 4.4E2 contains the 4.4-kb XbaI fragment demonstrated to have some promoter activity in a previous study (25). This construct was made in the promoterless pGL2basic vector. All other constructs were prepared by ligation of L1 fragments into the lucpA vector, which contains a luciferase/simian virus 40 poly(A) gene cassette. Exons are indicated by open and filled boxes (noncoding and coding sequences, respectively). (B) Activation of L1 gene constructs by Pax-6 in N2A cells. (C) Activation of L1 gene constructs by FBS and by BMP2 and BMP4. The values for luciferase activity were determined from six independent experiments, performed in duplicate.
N2A cells were cultured in DMEM containing 10% FBS. Cells were cotransfected with Pax-6 expression plasmid and L1 reporters as described (8). The reporter plasmid pCMVβ (CLONTECH) was cotransfected in all experiments to provide an internal reference standard of β-galactosidase activity to which luciferase activities were normalized. Values for luciferase activities were derived from six independent experiments performed in duplicate. Ng108 cells were maintained in DMEM with 10% FBS, 1% glutamine, and 1× hypoxanthine/aminopterin/thymidine media. For transfection and BMP treatment, 1 × 105 cells were seeded into 6-well dishes and cultured for 24 hr in serum-containing media, changed to a chemically defined media (CDM) (15, 19), incubated for 72 hr, and then transfected with L1-luciferase reporters as described (8). The media were replaced 24 hr later with either CDM, DMEM with 10% FBS, CDM with 10 ng/ml BMP2, or CDM with 10 ng/ml BMP4 and then incubated for an additional 72 hr. Cells were harvested and β-galactosidase and luciferase assays were performed as described (8). BMPs were provided by the Genetics Institute, Cambridge, MA.
To generate transgenic mice, L1lacZ and L1lacZΔHPD transgene fragments were excised from plasmids by using restriction enzymes XbaI and SnaBI. Each transgene fragment was injected into CB6 mouse zygotes by using standard microinjection techniques (20). Injected zygotes were transferred to the oviducts of pseudopregnant foster mothers. At day 11.5 of gestation, the mice were sacrificed, and F0 progeny were collected and analyzed for β-galactosidase expression either in whole-mount preparations or in sections as described (8). For each construct, 16 embryos were obtained; these sample sizes were picked to allow us to evaluate the contributions of independent integration events.
RESULTS
Barx2 and Pax-6 Bind to Different Sequences Within the HPD.
The HPD (Fig. 1B) contains two overlapping DNA binding motifs: an ATTA sequence that is characteristic of a binding site for homeodomain proteins (21, 22) and a sequence that matches the consensus binding site for the paired domain of Pax proteins (23, 24). We first examined binding of the HPD to seven different homeodomain and Pax proteins: HoxA1, Barx2, Otx1, Otx2, Pax-1, Pax-3, and Pax-6; only Barx2 and Pax-6 bound to the HPD (data not shown). To determine the sequences in the HPD that were required for binding to Barx2 and Pax-6, we analyzed the direct binding of 32P-labeled HPD probe, and three variants (H−, PD−, and HPD−) to these proteins (Figs. 1B and 2). The H− variant contained two bp substitutions that altered the ATTA homeodomain binding sequence, the PD− variant contained eight bp substitutions that disrupted the consensus binding sequence for the paired domain of Pax proteins, and the HPD− probe contained both sets of mutations. Unlabeled HPD, HD−, PD−, and HPD− also were included as competitors in binding reactions between Barx2 and Pax-6 and the 32P-labeled HPD probe. Antibody supershift/blockade experiments also were performed to demonstrate the specificity of binding.
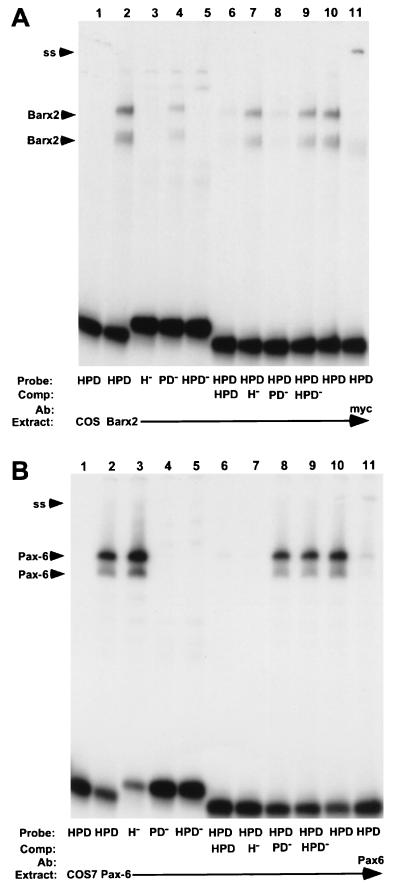
Figure 2.
Binding of Barx2 (A) and Pax-6 (B) produced in COS7 cells to the HPD, H−, PD−, or HPD− probes. The specific 32P-labeled probe, protein extract, unlabeled competitor, or antiserum used in gel shift experiments is indicated at the bottom. Binding reactions contained DNA probe and 5 μg of protein extract from either mock-, Barx2-, or Pax-6-transfected COS7 cells. Unlabeled HPD, H−, PD−, or HPD− competitors were included at 200-fold molar excess and 1 μl of anti-myc-tag or Pax-6 antisera were added at the beginning of binding reactions. Positions of supershifted Barx2 and Pax-6 complexes (SS) are indicated.
Barx2 formed two binding complexes with both the 32P-labeled HPD and PD− probes, but did not bind to H− and HPD− probes (Fig. 2A, lanes 2–5). Unlabeled HPD and PD−, but not H− and HPD− competitors blocked the formation of both Barx2 complexes with the 32P-labeled HPD probe (Fig. 2A, lanes 6–9). These results indicated that the ATTA motif is critical for binding of the homeodomain of Barx2 and that mutations in the paired domain recognition sequence do not diminish the ability of Barx2 to bind the HPD. An antibody to the myc tag present at the amino terminus of the Barx2 protein generated a supershifted complex when added to the binding reaction (Fig. 2A, lane 11), indicating that both of the complexes formed with the HPD contain Barx2.
Pax-6 produced two binding complexes with 32P-labeled HPD and H− probes, but not with PD− and HPD− probes (Fig. 2B, lanes 2–5). In competition experiments, only unlabeled HPD and H− (but not PD− or HPD−) competitors interfered with binding of Pax-6 to the HPD (Fig. 2B, lanes 6–9). These results demonstrate that the paired domain recognition sequence is necessary for Pax-6 binding to the HPD and that the ATTA motif is not required. A mAb to Pax-6 blocked the formation of both complexes and also produced a small amount of supershifted complex (Fig. 2B, lane 11), indicating that both complexes contain Pax-6. Collectively, these experiments show that the HPD contains distinct binding modules that can be recognized independently by homeodomain proteins and paired domain (Pax) proteins.
Pax-6 Activates the Expression of L1 Reporter Constructs in N2A Cells.
Different L1-luciferase reporter constructs (Fig. 3A) were tested for activity in N2A cells after cotransfection with a Pax-6 expression plasmid. The 4.4E2 plasmid contains 4.4 kb of the L1 gene that includes the HPD and is nearly identical to a construct previously shown to have some activity in these cells (25). Additional reporters were prepared containing a larger fragment that included exon 1 and the L1 promoter (8).
As shown in Fig. 3B, 4.4E2 produced low levels of luciferase activity that were not increased in cells on cotransfection with the Pax-6 expression plasmid. This result indicates that the region containing the HPD immediately upstream of exon 2 cannot activate expression of the reporter in response to Pax-6, probably because it does not contain a promoter that functions in this context. The L1–7 construct, containing the L1 promoter but not the first intron, also produced basal levels of luciferase activity and was not activated appreciably by Pax-6. In contrast, L1–10, which contains the L1 promoter and the first intron, and L1–11, which contains the region spanning the promoter to the fourth exon, showed basal levels of activity similar to L1–7, but were activated by Pax-6 approximately 4- and 5-fold, respectively (Fig. 3B). Deletion of the HPD in the construct L1–11ΔHPD resulted in a complete loss of Pax-6 activation and a lower basal level of expression. Overall, these results indicated that the HPD element is essential for Pax-6 activation of the L1 gene in N2A cells.
BMP2 and BMP4 Induce the Expression of L1 Gene Constructs in Ng108 Cells.
BMPs increase the synthesis of L1 mRNA and protein in the Ng108–15 neuroblastoma-glioma hybrid cell line (15, 19). We therefore sought to determine whether BMPs could activate our L1 gene constructs in these cells and to define the sequences required for activation by these factors. The activity of the L1 reporter constructs was examined in Ng108–15 cells under four different conditions: CDM (15), DMEM with FBS, CDM with 10 ng/ml BMP2, and CDM with 10 ng/ml BMP4. As shown in Fig. 3C, the L1–7 construct had low basal activity and showed no increase in activity on addition of FBS, BMP2, or BMP4. In contrast, L1–10 activity was increased 4- and 6-fold in Ng108 cells treated with FBS and BMPs, respectively, indicating that the first intron and exon 2 are required for activation by FBS and BMPs. L1–11 also was activated by FBS and BMP treatment (2- and 3.5-fold, respectively). This level of induction was reduced relative to L1–10, indicating that the region between exons 2 and 4 (which includes the NRSE) diminishes the overall activation of L1 expression by BMPs. L1–11ΔHPD was not activated by either FBS or BMPs, indicating that deletion of the HPD eliminates the ability of the L1 gene to respond both to serum and BMPs.
Function of the HPD in Transgenic Mice.
To test the function of the HPD in vivo, we examined the expression patterns of two L1 reporter constructs, L1lacZ and L1lacZΔHPD in transgenic mice. These constructs are identical to L1–11 and L1–11ΔHPD, respectively (Fig. 3A), except that they contain the Escherichia coli lacZ gene instead of a luciferase gene. We generated 16 transgenic Fo embryos carrying each construct. Each of these embryos represented an independent transgene integration event. The β-galactosidase expression patterns then were examined at 11.5 days of gestation in whole mounts and sections.
In general, embryos carrying the L1lacZ transgene showed a more consistent and intense level of expression in the central nervous system (CNS), whereas those that carried L1lacZΔHPD showed frequent loss of expression in rostral regions of the CNS, primarily in the telencephalon and mesencephalon. Three distinct telencephalon-mesencephalon expression patterns, designated TM1, TM2, and TM3, were observed in these studies (Fig. 4A). Embryos classified as TM1 showed strong expression in either the telencephalon or mesencephalon, those in the TM2 group showed moderate expression in either the telencephalon or mesencephalon, and embryos in the TM3 category showed no expression in either of these regions. As shown in Fig. 4A, the TM1 pattern was found more frequently in embryos carrying the L1lacZ reporter (10/16) than in embryos carrying the L1lacZΔHPD construct (2/16). Conversely, a greater proportion of L1lacZΔHPD embryos (9/16) showed no transgene expression in the telencephalon and mesencephalon (pattern TM3) relative to L1lacZ embryos (3/16).
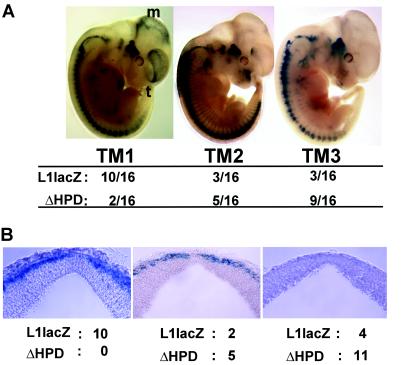
Figure 4.
(A) Whole-mount analysis of L1lacZ and L1lacZΔHPD expression patterns in transgenic mouse embryos staged at embryonic day 11.5. Sixteen animals were examined for each construct. Patterns of β-galactosidase expression in the telencephalon and mesencephalon were divided into three categories (TM1, TM2, and TM3) (numbers refer to relative frequencies of patterns; see Results for further explanation of these patterns). (B) Deletion of the HPD reduces the expression of β-galactosidase within the mesencephalon. Transverse sections (50 μm) were prepared from embryonic day 11.5 transgenic embryos. The numbers of L1lacZ and L1lacZΔHPD embryos showing strong (Left), intermediate (Center), or no (Right) expression of β-galactosidase in the marginal zone of the mesencephalon are indicated below each panel.
Examination of β-galactosidase expression in transverse sections of all of these embryos confirmed our observations that deletion of the HPD resulted in a loss of β-galactosidase expression in the telencephalon and mesencephalon. The most dramatic loss of β-galactosidase expression occurred in the mesencephalon. None of the L1lacZΔHPD embryos showed the intense staining in the mesencephalon that was observed in the majority (10/16) of L1lacZ embryos (Fig. 4B, Left). Although the majority of L1lacZΔHPD embryos (11/16) showed no expression in the mesencephalon, this lack of expression was infrequent (4/16) in embryos carrying the L1lacZ transgene (Fig. 4B, Right). Even the strongest pattern of β-galactosidase expression observed in L1lacZΔHPD embryos was not comparable in intensity to the level of expression observed in L1lacZ embryos (Fig. 4B, compare Center and Left).
The L1lacZ transgene also was expressed more frequently in the hind brain than the L1lacZΔHPD transgene (11/16 versus 7/16 embryos) although the intensity of β-galactosidase expression in both L1lacZ and L1lacZΔHPD was comparable (data not shown). The two transgenes were expressed in the spinal cord with approximately equal frequency (11/16 versus 10/16) and identical intensity. In contrast to the loss of expression observed in the rostral CNS, all L1lacZ and L1lacZΔHPD embryos showed expression of β-galactosidase in cranial and dorsal root ganglia (16/16), suggesting that deletion of the HPD had no effect on L1 expression in the peripheral nervous system (data not shown).
DISCUSSION
We have characterized the role of a binding site for homeodomain and Pax proteins, designated the HPD, in the regulation of L1 expression in vitro and in vivo. The HPD element is composed of two consensus transcription factor binding motifs that are recognized by homeodomain and Pax proteins, respectively. The HPD was found to be a regulatory target of the transcription factor Pax-6 and was necessary for induction of L1 expression by two members of the BMP family, BMP2 and BMP4. Deletion of the HPD from an L1 reporter construct in transgenic mice resulted in a loss of L1 expression in the rostral CNS. These studies indicate that the HPD is necessary to activate L1 expression in neural tissues, and it is also likely to be a target of BMP signaling during development.
Of the seven homeodomain and Pax proteins that were examined for their ability to bind to the HPD, only two, Barx2 and Pax-6, bound specifically. We examined the binding of Barx2 and Pax-6 to mutated versions of the HPD. Mutations in the HPD that affected the binding of one protein could be generated without affecting the binding of the other, indicating that the homeodomain and paired domain recognition modules can operate independently. The modular structure of the HPD thus is consistent with the notion that this element could receive regulatory signals from two different classes of transcription factors.
Although the HPD element had been demonstrated to bind to Pax-6 in a previous study (16) the role of the element or Pax-6 in regulating L1 expression could not be established because the true promoter (8) had not yet been identified. In the present study, we investigated L1 expression in the context of this promoter and sequences from the first intron of the gene. Expression from an L1 construct containing the region extending from the promoter to the second exon was increased on cotransfection of Pax-6 in N2A cells. Deletion of the HPD in this construct abolished the response, indicating that the HPD is essential for activation of L1 gene expression by Pax-6 in neural cells.
In connection with activation by homeodomain proteins, we showed in a previous study (17) that L1 reporters containing the HPD were activated by Barx2 in a fibroblast cell line (NIH3T3). In the absence of Barx2, however, L1 constructs lacking the HPD had significantly greater luciferase activity than those having the HPD, indicating that in NIH3T3 cells, the HPD acts as a silencer. Furthermore, cotransfection of Barx2 together with L1 constructs lacking the HPD resulted in a dramatic repression of luciferase activity. These findings underline the need to explore more fully the different cellular contexts in which the HPD acts either as an enhancer or silencer and to determine the particular homeodomain and Pax proteins that carry out these functions.
A number of experiments have indicated that expression of L1 could be induced in cell culture on treatment with BMP2 and BMP4 (15, 19). We therefore explored the role of BMP2 and BMP4 in regulating L1 promoter activity in neural cells. We found that the expression of gene constructs containing the promoter and the first intron of the L1 gene was induced by BMP treatment of Ng108–15 cells. Deletion of the HPD element prevented this induction, indicating that the HPD was essential for activation of L1 expression by BMPs. These experiments therefore indicate that the HPD and the proteins to which it binds are key components in the induction of L1 expression by BMPs. In accord with these results, other investigators have demonstrated that BMPs regulate the expression of several homeodomain and Pax proteins, including Msx1 (10), Msx2 (26), Mix1 (12), Xom (11), Pv1 (13), and Barx1 (27). The BMP antagonist Noggin, which functions by binding to BMPs and blocking their interaction with BMP receptors, can up-regulate Pax-1 expression (28). These experiments therefore suggest that there are several transcription factors whose activity may be influenced by BMP signaling and which might be involved in the regulation of L1 expression by binding to the HPD.
Our previous findings that deletion of the NRSE in the L1 transgene led to ectopic activation of L1 expression in several non-neural nontissues that require BMP signaling for morphogenesis (8) suggest that the NRSE together with its binding factor NRSF/REST (29) may block the activation of L1 gene expression by BMPs. It therefore will be important in future studies to determine whether NRSF/REST is part of a mechanism that silences BMP activation of L1 gene expression, and if so, whether it functions independent of or together with particular homeodomain or Pax proteins that bind to the HPD.
Given the results in vitro, it was important to examine the influence of the HPD on expression in vivo. To accomplish this, we generated transgenic mice carrying either a wild-type L1 reporter construct (L1lacZ) or a construct lacking the HPD element (L1lacZΔHPD) and examined β-galctosidase expression from these transgenes on a background of random genomic integration sites. Deletion of the HPD reduced both the level and frequency of cases in which L1-β-galactosidase reporter constructs were expressed in the telencephalon and mesencephalon. Therefore we conclude that the HPD functions as an enhancer in the forebrain and midbrain.
The function of the HPD as an enhancer at a particular tissue site necessarily depends on the particular homeodomain and Pax proteins that bind to the HPD. One Pax protein, Pax-6, is important for proper morphogenesis of the telencephalon (30). Moreover, in the Small eye mouse, which produces a truncated form of Pax-6 (31), the cytoarchitecture of the cortex is disrupted and the pattern of L1 expression is abnormal (32). Our combined observations that Pax-6 activates L1 expression via the HPD and that deletion of the HPD leads to a loss of L1 expression in the telencephalon suggests that Pax-6 is a key regulator of L1 gene expression. In the mesencephalon where Pax-6 is not expressed, other transcription factors are likely to be important for regulating L1 expression. Although Pax-3 shows prominent expression in the mesencephalon and would be an attractive candidate for the regulation of L1 via the HPD in this brain region, we did not detect any binding of Pax-3 to the HPD in vitro. However, Pax-2, Pax-5, and En-2, which are early determinants of mesencephalon development (33), also may regulate L1 expression via the HPD and deserve further exploration.
Further elucidation of the role of the HPD in determining the pattern of neural and non-neural L1 expression may be derived in future experiments in which the homeodomain paired domain recognition sequences are independently mutated. This should allow us to examine the relative contributions of homeodomain and Pax transcription factors, such as those mentioned above, in the spatial regulation of L1 expression via the HPD. Moreover, this approach may provide some clues as to the identity of the factors that mediate the induction of L1 expression by BMPS. It also may be worthwhile to consider the targeted mutagenesis of the HPD element within the native L1 gene by homologous recombination. This may disrupt L1 expression in the mesencephalon and telencephalon, leading to aberrant neural morphogenesis and will provide insight into how the HPD functions in its normal chromosomal environment.
L1 is just one of a number of CAMs that must be considered in relating homeobox and Pax gene activity to morphogenesis. We have shown that the genes for several CAMs are regulated by homeodomain and Pax proteins (34). For example, the mouse N-CAM gene contains binding sites for Hox and Pax proteins that are necessary for the establishment and maintenance of N-CAM expression in the developing spinal cord (35, 36). Binding sites for Hox and Pax proteins have been identified within the regulatory regions of other neurally expressed CAMs, including Ng-CAM, F3, and axonin-1 (37, 38), suggesting that these CAMs also might be regulatory targets of Hox and Pax transcription factors. In several cases, the patterns of Hox and Pax gene expression in the developing brain correlate with the morphological tissue borders that define segments of developing neural tissue known as neuromeres (39, 40). Neighboring neuromeres have different adhesive properties, suggesting that sorting out of the cells in these compartments may result from the expression of different repertoires of CAMs (32, 41).
Overall, the present findings on L1 indicate that the HPD binds to both homeodomain proteins and Pax proteins, responds to signals from BMPs, and is an important regulator of L1 gene expression in vivo. It will be necessary in future experiments to correlate the expression patterns of L1lacZ and L1lacZΔHPD transgenes with the expression of BMPs, BMP receptors, and the homeodomain and Pax transcription factors that bind to the HPD. The results should allow us to understand better how the regulation of L1 expression by BMPs acting via homeodomain and Pax proteins could influence the patterning of tissues both within and outside the nervous system.
Acknowledgments
We are grateful to Peter Ballance, Jill Cleary, Nicole Son, and Dana Vollmer for excellent technical assistance. We thank Drs. Kathryn Crossin, Bruce Cunningham, Joe Gally, and Vince Mauro for helpful suggestions and critical reading of the manuscript. This work was supported by Public Health Service Grants NS34493 (to F.S.J.) and HD33576 (to G.M.E.) and a grant from the G. Harold and Leila Mathers Charitable Trust. G.M.E. is a consultant to Becton Dickinson.
ABBREVIATIONS
- BMP
bone morphogenetic protein
- CAM
cell adhesion molecule
- HPD
homeodomain and paired domain binding site
- CNS
central nervous system
- CDM
chemically defined media
- NRSE
neuron restrictive silencer element
References
- 1.Edelman G M, Crossin K L. Annu Rev Biochem. 1991;60:155–190. doi: 10.1146/annurev.bi.60.070191.001103. [DOI] [PubMed] [Google Scholar]
- 2.Stallcup W B, Beasley L L, Levine J M. J Neurosci. 1985;5:1090–1101. doi: 10.1523/JNEUROSCI.05-04-01090.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miragall F, Kadmon G, Husmann M, Schachner M. Dev Biol. 1988;129:516–531. doi: 10.1016/0012-1606(88)90397-1. [DOI] [PubMed] [Google Scholar]
- 4.Moos M, Tacke R, Scherer H, Teplow D, Fruh K, Schachner M. Nature (London) 1988;334:701–703. doi: 10.1038/334701a0. [DOI] [PubMed] [Google Scholar]
- 5.Wong E V, Kenwrick S, Willems P, Lemmon V. Trends Neurosci. 1995;18:168–172. doi: 10.1016/0166-2236(95)93896-6. [DOI] [PubMed] [Google Scholar]
- 6.Liljelund P, Ghosh P, van den Pol A N. J Biol Chem. 1994;269:32886–32895. [PubMed] [Google Scholar]
- 7.Dodd J, Jessell T M. Science. 1988;242:692–699. doi: 10.1126/science.3055291. [DOI] [PubMed] [Google Scholar]
- 8.Kallunki P, Edelman G M, Jones F S. J Cell Biol. 1997;138:1343–1354. doi: 10.1083/jcb.138.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang S J, Hoodless P A, Lu Z, Breitman M L, McInnes R R, Wrana J L, Buchwald M. Development (Cambridge, UK) 1998;125:1877–1887. doi: 10.1242/dev.125.10.1877. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki A, Ueno N, Hemmati-Brivanlou A. Development (Cambridge, UK) 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- 11.Ladher, R., Mohun, T., Smith, J. & Snape, A. (1996) Development (Cambridge, U.K.)122, Suppl., 2385–2394. [DOI] [PubMed]
- 12.Mead P E, Brivanlou I H, Kelley C M, Zon L I. Nature (London) 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- 13.Ault K T, Dirksen M-L, Jamrich M. Dev Biol. 1996;93:6415–6420. doi: 10.1073/pnas.93.13.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pituello F, Yamada G, Gruss P. Proc Natl Acad Sci USA. 1995;92:6952–6956. doi: 10.1073/pnas.92.15.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perides G, Hu G, Rueger D C, Charness M E. J Biol Chem. 1993;268:25197–25205. [PubMed] [Google Scholar]
- 16.Chalepakis G, Wijnholds J, Giese P, Schachner M, Gruss P. DNA Cell Biol. 1994;13:891–900. doi: 10.1089/dna.1994.13.891. [DOI] [PubMed] [Google Scholar]
- 17.Jones F S, Kioussi C, Copertino D W, Kallunki P, Holst B H, Edelman G M. Proc Natl Acad Sci USA. 1997;94:2632–2637. doi: 10.1073/pnas.94.6.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copertino D W, Edelman G M, Jones F S. Proc Natl Acad Sci USA. 1997;94:1846–1851. doi: 10.1073/pnas.94.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perides G, Safran R M, Downing L A, Charness M E. J Biol Chem. 1994;269:765–770. [PubMed] [Google Scholar]
- 20.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Habor Lab. Press; 1994. [Google Scholar]
- 21.Hoey T, Levine M. Nature (London) 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- 22.Desplan C, Theis J, O’Farrell P H. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zannini M, Francis-Lang H, Plachov D, DiLauro R. Mol Cell Biol. 1992;12:4230–4241. doi: 10.1128/mcb.12.9.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czerny T, Schaffner G, Busslinger M. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 25.Kohl A, Giese K P, Mohajeri M H, Montag D, Moos M, Schachner M. J Neurol Res. 1992;32:167–177. doi: 10.1002/jnr.490320206. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari D, Lichtler A C, Pan Z Z, Dealy C N, Upholt W B, Kosher R A. Dev Biol. 1998;197:12–24. doi: 10.1006/dbio.1998.8880. [DOI] [PubMed] [Google Scholar]
- 27.Tucker A S, Matthews K L, Sharpe P T. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 28.McMahon J A, Takada S, Zimmerman L B, Fan C M, Harland R M, McMahon A P. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenherr C J, Anderson D J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 30.Stoykova A, Fritsch R, Walther C, Gruss P. Development (Cambridge, UK) 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- 31.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson J M, Prosser J, Jordan T, Hastie N D, VanHeyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 32.Caric D, Gooday D, Hill R E, McConnell S K, Price D J. Development (Cambridge, UK) 1997;124:5087–5096. doi: 10.1242/dev.124.24.5087. [DOI] [PubMed] [Google Scholar]
- 33.Joyner A L. Trends Genet. 1996;12:15–19. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- 34.Edelman G M, Jones F S. Brain Res Rev. 1998;26:337–352. doi: 10.1016/s0165-0173(97)00034-9. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Jones F S, Krushel L A, Edelman G M. Proc Natl Acad Sci USA. 1996;93:1892–1896. doi: 10.1073/pnas.93.5.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holst B D, Goomer R S, Wood I C, Edelman G M, Jones F S. J Biol Chem. 1994;269:22245–22252. [PubMed] [Google Scholar]
- 37.Kallunki P, Jenkinson S, Edelman G M, Jones F S. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 38.Buttiglione M, Cangiano G, Goridis C, Gennarini G. Mol Brain Res. 1995;29:297–309. doi: 10.1016/0169-328x(94)00262-d. [DOI] [PubMed] [Google Scholar]
- 39.Puelles L, Rubenstein J L. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 40.Bulfone A, Puelles L, Porteus M H, Frohman M A, Martin G R, Rubenstein J L R. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingate R J, Lumsden A. Development (Cambridge, UK) 1996;122:2143–2152. doi: 10.1242/dev.122.7.2143. [DOI] [PubMed] [Google Scholar]