Abstract
RalBP1, a multifunctional protein implicated in cancer cell proliferation, radiation and chemoresistance and ligand dependent receptor internalization, is upregulated in bladder cancer and is a downstream effector of RalB, a GTPase associated with metastasis. RalBP1 can be regulated by phosphorylation by protein kinase C (PKC). No studies have comprehensively mapped RalBP1 phosphorylation sites or whether RalB affects these. We identified fourteen phosphorylation sites of RalBP1 in human bladder carcinoma UMUC-3 and embryonic kidney derived 293T cells. The phosphorylated residues are concentrated at the N-terminus. Ten of the first 100 amino acids of the primary structure were phosphorylated. Nine were serine residues, and one a threonine. We evaluated the effect of RalB overexpression on RalBP1 phosphorylation and found the largest change in phosphorylation status at S463 and S645. Further characterization of these sites will provide novel insights on RalBP1 biology, its functional relationship to RalB and possible avenues for therapeutic intervention.
Keywords: RalBP1, phosphorylation, Ral GTPase
INTRODUCTION
RalBP1, a multifunctional protein, plays a role in cell signaling [1; 2; 3; 4; 5], ligand dependent receptor internalization [6; 7; 8] and xenobiotic defense mechanisms, putatively as a membrane bound ATP dependent pump [9; 10]. The latter is particularly important since RalBP1 plays a role in cancer cell chemoresistance [11]. Originally, this protein was cloned as a GTP-dependent interaction partner to RalA, a small GTPase downstream of Ras with importance for human carcinogenesis [2]. RalBP1 also binds directly to RalB [12], a homologous protein associated with cancer metastasis [3].
RalBP1 shares sequence similarity with GAP proteins capable of activating the GTPase activity of members of the Rho/Rac family of GTPases. RalBP1 activates the GTPase activity of Cdc42 and, to a lesser extent, Rac1 but not RhoA, Ras, or Ral [13]. Since RalBP1 directly binds RalB and may affect Rac1 and Cdc42 activity, two proteins involved in cell migration [14], RalBP1 may be critical in tumor invasion and metastasis. Moreover, since RalBP1 is specifically expressed in bladder cancer more than other human tumors [15], the functional relevance of this protein may be even more important in bladder cancer.
Despite the important regulatory role of phosphorylation in cell signaling, little is known about RalBP1. Recent reports on RalBP1 regulation by Protein Kinase C alpha (PKCalpha) sequence analysis indicated four putative PKCalpha phosphorylation sites [16]. In vitro analysis of the four amino acids (S118, T297, S353 and S509) indicates that T297 is by far the most abundantly phosphorylated site [17]. PKC knockdown does not affect doxorubicin sensitivity of mouse embryonic fibroblasts from a RalBP1−/− mouse but increases that of RalBP1+/+ derived cells [18], further indicating a regulatory role of PKC for RalBP1.
In summary, RalBP1 is an important signaling intermediary in cancer, affecting cell migration and metastasis and chemotherapeutic resistance. We sought to comprehensively map the phosphorylation sites of RalBP1 using mass spectroscopic analysis and determine whether RalB, a major upstream binding partner of RalBP1, affects these.
MATERIALS AND METHODS
Cell culture, cDNA constructs and transfection procedures
UMUC-3 cells were cultured as described [19]. 293T cells were obtained from American Type Culture Collection (Rockville, MD) and cultured in DMEM with 2 mmol/L L-glutamine, 4.5g/L D-glucose and 10% fetal bovine serum (FBS). Plasmid transfections were performed with FuGENE according to manufacturer’s instructions (Roche, Basel, Switzerland). Full length N-terminally tagged HA-RalBP1 was obtained from GeneCopoeia (Germantown, MA, USA). Ral constructs have previously been described [19].
Western Blots
Cells were harvested in modified RIPA lysis buffer (50mM Tris HCl pH 7.2, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.5% Sodium Deoxycholate, 0.1% SDS, 1% Igepal) and supplemented with protease and phosphatase inhibitor (Sigma, St. Louis, MO, USA). The lysates were incubated for 20 minutes before un-dissolved residues were spun down. Antibodies were purchased from the following manufacturers: β-Tubulin: Calbiochem, (Darmstadt, Germany); HA.11: Covance (Denver, PA, USA); Phospho-Threonine and Phospho-Tyrosine (P-Tyr-100): Cell Signaling (Beverly, MA, USA); RalBP1 Abnova (Taipei City, Taiwan)
Immunoprecipitation and Mass Spectrometry (MS) analysis
18μg of DNA and 56μl FuGene was used to transfect a 70% confluent 150mm plate and incubated for 24 hours before harvest in 2ml lysis buffer (see above) including protein and phosphatase inhibitors (Sigma, St. Louis, MO, USA)). Lysates were incubated for 2 hours with covalently conjugated anti-HA agarose beads (Sigma, St. Louis, MO, USA) and washed in lysis buffer four times. Precipitates were analyzed by western blotting as described above. For MS analysis, precipitates were separated by SDS-PAGE and stained with colloidal Coomassie blue stain. Briefly, gels were treated 2× 15 minutes in fix solution (10% Acetic acid, 40% Methanol) before overnight staining in Colloidal Coomassie Stain solution (80ml stock solution (1.5mM Coomassie Brilliant Blue R250, 3% v/v H3PO4, 950mM (NH4)2SO4) diluted with 20ml Methanol). The protein band migrating at the appropriate size as determined by comparison to Kaleidoscope weight standards (BioRad, Hercules, CA, USA) was excised. The gel piece was transferred to a siliconized tube, washed and destained in 200 μL 50% methanol overnight and subsequently dehydrated in acetonitrile, rehydrated in 30 μL of 10 mM dithiolthreitol in 0.1 M ammonium bicarbonate and reduced at room temperature for 30 minutes. DTT was removed and the sample alkylated in 30 μL 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 30 minutes. The reagent was removed, the gel pieces dehydrated in 100 μL acetonitrile, then rehydrated in 100 μL 0.1 M ammonium bicarbonate. A final dehydration step was performed in 100 μL acetonitrile, the acetonitrile removed and the pieces completely dried by vacuum centrifugation. The gel pieces were rehydrated in 20 ng/μL (Trypsin and Arg-C) in 50 mM ammonium bicarbonate buffer at 4°C for 10 minutes. Any excess enzyme solution was removed and 20 μL 50 mM ammonium bicarbonate added. The sample was digested overnight at 37°C and the peptides extracted from the polyacrylamide in two 30 μL aliquots of 50% acetonitrile/5% formic acid. These extracts were combined and evaporated to 15 μL for MS analysis.
The LC-MS system was a Finnigan LTQ-FT mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed 8 cm × 75 um id Phenomenex Jupiter 10 um C18 reversed-phase capillary column. 0.5–5 μL volumes of extract were injected and peptides eluted from the column by acetonitrile/0.1 M acetic acid gradient at a flow rate of 0.25 μL/min. The nanospray ion source was operated at 2.8 kV. The digest was analyzed using the double play capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in sequential scans. This analysis produces approximately 2000 CAD spectra of ions ranging in abundance over several orders of magnitude. Not all CAD spectra are derived from peptides.
The data were analyzed by database searching using the Sequest search algorithm against Human International Protein Index (IPI) and then RalBP1.
Computational analysis of phosphorylation sites
Computational analysis was performed using the Eukaryotic Linear Motif (ELM) Resource for Functional Sites in Proteins available at http://elm.eu.org/.
RESULTS
RalBP1 is phosphorylated at Threonine residues and this is not altered by serum deprivation
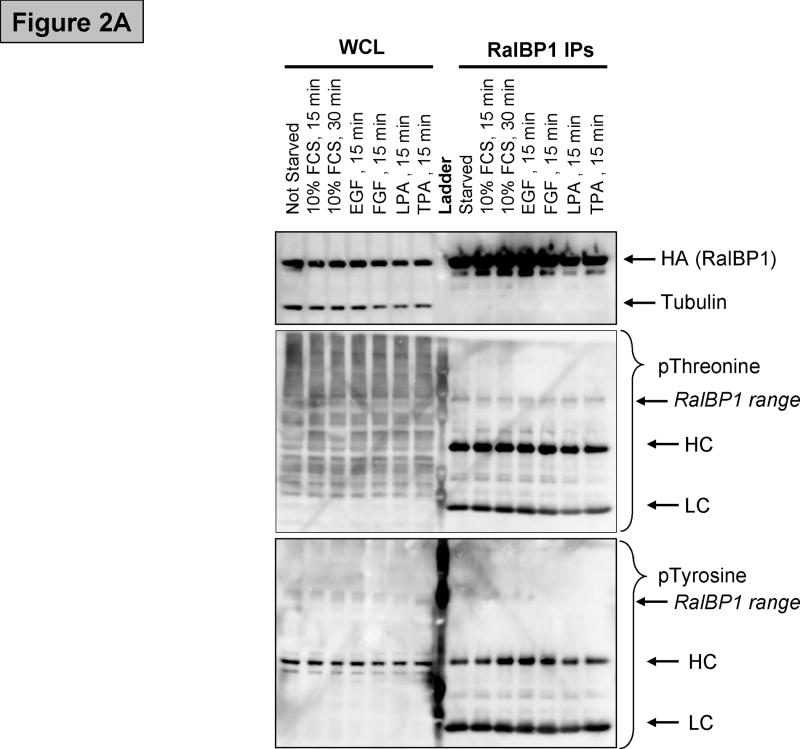
To evaluate the precipitation and detection of RalBP1 protein, the expression construct was transfected into 293T cells and harvested the following day. RalBP1 was detected by anti-HA or anti-RalBP1 antibody in the whole cell lysate (Figure 1). Enough antigen was readily precipitated by the HA antibody to present a clear band in a coomassie blue stained gel. No major contaminating bands were obvious. Western blot with anti-phospho-Tyrosine (data not shown) and anti-phospho-Threonine (Figure 1) antibodies indicated that RalBP1 is phosphorylated at Threonine residues but not at Tyrosine residues. To examine how phosphorylation was regulated by extracellular stimuli, several common phosphorylation cues were investigated. Cells were transfected as previously described and deprived of serum in their growth media for 24 hours, then stimulated with 10% FCS, 50ng/ml EGF, 50ng/ml FGF, 5uM LPA or 200nM TPA for fifteen minutes. Cells were harvested, HA-RalBP1 was precipitated and precipitates were analyzed by SDS-Page with subsequent western blotting. No significant changes in phosphorylation levels between serum deprived and stimulated cells was detected utilizing anti phospho-Threonine antibodies (Figure 2A).
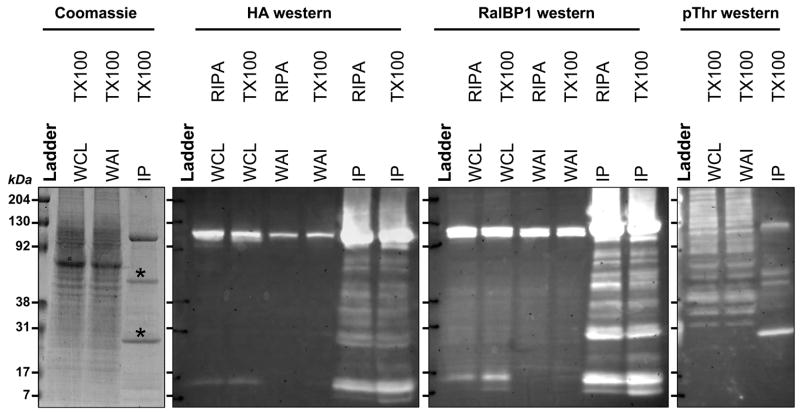
Figure 1.
RalBP1 was precipitated from 293T cells transiently transfected with RalBP1-HA and detected directly with coomassie blue staining or indirectly through anti-HA antibodies, anti-RalBP1 antibodies or anti phosphor-threonine (pThr) antibodies. Two solubilization protocols were evaluated, RIPA buffer or TritonX-100 containing buffer. Antigen was detected before precipitation in whole cell lysate (WCL) and in whole cell lysate after precipitation (WAI). The precipitate was boiled and loaded (IP), and enrichments and purification were observed as depicted. (*) Antibody fragments.
Figure 2.
(A) Western blot showing RalBP1 immunoprecipitation (IP) of cells transfected with RalBP1-HA, serum-starved for 24 hours and stimulated as indicated. Analysis of whole cell lysates (WCL) indicates that equal amounts of RalBP1-HA were expressed in cells. Comparable levels of RalBP1-HA were precipitated, as indicated by HA-western blot. No differences in phosphorylation were detected by phosphor-Threonine or Tyrosine specific antibodies. Heavy and light chains (HL and LC) are marked accordingly. (B) Primary sequence of RalBP1 showing peptides detected in mass spectroscopic analysis (yellow). 406 of 655 amino acids, 62%, were covered. Fourteen sites were detected (bold). Two sites were ambiguous. It was unclear whether S116 or S118 was phosphorylated or whether it was S252 or T253 that was modified. 29 of the 41 Serine residues, 17 of 26 Threonine and 8 of 12 Tyrosine residues were covered.
RalBP1 is primarily phosphorylated at the N-terminus
To determine the residues phosphorylated on RalBP1, 293T cells were transfected with HA-RalBP1 and precipitated as described. A strong coomassie blue stained band at the correct molecular weight was excised from the gel and examined by mass spectroscopy. This detected RalBP1-derived peptides covering 60% of the total protein (Figure 2B). Fourteen separate phosphorylated residues were found (Table 1). Ten of the fourteen residues are located within the first 100 amino acids of the protein and thirteen of the fourteen are Serine residues. To verify these results, an identical experiment was carried out in the bladder cancer cell line UMUC-3. All fourteen residues detected in 293T were also found in UMUC-3 (Table 1).
TABLE 1.
Phosphorylated amino acid residues in RalBP1 and computationally derived putative kinases and binding proteins
| Cell line | Putative Phosphorylation Site by ELM | ||
|---|---|---|---|
| 293T | UMUC-3 | Potential Kinase | Potential Binding site |
| S11 | S11 | MAPK, CK2 | WW4 |
| T27 | T27 | MAPK | WW4 |
| S29 | S29 | CK2 | 14-3-3 |
| S30 | S30 | ||
| S34 | S34 | CDK, MAPK | WW4 |
| S48 | S48 | MAPK | WW4 |
| S62 | S62 | CK2 | |
| S92 | S92 | CK2 | |
| S93 | S93 | ||
| S99 | S99 | CDK, MAPK | WW4 |
| S116 or S1181 | S116 or S118 | PKC (S118)2 | |
| S252 or T2531 | S252 or T253 | ||
| S463 | S463 | ||
| S645 | S645 | ||
Located too close to determine which was phosphorylated.
S118 was previously reported [17].
Several phosphorylated sites within the first hundred amino acids were observed within the same peptide. Figure 3 shows the different distinct peptides detected with more than one phosphorylated residue. Only peptides covering the T27-S34 phosphorylation sites were also void of phosphorylation modifications. Only the S62 and S92/S93 sites were not detected in an unmodified form. Phosphorylation at T27 was not detected in peptides without pS30. Phosphorylation at S48 (pS48) was not detected in peptides without pS62. pS99 was not detected in peptides not phosphorylated at S92 and S93, and these two residues were always simultaneously modified. The same observations were made in both cell lines.
Figure 3.
In three different regions several peptides were phosphorylated at multiple sites. Detected phosphorylations are indicated in red; peptides detected are written out. In the first region, only one peptide was detected with phosphorylation at T27. Most S29, S30 and S34 phosphorylation patterns were detectable, with one exception. S29 was never phosphorylated together with S34 alone. In the second region, pS48 was only detected in peptides with pS62 peptides. No peptides were detected without S62 phosphorylation. The third region where more than one residue was detected to be phosphorylated contained the S92, S93, S99 sites. No peptides without S92 and S93 phosphorylation were detected. These observations are identical between the UMUC-3 and 293T cell lines.
Computational analysis of putative kinases responsible for RalBP1 phosphorylation
To identify kinase candidates and putative kinase-regulated binding sites responsible for the phosphorylation sites, a domain analysis was carried out utilizing the Eukaryotic Linear Motif Resource for Functional Sites in Proteins [20]. Mitogen Activated Protein Kinases (MAP Kinases), Cyclin Dependent Kinases (CDK) and Casein Kinase 2 (CK2) were among the identified kinase candidates. One putative 14-3-3 biding site at residue S29 and four phosphorylated WW4 domains were detected (Table 1).
RalB overexpression increases RalBP1 phosphorylation at the C-terminus
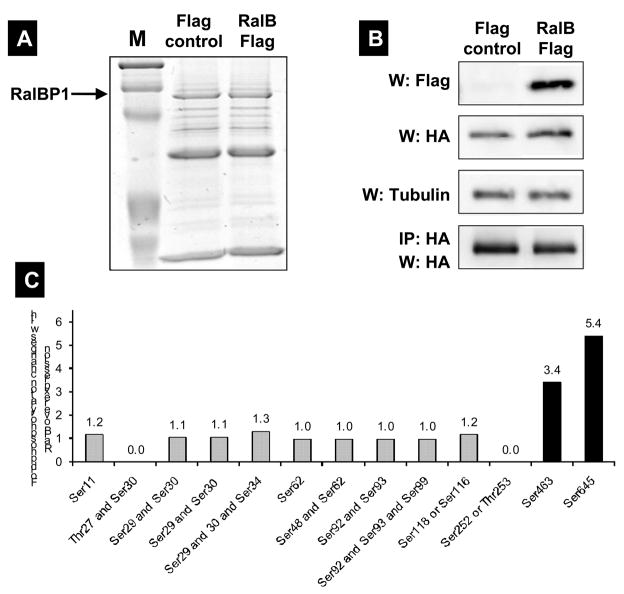
To determine whether RalB overexpression influences the phosphorylation status of RalBP1, a similar experiment to the one above was carried out. Wild type RalB or control vector was co-transfected with HA-RalBP1 in 293T cells. RalB expressed at high levels and RalBP1 could be readily precipitated and detected. Twelve phosphorylation sites were again detected. Peptides for T27 and S252/T253 were not detected in this data set. While the N-terminus is heavily phosphorylated, none of these sites showed significant change in phosphorylation status with RalB. The Ral interaction domain is much closer to the C-terminus, where the largest change in phosphorylation status was observed at sites S463, merely ten residues from the C-terminus at S645. These sites show a 3.5 and a 5.5 fold higher phosphorylation level in cells overexpressing RalB compared to vector control cells. All other sites show smaller changes between RalB-overexpressing cells and controls and are deemed too small to be significant (Figure 4).
Figure 4.
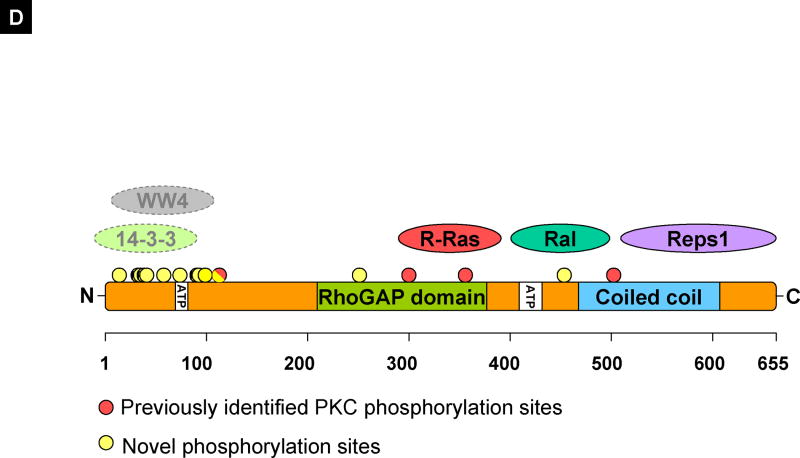
(A) RalBP1 was precipitated from 293T cells transiently transfected with RalBP1-HA with Flag control vector or RalB-Flag and detected with coomassie blue staining. (B) Western (W) blot or IP for Flag or HA indicating expression of RalBP1-HA and RalB-Flag in corresponding samples. (C) Phosphorylation changes in RalBP1-HA in 293T cells overexpressing wild type RalB. Only sites S463 and S645 show significant change in phosphorylation levels between mock and RalB-transfected cells. (D) RalBP1 structural domains include RhoGAP and the coiled coil at aa 207–373 and 469–610 respectively (http://elm.eu.org/). ATP binding sites (aa 65–80 or 415–448) [16]. Putative interaction partners are schematically depicted. Previously suggested PKC phosphorylation sites are in red and include 118S, 297T, S353 and S509 [17]. Phosphorylation sites described here are in yellow and include S11, T27, S29, S30, S34, S48, S62, S92, S93, S99, S116 or S118, S252 or T253 and S463. S118 is indicated as half red and half yellow. Putative binding proteins 14-3-3 and WW4-binding proteins shown in computationally assigned locations.
DISCUSSION
To broaden the understanding of the multifunctional protein RalBP1 we undertook a systematic approach to identify and analyze its post-translational modification. None of the previously identified PKC phosphorylation sites S118, T297, S353 and S509 [17] were conclusively identified. In our data only S118 was tentatively verified. It was not possible to determine whether S116 or S118 was phosphorylated in that specific peptide. S509 and S353 were unmodified in our cell lines. The most strongly modified PKC-alpha residue, T297 [17], was not covered by our analysis.
The primary sequence was further analyzed in silico by publicly available protein domain identification algorithms for the potential impact of phosphorylation or the potential kinase that modified that particular site. The first molecule identified as a phosphorylation-dependent interaction partner was 14-3-3. The consensus site [RHK][STALV].[ST].[PESRDIF] matches the S30 site 26-RTPSSE-31 and indicates that this might be an important interaction site for this family of signaling molecules. 14-3-3 molecules are the archetypical scaffold proteins that organize signaling complexes and have been found to interact with more than 200 different molecules [21]. 14-3-3 modifies the function of its targets by changing their activity, intracellular localization and the composition of larger complexes that they associate with. They exert these functions mainly via phospho-serine/threonine dependent protein-protein interactions. The identification of a 14-3-3 interaction motif in the N-terminus of RalBP1 is exciting, since it implies that this part of the protein may have a crucial role in determining its molecular context.
WW4 domains have a small recognition motif consisting of a phosphorylated serine or threonine followed by a proline. The S11, S34, S48 and S99 motifs are all potentially WW4 binding motifs [22; 23]. WW4 domains are similar to 14-3-3 interaction domains, with a protein-protein interaction domain dependent on phosphorylation. Interestingly, none of these sites are constitutively phosphorylated in the N-terminus (Figure 3) which may indicate that phosphorylation of these sites could modulate RalBP1 function.
Sites for Cyclin dependent Kinases, Casein Kinases and praline-directed Mitogen Activated Protein Kinases all are integral in the control of the cell cycle. These observations provide little information about the potential roles for these modifications of RalBP1 but do provide a list of plausible suspects that could be revisited as more data becomes available. Although the MAPK sites are at hand, the consensus sites [ST]P have little significance if other docking domains are not at hand. No specific docking domains for MAP Kinases were found in the protein, although several Lysine and Arginine-rich domains were identified, previously noted as important for MAPK substrate docking that can be located in a distant region of the protein [24].
Some sites were close enough to determine whether they were modified in certain patterns. These patterns might indicate that the phosphorylation of one site depends on another site. For instance S62 phosphorylation might be mandatory for S48 to become phosphorylated. However, this might also indicate that S62 is constitutively phosphorylated, whereas S48 is regulated by an as-yet-unknown mechanism. The same arguments might be made for the other sites depicted in Figure 3. The analysis of these regions does, however, indicate which sites are more frequently unmodified compared to neighboring sites.
Finally, cells overexpressing RalB and mock transfected cells were compared. In this analysis changes at two sites were detected. Especially exciting is the significant change in phosphorylation at site S463 as this is within the smallest required sequence [25] to bind Ral to RalBP1 (Figure 4). The largest caveat is the low level of phosphorylation at this site, which makes this observation less sure. However, our previous studies showed that less than 5% of wildtype Ral is bound to RalBP1 in even the most favorable situations [19]. In a situation where Ral binding to RalBP1 promotes phosphorylation, or RalBP1 phosphorylation at this site allows binding, a low level of phosphorylation could be expected. The observed phosphorylation changes at the S645 site are robust, and observed on more peptides, which solidifies the observation. S645 is located very close to the C-terminus and could have bearing on larger tertiary structure modifications of the protein and cellular localization, or involve other sites, like the binding domain for Ral.
In summary, we present the first systematic analysis of RalBP1 phosphorylation and describe several novel phosphorylation sites. These studies identify the N-terminus of the protein as the region most strongly modified by phosphorylation. The N-terminus has not yet been designated for any specific function, and strong post-translational modification in this area is therefore intriguing. Finally, we have tantalizing evidence of RalB-induced phosphorylation. These preliminary data open up an exciting area of study that will shed light on the regulation of RalBP1 downstream signaling events.
Acknowledgments
We acknowledge Drs. Mike Harding, Gary Oxford and Hong Wang for expert advice and Asoka Galpotthawela and Chuck Owens for expert technical assistance. We thank the W.M. Keck Biomedical Mass Spectrometry Laboratory and Nicholas Sherman, Kristina Nelson and Andrea Bradfield. The W.M. Keck Biomedical Mass Spectrometry Laboratory and University of Virginia Biomedical Research Facility are funded by a grant from the University of Virginia Pratt Committee. This work was supported by NIH CA075115 grant to D.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Camonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends CellBiol. 2005;15:327–32. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 3.Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Bidere N, Staudt D, Cubre A, Orenstein J, Chan FK, Lenardo M. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Mol Cell Biol. 2006;26:3505–13. doi: 10.1128/MCB.26.9.3505-3513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morinaka K, Koyama S, Nakashima S, Hinoi T, Okawa K, Iwamatsu A, Kikuchi A. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene. 1999;18:5915–22. doi: 10.1038/sj.onc.1202974. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, Iwamatsu A, Kishida S, Kikuchi A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. Embo J. 1999;18:3629–42. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, Camonis JH. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113( Pt 16):2837–44. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 9.Awasthi S, Singhal SS, Srivastava SK, Torman RT, Zimniak P, Bandorowicz-Pikula J, Singh SV, Piper JT, Awasthi YC, Pikula S. ATP-Dependent human erythrocyte glutathione-conjugate transporter. I. Purification, photoaffinity labeling, and kinetic characteristics of ATPase activity. Biochemistry. 1998;37:5231–8. doi: 10.1021/bi972130z. [DOI] [PubMed] [Google Scholar]
- 10.Awasthi S, Singhal SS, Pikula S, Piper JT, Srivastava SK, Torman RT, Bandorowicz-Pikula J, Lin JT, Singh SV, Zimniak P, Awasthi YC. ATP-Dependent human erythrocyte glutathione-conjugate transporter. II. Functional reconstitution of transport activity. Biochemistry. 1998;37:5239–48. doi: 10.1021/bi972131r. [DOI] [PubMed] [Google Scholar]
- 11.Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, Bandorowicz-Pikula J, Singh SV, Zimniak P, Awasthi YC. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–34. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- 12.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–7. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 13.Park SH, Weinberg RA. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–55. [PubMed] [Google Scholar]
- 14.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 15.Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–93. [PubMed] [Google Scholar]
- 16.Awasthi S, Sharma R, Singhal SS, Zimniak P, Awasthi YC. RLIP76, a novel transporter catalyzing ATP-dependent efflux of xenobiotics. Drug Metab Dispos. 2002;30:1300–10. doi: 10.1124/dmd.30.12.1300. [DOI] [PubMed] [Google Scholar]
- 17.Singhal SS, Yadav S, Singhal J, Drake K, Awasthi YC, Awasthi S. The role of PKCalpha and RLIP76 in transport-mediated doxorubicin-resistance in lung cancer. FEBS Lett. 2005;579:4635–41. doi: 10.1016/j.febslet.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Singhal SS, Wickramarachchi D, Singhal J, Yadav S, Awasthi YC, Awasthi S. Determinants of differential doxorubicin sensitivity between SCLC and NSCLC. FEBS Lett. 2006;580:2258–64. doi: 10.1016/j.febslet.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 19.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 20.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–30. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–13. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–43. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 23.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–8. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 24.Smith JA, Poteet-Smith CE, Malarkey K, Sturgill TW. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J Biol Chem. 1999;274:2893–8. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–4. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- 26.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J Cell Biol. 2006;174:877–88. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]