Abstract
The evolutionary rate of mitochondrial DNA (mtDNA) is highly variable across lineages in animals, and particularly in mammals. This variation has been interpreted as reflecting variations in metabolic rate: mitochondrial respiratory activity would tend to generate mutagenic agents, thus increasing the mutation rate. Here we review recent evidence suggesting that a direct, mechanical effect of species metabolic rate on mtDNA evolutionary rate is unlikely. We suggest that natural selection could act to reduce the (somatic) mtDNA mutation rate in long-lived species, in agreement with the mitochondrial theory of ageing.
Keywords: ageing, mutation, substitution, soma, antagonistic pleiotropy, reactive oxygen species
1. Introduction
The rate at which DNA and proteins accumulate substitutions is an important aspect of molecular evolution. Substitution rates reflect the combined effects of mutation, natural selection and genetic drift, the respective roles of which have been sharply debated for decades. The so-called ‘molecular clock’ hypothesis, which states that the rate of molecular evolution is constant in time and across lineages, is only rarely met: some species evolve significantly faster than others as far as molecules are concerned (Bromham & Penny 2003). The causes of these variations are still largely mysterious. Mitochondrial DNA (mtDNA) has received particular attention as far as evolutionary rates are concerned. Mitochondrial evolution appears to be especially capricious. Sudden increases or decreases in the substitution rate have been reported in various lineages (Spradling et al. 2001; Nabholz et al. 2008). Given the popularity of mtDNA as a marker of population and species history, understanding the dynamics of this genome is of primary interest to the evolutionary community.
Several studies have reported a negative relationship between mtDNA substitution rate and species body mass: mtDNA tends to evolve faster in small organisms (Martin & Palumbi 1993; Bromham et al. 1996). This is true of both non-synonymous (amino acid changing) and synonymous substitutions (Gillooly et al. 2007), which is indicative of a mutational mechanism—if only selection on proteins was at work, no effect would be expected at third-codon positions. The relationship could partly result from differences in generation time: assuming a constant per-generation mutation rate, species with a shorter generation time should undergo a higher per-year mutation rate. This hypothesis, however, cannot explain the whole pattern. The generation time effect must affect the mitochondrial and nuclear genomes to a similar extent; variations in substitution rate across species, however, are much stronger for mtDNA than for nuclear DNA, at least in mammals (Nabholz et al. 2008).
2. Oxidative damages
An alternative explanatory model invokes variations in metabolic rate across species. Mitochondrial respiratory activity generates reactive oxygen species (ROS) as a by-product of membrane electron transport. These oxidative molecules are known to be mutagenic: they damage DNA by oxidizing nucleotides. Owing to its cytological location, mtDNA is the main target of these replication-independent mutations. Differences in mtDNA mutation rates across species could therefore reflect differences in metabolic rate, which is inversely related to body mass (Martin & Palumbi 1993). This hypothesis was recently championed by Gillooly et al. (2005) who modelled metabolic rate as a function of body mass and temperature, and reported a quasi-perfect match between mtDNA substitution rate and predicted metabolic rate.
The metabolic rate hypothesis, however, is not devoid of problems. First, it should be recalled that only germ-line mutations, which are transmitted to the next generation, are relevant from an evolutionary viewpoint. It is currently unclear whether the metabolic rate in female germ-line cells is correctly predicted by the metabolic rate in somatic tissues, for which we have measurements (Lanfear et al. 2007). Second, the metabolic rate hypothesis is difficult to reconcile with the existence of site-specific mutation hotspots in the mitochondrial genome (Galtier et al. 2006)—how could chemical mutagens hit specific nucleotide positions, but not their immediate neighbours? Third, the empirical evidence does not always conform to the predictions of the metabolic rate hypothesis (Lanfear et al. 2007). Finally, from a physiological point of view, the relationship between metabolic rate, ROS production and mtDNA mutation is far from clear, as we now discuss in more detail.
3. Mitochondrial mutation and ageing
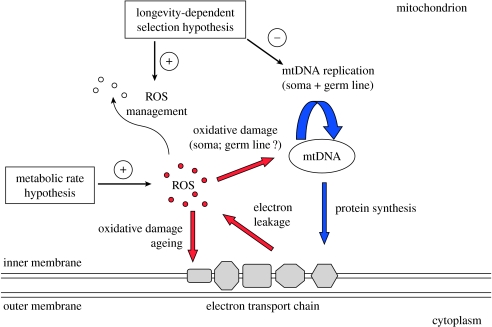
Most of the literature about ROS production and mtDNA mutation has been achieved in the context of ageing studies. Harman (1957) introduced the free radical theory of ageing, which states that the ROS generated throughout the lifespan of an organism cause its senescence. In its modern form, the mitochondrial theory of ageing posits that somatic mutations occurring in mtDNA, by affecting the efficiency of respiratory proteins, result in an increase in the rate of ROS production, which in turn increases the mutation rate—a vicious circle eventually causing ageing (figure 1). This model has been supported by various observations, including the inverse relationship between mitochondrial oxidative damage marker concentration and maximum lifespan in mammals (Barja & Herrero 2000), and the report of increased mtDNA mutation load in aged cells (Kujoth et al. 2007).
Figure 1.
Major structures, processes and evolutionary forces involved in mitochondrial ageing.
Closer examination of the evidence suggests that the situation might be more complex. Recently, two distinct groups have designed transgenic mice, in the genome of which the native mtDNA polymerase had been replaced by a modified, low-fidelity enzyme. These mtDNA-mutator mice showed strongly accelerated senescence, confirming the implications of mtDNA mutations in ageing (Trifunovic et al. 2004), but no increase in ROS production (Kujoth et al. 2005). In this case, ageing was apparently not caused by a positive feedback loop between mtDNA mutations and ROS production. ROS, furthermore, are not only a poison: they play a functional signalling role in healthy cells (Linnane et al. 2007), and could be under homeostatic control, irrespective of tissue preservation and ageing. Another (indirect) argument comes from the analysis of membrane lipid composition. In mammals and birds, a strong, negative correlation was reported between maximum lifespan and the proportion, among lipids, of n-3 polyunsaturated fatty acids (Hulbert et al. 2007). n-3 Polyunsaturated acyl chains are most sensitive to peroxidation, suggesting that lipid composition could have adapted to elongated lifespan. Lipid composition, however, is not significantly correlated to the mass-specific metabolic rate in mammals (Hulbert et al. 2007).
4. The longevity hypothesis
This rapid review cast doubts on the simplistic, mechanical relationship between metabolic rate, ROS production, mtDNA mutation and ageing we outline above. One suggestion, however, is that long-lived mammals and birds may have adapted to an increased lifespan by evolving macromolecular components more resistant to oxidative damages (Pamplona & Barja 2007; Min & Hickey 2008; Moosmann & Behl 2008). This conclusion points to a new hypothesis potentially explaining the variations of mtDNA substitution rate across species—longevity-dependent selection. According to this model, natural selection would act to reduce the mitochondrial mutation rate in long-lived species: a low somatic mutation rate would be required to achieve long lifespan. The low mtDNA mutation rate in long-lived species would not be a mechanical consequence of their reduced mass-specific metabolic rate, but rather a selected character (figure 1).
This hypothesis was first introduced by Samuels (2004) who reported a depletion of oligonucleotide direct repeats in the mitochondrial genomes of long-lived mammals. Direct repeats are known to be mutagenic. Their reduced number probably decreases the somatic mtDNA mutation rate in long-lived animals, which could be advantageous. Taking a phylogenetic approach, Nabholz et al. (2008) showed that the neutral mtDNA substitution rate is quite variable across the species of short-lived mammals, but constrained to low values in long-lived mammals, consistent with the longevity hypothesis. Correlation of mtDNA substitution rate and maximum lifespan was found to be stronger than that of either body mass or generation time (Nabholz et al. 2008; Welch et al. 2008).
Broad-scale comparisons can also help discriminate between the metabolic rate and the longevity models. Birds live longer, but have a higher mass-specific metabolic rate than mammals of similar body mass. Nabholz et al. (in press) showed that, on average, mtDNA substitution rate is lower in birds than mammals, favouring the longevity rather than the metabolic rate model. Cnidarians also provide interesting insights. The mtDNA substitution rate is very low in anthozoans (sea anemones, corals), which lack a mortal soma, and whose clonal colonies can live for hundreds of years (Hellberg 2006). Senescence is probably strongly counter-selected in these cells. Medusozoans (jellyfish, hydroids), by contrast, have a typical metazoan life cycle, with differentiated gonads in short-lived medusa; their mtDNA substitution rate is much higher than that of anthozoans (Huang et al. 2008), consistent with the longevity hypothesis. Similarly, plants, which lack a differentiated somatic line, generally harbour a low mitochondrial substitution rate (Wolfe et al. 1987), although plant mitochondria typically operate in the oxygen-rich environment resulting from photosynthesis (Noctor et al. 2007).
5. Mitochondrial DNA and the evolution of senescence
Theoretical evolutionary biologists have been interested in the ultimate reasons for senescence evolution: why should the fitness of an individual decrease during its lifespan? The main models that were proposed to explain this apparent paradox invoke late-onset mutations (Hughes & Reynolds 2005). Deleterious mutations are less strongly counter-selected if they are expressed in late stages of life, because their impact on the life-average reproductive success is weaker. Such mutations have a higher probability of fixation than early-onset deleterious mutations. The accumulation of many late-onset mutations, each of weak effect, could explain senescence (mutation accumulation model). The effect is more pronounced for mutations that are advantageous in early stages of life, but deleterious in older individuals. Natural selection favours the fixation of such mutations if their life-average effect is beneficial (antagonistic pleiotropy model, Williams 1957). The relative contribution of these two kinds of mutation to senescence evolution has been debated, but few empirical data are available.
Being somatic, the mtDNA mutations invoked by the mitochondrial theory of ageing do not correspond to these two models. They do not affect heritable traits, and cannot be directly selected against. However, germ-line mutations that could modulate the mtDNA somatic mutation rate would potentially be under the influence of natural selection—our working hypothesis. According to this model, a mutation that increases the mtDNA mutation rate would be deleterious in late stages of life, when somatic mutations have had time to affect the mitochondrial proteome of a large number of cells, and induce substantial oxidative damage. If uncovered, such mutations would provide evidence for the ‘mutation accumulation’ model of senescence evolution.
Confirmation of the longevity-dependent selective model would require the identification of modifiers of the mtDNA mutation rate and the investigation of their influence on species longevity. The most obvious candidates are genes controlling the fidelity of mtDNA replication (e.g. DNA polymerase gamma), genes involved in the regulation of the mitochondrial metabolism and redox status (e.g. coupling/uncoupling proteins) and antioxidant genes regulating the amount of cellular ROS (e.g. superoxide dismutase; table 1). We suggest that molecular evolutionary analyses of these genes in long-lived versus short-lived mammals or birds would be worthwhile.
Table 1.
Candidate genes potentially involved in the control of mtDNA mutation rate and ageing.
| gene | protein product | function/process | involvement in mtDNA mutation and ageing |
|---|---|---|---|
| POLG | mtDNA polymerase | mtDNA replication and maintenance | protein defects leads to accumulation of mtDNA mutations |
| POLG2 | Pol γ accessory subunit | mtDNA replication and maintenance | protein defects lead to accumulation of mtDNA mutations |
| TFAM | transcription factor A | mtDNA replication and maintenance | protein defects increase exposure of mtDNA to ROS |
| TWINKLE (PEO1) | mitochondrial helicase | mtDNA replication and maintenance | protein defects lead to multiple DNA deletions |
| ANT1 (SLC25A4) | ADP/ATP translocase 1 | ROS production | under-expression stalls electron transport chain and increases ROS production |
| UCPs | mitochondrial uncoupling proteins UCP1,UCP2, UCP3 | ROS production | protein defects may increase ROS production |
| SOD2 | superoxide dismutases [Mn] | protection against oxidative damage | protein defects increase oxidative stress |
| MPV17I | peroxisomal membrane protein family | protection against oxidative damage | downregulation leads to increased ROS damage |
| HTRA2 | serine protease HTRA2 | protection against oxidative damage | downregulation leads to increased ROS damage and apoptosis |
| FOXO3A | forkhead box protein O3 | protection against oxidative damage | protein defects increase ROS and trigger apoptosis upon cellular stress |
| PINK1 | serine/threonine-protein kinase | protection against oxidative damage | protein defects triggers mitochondrial dysfunction and apoptosis |
Acknowledgements
This work was supported by Agence Nationale de la Recherche project 06-BIOS-0001-01. This is contribution ISEM 2009_011.
Footnotes
One contribution of 11 to a Special Feature on ‘Whole organism perspectives on understanding molecular evolution’.
References
- Barja G., Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- Bromham L., Penny D. The modern molecular clock. Nat. Rev. Genet. 2003;4:216–224. doi: 10.1038/nrg1020. doi:10.1038/nrg1020 [DOI] [PubMed] [Google Scholar]
- Bromham L., Rambaut A., Harvey P.H. Determinants of rate variation in mammalian DNA sequence evolution. J. Mol. Evol. 1996;43:610–621. doi: 10.1007/BF02202109. doi:10.1007/BF02202109 [DOI] [PubMed] [Google Scholar]
- Galtier N., Enard D., Radondy Y., Bazin E., Belkhir K. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 2006;16:215–222. doi: 10.1101/gr.4305906. doi:10.1101/gr.4305906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J.F., Allen A.P., West G.B., Brown J.H. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc. Natl Acad. Sci. USA. 2005;102:140–145. doi: 10.1073/pnas.0407735101. doi:10.1073/pnas.0407735101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J.F., McKoy M.W., Allen A.P. Effects of metabolic rate on protein evolution. Biol. Lett. 2007;3:655–659. doi: 10.1098/rsbl.2007.0403. doi:10.1098/rsbl.2007.0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1957;2:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hellberg M.E. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol. Biol. 2006;6:24. doi: 10.1186/1471-2148-6-24. doi:10.1186/1471-2148-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Meier R., Todd P.A., Chou L.M. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J. Mol. Evol. 2008;66:167–174. doi: 10.1007/s00239-008-9069-5. doi:10.1007/s00239-008-9069-5 [DOI] [PubMed] [Google Scholar]
- Hughes K.A., Reynolds R.M. Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 2005;50:421–445. doi: 10.1146/annurev.ento.50.071803.130409. doi:10.1146/annurev.ento.50.071803.130409 [DOI] [PubMed] [Google Scholar]
- Hulbert A.J., Pamplona R., Buffenstein R., Buttemer W.M. Life and death: metabolic rate, membrane composition and life span of animals. Physiol. Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. doi:10.1152/physrev.00047.2006 [DOI] [PubMed] [Google Scholar]
- Kujoth G.C., et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. doi:10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Kujoth G.C., Bradshaw P., Haroon S., Prolla T. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:161–173. doi: 10.1371/journal.pgen.0030024. doi:10.1371/journal.pgen.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Thomas J., Welch J., Brey T., Bromham L. Metabolic rate does not calibrate the molecular clock. Proc. Natl Acad. Sci. USA. 2007;104:15388–15393. doi: 10.1073/pnas.0703359104. doi:10.1073/pnas.0703359104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane A.W., Kios M., Vitetta L. Healthy aging: regulation of the metabolome by cellular redox modulation and prooxidant signaling systems: the essential roles of superoxide anion and hydrogen peroxide. Biogerontology. 2007;8:445–467. doi: 10.1007/s10522-007-9096-4. doi:10.1007/s10522-007-9096-4 [DOI] [PubMed] [Google Scholar]
- Martin A.P., Palumbi S. Body size, metabolic-rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. doi:10.1073/pnas.90.9.4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X.J., Hickey D.A. An evolutionary footprint of age-related natural selection in mitochondrial DNA. J. Mol. Evol. 2008;67:412–417. doi: 10.1007/s00239-008-9163-8. doi:10.1007/s00239-008-9163-8 [DOI] [PubMed] [Google Scholar]
- Moosmann B., Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Nabholz B., Glémin S., Galtier N. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol. Biol. Evol. 2008;25:120–130. doi: 10.1093/molbev/msm248. doi:10.1093/molbev/msm248 [DOI] [PubMed] [Google Scholar]
- Nabholz, B., Glémin, S. & Galtier, N. In press. The erratic mitochondrial clock: variations of mutation rate, not population size, explains mtDNA diversity patterns in mammals and birds. BMC Evol. Biol. [DOI] [PMC free article] [PubMed]
- Noctor G., De Paepe R., Foyer C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007;12:125–134. doi: 10.1016/j.tplants.2007.01.005. doi:10.1016/j.tplants.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Pamplona R., Barja G. Highly resistant macromolecular components and low rate of generation of endogeneous damage: two key traits of longevity. Ageing Res. Rev. 2007;6:189–210. doi: 10.1016/j.arr.2007.06.002. doi:10.1016/j.arr.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Samuels D.C. Mitochondrial DNA repeats constrain the life span of mammals. Trends Genet. 2004;20:226–229. doi: 10.1016/j.tig.2004.03.003. doi:10.1016/j.tig.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Spradling T., Hafner M., Demastes J. Differences in rate of cytochrome-b evolution among species of rodents. J. Mammal. 2001;82:65–80. doi:10.1644/1545-1542(2001)082<0065:DIROCB>2.0.CO;2 [Google Scholar]
- Trifunovic A., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. doi:10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Welch J.J., Bininda-Emonds O.R., Bromham L. Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol. Biol. 2008;8:53. doi: 10.1186/1471-2148-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.C. Pleiotropy, natural selection and the evolution of senescence. Evolution. 1957;11:398–411. doi:10.2307/2406060 [Google Scholar]
- Wolfe K.H., Sharp P.M., Li W.H. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl Acad. Sci. USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. doi:10.1073/pnas.84.24.9054 [DOI] [PMC free article] [PubMed] [Google Scholar]