Abstract
We analysed over 8 million base pairs of bacterial artificial chromosome-based sequence alignments of four Old World monkeys and the human genome. Our findings are as follows. (i) Genomic divergences among several Old World monkeys mirror those between well-studied hominoids. (ii) The X-chromosome evolves slower than autosomes, in accord with ‘male-driven evolution’. However, the degree of male mutation bias is lower in Old World monkeys than in hominoids. (iii) Evolutionary rates vary significantly between lineages. The baboon branch shows a particularly slow molecular evolution. Thus, lineage-specific evolutionary rate variation is a common theme of primate genome evolution. (iv) In contrast to the overall pattern, mutations originating from DNA methylation exhibit little variation between lineages. Our study illustrates the potential of primates as a model system to investigate genome evolution, in particular to elucidate molecular mechanisms of substitution rate variation.
Keywords: Old World monkeys, substitution rates, male mutation bias, comparative genomics
1. Introduction
The genomics of humans and apes (hominoids), particularly of humans and chimpanzees, is an active field of research. Here, we propose that Old World monkeys offer an excellent genomic model system, complementary to hominoids for several reasons. Old World monkeys are the closest outgroup to hominoids. Divergence times among some Old World monkeys are comparable to those among well-known hominoids. For example, the rhesus macaque and baboon share a similar divergence time (6–8 Myr ago) to that of humans and chimpanzees (Delson et al. 2000; Steiper et al. 2004, although see Raaum et al. (2005) for an alternative, older estimate). A large amount of genomic resources are available from Old World monkeys. Furthermore, since some Old World monkeys, especially the rhesus macaque and baboon, served as non-human primate model systems for biomedical studies (e.g. Higley et al. 1991), comparative genomic analyses will complement functional data.
As the first step to developing a genomic model system, we present here the comparative genomic analyses of several Old World monkeys. We generated a large amount of sequence alignments between four Old World monkeys (macaque, baboon, grivet and colobus monkey) and the human. From these, we obtained estimates of genomic divergence among these species. We also compared substitution rates (also referred to as ‘evolutionary rates’) of the X-chromosome and autosomes, and between different Old World monkey lineages. Finally, we investigated whether mutations that mainly originate from DNA methylation exhibit different patterns of evolutionary rate variation compared with other mutations, as suggested recently (Kim et al. 2006).
2. Material and methods
We downloaded bacterial artificial chromosome (BAC) sequences of the following species from GenBank in April 2008: 443 baboon (Papio anubis and Papio hamadryas) BACs; 301 black-and-white colobus monkey (Colobus guereza, referred to as ‘colobus monkey’ here) BACs; and 305 grivet (Chlorocebus aethiops) BACs. Pairwise alignments of human and rhesus macaque (rheMac2), chimpanzee (panTro1) and orangutan (ponAbe2) were downloaded from the UCSC genome browser. Alignment of these sequences and annotation followed methods in Elango et al. (2008) (see the electronic supplementary material).
To analyse substitution rates at CpG and non-CpG sites, we assembled alignments of human–chimpanzee–orangutan–baboon–macaque–colobus monkey. The orangutan and colobus monkey are used as outgroups to the human–chimpanzee and baboon–macaque pairs, respectively. CpG sites (sites that were likely to have been a part of CpG dinucleotide) and non-CpG sites are identified using the parsimony method as in Meunier & Duret (2004) and Elango et al. (2008). C to T (or G to A) transitions are then counted for CpG and non-CpG sites separately. Only non-coding, non-repetitive sites are used for this analysis.
3. Results
(a) Genomic divergence among Old World monkeys and humans
We assembled alignments of 8.28 million base pairs over 20 autosomes (excluding chromosomes 3 and 17) and the X-chromosome of four Old World monkeys (macaque–baboon–colobus–grivet) and the human reference sequence (electronic supplementary material, table 1). Genomic divergence estimates from whole data are given in table 1. Results from non-coding sequences (with or without repetitive sequences) are shown in the electronic supplementary material. Overall, repetitive sequences accumulate more mutations than non-repetitive sequences and divergence estimates from data including repetitive sequences are usually greater than estimates from those without repetitive sequences. For instance, the divergence between humans and rhesus macaques from the whole data (including autosomes and the X-chromosome) is 6.19 per cent. Non-coding sequences including repetitive sequences show greater divergence (6.36%). Divergence estimated from non-coding, non-repetitive portions is 5.73 per cent for the same species pair. Other pairwise comparisons showed similar trends.
Table 1.
Divergence among the Old World monkeys and humans estimated from whole alignments. Below and above diagonals are autosomal and X-chromosome divergence, respectively.
| baboon | macaque | grivet | colobus | human | |
|---|---|---|---|---|---|
| baboon | — | 0.0106 (±1.23×10−4) | 0.0153 (±1.48×10−4) | 0.0270 (±1.98×10−4) | 0.0538 (±2.85×10−4) |
| macaque | 0.0156 (±0.95×10−4) | — | 0.0164 (±1.54×10−4) | 0.0281 (±2.02×10−4) | 0.0543 (±2.86×10−4) |
| grivet | 0.0223 (±0.95×10−4) | 0.0233 (±0.56×10−4) | — | 0.0280 (±2.02×10−4) | 0.0547 (±2.88×10−4) |
| colobus | 0.0330 (±0.96×10−4) | 0.0340 (±0.69×10−4) | 0.0340 (±0.69×10−4) | — | 0.0564 (±2.92×10−4) |
| human | 0.0617 (±0.94×10−4) | 0.0627 (±0.95×10−4) | 0.0631 (±0.95×10−4) | 0.0640 (±0.96×10−4) | — |
The genetic divergences among the four Old World monkey species mirror those between well-studied hominoids: the rhesus macaque and baboon are approximately 1.35 per cent divergent in the non-coding, non-repetitive portions of the data. The divergence between the grivet and the baboon–macaque pair is approximately 2 per cent. All three Old World monkeys are approximately 3 per cent different from the colobus monkey. Thus, divergences between the baboon–macaque–grivet–colobus monkey closely resemble those between the human–chimpanzee–gorilla–orangutan (e.g. Chen & Li 2001), even though the actual divergence times between these four species may differ from those between the hominoids (e.g. Raaum et al. 2005).
(b) Low male mutation bias in Old World monkeys compared with hominoids
In primates, in particular between humans and chimpanzees, the X-chromosome exhibits lower divergence than autosomes (Nachman & Crowell 2000). While a commonly cited explanation for such phenomena is the ‘male-driven evolution’ hypothesis (Li et al. 2002), some studies propose that lineage-specific evolutionary forces such as hybridization may be responsible (Patterson et al. 2006). Here, we examined the divergence of the X-chromosomes and autosomes separately (table 1). In all comparisons, regardless of the type of sites used, the X-chromosome shows lower divergence than the autosomes.
We estimated the male-to-female mutation ratio, denoted as αm, from the comparison of the divergence on the X-chromosome (X) to autosomal divergence (A), following Miyata et al. (1987), as (table 2).
Table 2.
Male-to-female mutation ratio (αm) estimated by comparing the X-chromosome and autosomal divergence. The 95% CIs using bootstrapping are shown.
| baboon | macaque | grivet | colobus | |
|---|---|---|---|---|
| macaque | −32.6 [−872.8, 728.49] | — | ||
| grivet | 17.0 [−228.2, 310.9] | 17.3 [−5242, 5462] | — | |
| colobus | 3.13 [2.96, 3.88] | 3.35 [2.95, 3.87] | 3.16 [3.06, 4.07] | — |
| human | 2.04 [1.91, 2.25] | 2.14 [1.96, 2.32] | 2.13 [1.97, 2.33] | 2.03 [1.82, 2.15] |
Between closely related Old World monkey species, αm estimates fluctuated greatly and the confidence intervals included zero (e.g. between the macaque and baboon). It is likely that ancestral polymorphism between closely related Old World monkeys hinders accurate estimations of αm (e.g. Makova & Li 2002). Still, it is interesting to note that the mean αm from all pairwise comparisons is approximately 2, much less than the earlier estimates of 5–6 in primates (Shimmin et al. 1993). Even when we restrict our comparisons to the comparisons between humans and Old World monkeys (instead of between different Old World monkeys) to avoid confounding effects of ancestral polymorphism, the mean αm is approximately 2. Thus, the male mutation bias in Old World monkeys appears to be lower than that in hominoids.
(c) Evolutionary rate variation between Old World monkey lineages
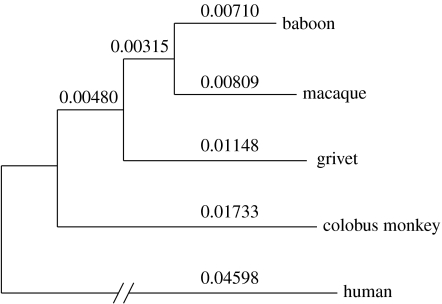
We analysed evolutionary rate variation among Old World monkeys using the relative rate test (Wu & Li 1985), with the human as an outgroup. Figure 1 depicts the branch lengths estimated from the total data. It is known that substitution rates vary significantly among hominoids (Elango et al. 2006). Here, we find that evolutionary rate variation is even more conspicuous in Old World monkeys than in hominoids. Interestingly, the baboon lineage appears to be evolving more slowly than the other three Old World monkey branches, regardless of the nature of the dataset used (p<10−5 in all comparisons using the relative rate test). For example, the rate difference between the macaque and baboon lineages ranges between 5 (non-coding, non-repetitive dataset) and 14 per cent (total data).
Figure 1.
Phylogenetic relationship and branch lengths (roughly drawn to scale) among the five species analysed using the neighbour-joining method (Saitou & Nei 1987). The estimated numbers of substitutions per site are shown above each branch.
(d) Contrasting patterns of molecular evolution of transitions at CpG and non-CpG sites
Most mutations are considered to occur by errors in DNA replication. Consequently, species that undergo more DNA replication in a given amount of time may accumulate more mutations compared with those that undergo fewer replication events. This is the reasoning underlying the so-called ‘generation-time effect’ (Yi et al. 2002).
However, mutations at CpG dinucleotides often originate from DNA methylation, rather than replication error. Since DNA methylation is not restricted solely to the DNA replication process, it was proposed that mutations at CpG sites may accumulate relatively constantly over time (Kim et al. 2006). Here, we revisited this question, with a larger amount of data and closer outgroups.
We compared substitution rates between the human–chimpanzee pair with that between the baboon–macaque pair. The absolute divergence times within the two pairs are similar (Delson et al. 2000; Steiper et al. 2004), while evolutionary rates are generally greater in Old World monkey lineages, due to the generation–time effect (Yi et al. 2002; Steiper et al. 2004). Indeed, when only non-CpG sites are analysed, the baboon–macaque pair has accumulated approximately 30 per cent more substitutions than the human–chimpanzee pair (table 3), well in accord with the known degree of rate difference (Yi et al. 2002; Steiper et al. 2004). When only CpG sites are analysed, however, the two pairs have accumulated similar numbers of substitutions (table 3), for both repetitive and non-repetitive sites. Our results thus support the proposal that transitions at CpG sites exhibit relative time dependency (Kim et al. 2006).
Table 3.
Transitions at CpG and non-CpG sites exhibit qualitatively different patterns of molecular evolution. Divergences between human–chimpanzee (KHC) and between macaque–baboon (KMB) are shown separately. The 95% CIs of KMB/KHC are obtained from bootstrapping 1000 times.
| repetitive | non-repetitive | total | ||||
|---|---|---|---|---|---|---|
| CpG | non-CpG | CpG | non-CpG | CpG | non-CpG | |
| KHC | 10.1 | 0.75 | 7.31 | 0.63 | 8.39 | 0.67 |
| (× 10−2) | ||||||
| KMB | 9.2 | 0.98 | 8.11 | 0.82 | 8.50 | 0.87 |
| (× 10−2) | ||||||
| KMB/KHC | 0.915 | 1.31 | 1.109 | 1.29 | 1.01 | 1.29 |
| [95% CI] | [0.80, 1.03] | [1.20, 1.43] | [0.99, 1.22] | [1.20, 1.38] | [0.93, 1.09] | [1.22, 1.36] |
4. Discussion
This study illustrates that non-hominoid primate genomics are readily available to provide a useful genomic model system complementary to hominoids. By comparing evolutionary rate variation in Old World monkeys with those from hominoids, we can distinguish common patterns of primate genome evolution from those specific to certain lineages.
Male-driven evolution can reduce the divergence of the X-chromosomes compared with autosomes. The male-to-female mutation ratio (αm) in primates is considered to be approximately 5, mostly based upon comparisons between humans and chimpanzees (Shimmin et al. 1993; Li et al. 2002). We showed that between humans and Old Word monkeys, αm is lower than earlier estimates. A previous study also noted that αm between humans and macaques is lower than 5 (RMGSAC 2007). Therefore, the degree of male mutation bias appears to be variable, even within primates. Male mutation bias may be correlated with generation time, because the relative contributions of male germlines towards the mutational pool should increase with generation times (Bartosch-Harald et al. 2003). Our observation is in accord with this view. However, the extent of variation of male mutation bias and causative mechanisms underlying the observed variation should be investigated further.
Evolutionary rates vary greatly between different Old World monkeys. Human substitution rates are known to be lower than chimpanzee rates (Elango et al. 2006). We observe a comparable phenomenon between baboons and macaques. In fact, baboons appear to evolve more slowly than the other three branches in Old World monkeys. Such observations pose at least two immediate questions. First, what caused the reported substitution rate differences? Life-history traits may influence evolutionary rates, but it is not clear which are the main determinants of rate variation between the Old World monkeys. Detailed analyses of evolutionary rates between lineages, while controlling for specific life-history traits, will allow us to evaluate the role of different life-history variables on genome evolution. Second, the fact that substitution rates vary substantially and significantly between even closely related lineages make us ask whether we can confidently infer divergence times and other evolutionary events from molecular data. On the one hand, methods incorporating rate changes need to be developed urgently. On the other hand, the observation that CpG sites exhibit relatively constant molecular evolution compared with non-CpG sites (Kim et al. 2006, this study) may provide an alternative means to infer past evolutionary events.
Acknowledgements
This study is supported by the Blanchard-Milliken Faculty Fellowship, the Alfred P. Sloan Research Fellowship and an NSF grant (BCS-0751481) to S.V.Y.
Footnotes
One contribution of 11 to a Special Feature on ‘Whole organism perspectives on understanding molecular evolution’.
Supplementary Material
References
- Bartosch-Harald A., Berlin S., Smith N.G.C., Moller A.P., Ellegren H. Life history and the male mutation bias. Evolution. 2003;57:2398–2406. doi: 10.1554/03-036. doi:10.1554/03-036 [DOI] [PubMed] [Google Scholar]
- Chen F.C., Li W.-H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. doi:10.1086/318206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delson E., Tattersall I., Van Couvering J.A., Brooks A.S. Cercopithecina. In: Delson E., Tattersall I., Van Couvering J.A., Brooks A.S., editors. Encyclopedia of human evolution and prehistory. Garland; New York, NY: 2000. pp. 166–171. [Google Scholar]
- Elango N., Thomas J.W., Program N.C.S., Yi S. Variable molecular clocks in hominoids. Proc. Natl Acad. Sci. USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. doi:10.1073/pnas.0510716103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N., Kim S.-H., Vigoda E., Yi S., NISC Program Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLoS Comput. Biol. 2008;4:e1000015. doi: 10.1371/journal.pcbi.1000015. doi:10.1371/journal.pcbi.1000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley J.D., Hasert M.F., Suomi S.J., Linnoila M. Nonhuman primate model of alcohol abuse: effects of early experience, personality, and stress on alcohol consumption. Proc. Natl Acad. Sci. USA. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. doi:10.1073/pnas.88.16.7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Elango N., Warden C.W., Vigoda E., Yi S. Heterogenous genomic molecular clocks in primates. PLoS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. doi:10.1371/journal.pgen.0020163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-H., Yi S., Makova K. Male-driven evolution. Curr. Opin. Genet. Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. doi:10.1016/S0959-437X(02)00354-4 [DOI] [PubMed] [Google Scholar]
- Makova K.D., Li W.-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. doi:10.1038/416624a [DOI] [PubMed] [Google Scholar]
- Meunier J., Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. doi:10.1093/molbev/msh070 [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harbor Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Nachman M.W., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Richter D.J., Gnerre S., Lander E.S., Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. doi:10.1038/nature04789 [DOI] [PubMed] [Google Scholar]
- Raaum R.L., Sterner K.N., Noviello C.M., Stewart C.-B., Disotell T.R. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. doi:10.1016/j.jhevol.2004.11.007 [DOI] [PubMed] [Google Scholar]
- RMGSAC (Rhesus Macaque Genome Sequencing and Analysis Consortium) Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. doi:10.1126/science.1139247 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shimmin L.C., Chang B.H.J., Li W.-H. Male-driven evolution in DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. doi:10.1038/362745a0 [DOI] [PubMed] [Google Scholar]
- Steiper M.E., Young N.M., Sukrarna T.Y. Genomic data support the hominoid slowdown and an early Oligocene estimate for the hominoid–cercopithecoid divergence. Proc. Natl Acad. Sci. USA. 2004;101:17 021–17 026. doi: 10.1073/pnas.0407270101. doi:10.1073/pnas.0407270101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-I., Li W.-H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc. Natl Acad. Sci. USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. doi:10.1073/pnas.82.6.1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S., Ellsworth D.L., Li W.-H. Slow molecular clocks in Old World monkeys, apes, and humans. Mol. Biol. Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.