Abstract
Many genes show different expression levels in males and females, and these form the basis of sexually dimorphic phenotypes. Sex-biased genes experience accelerated rates of protein evolution, which has been attributed to sexual selection. However, it is possible that the increased rates of molecular evolution, and more importantly the sex-biased gene expression pattern itself, are due to decreased selective constraint. This notion may explain many of the patterns associated with sex-biased gene expression, and changes how we should view the role of natural and sexual selection in relation to these genes.
Keywords: gene expression, positive selection, relaxed constraint
1. Introduction
In many animals, there are marked differences between the sexes, ranging from the gonad to dimorphic somatic phenotypes that function in mate choice as well as post-reproductive strategies. These sex-specific phenotypes are encoded by a shared underlying genome through differential transcription and translation levels, and interest in this type of gene expression has recently increased as studies have indicated that a large proportion of the transcriptome is sex biased in many animals (Ranz et al. 2003; Yang et al. 2006; Mank et al. 2008a).
Sex-biased loci in general exhibit an accelerated rate of protein evolution, and many authors (including ourselves) have posited that this results from the powerful pressures of sexual selection (Zhang et al. 2004; Pröschel et al. 2006; Ellegren & Parsch 2007; Mank et al. 2007). Clearly, positive selection is a driver in the evolution of male reproductive proteins associated with sperm competition and fertilization (Zhang et al. 2004; Begun & Lindfors 2005); however, sexual selection has been invoked to explain the accelerated rates of evolution for broader categories of sex-biased genes as well (Zhang et al. 2004; Pröschel et al. 2006). The link is seductive, as a subset of sex-biased genes presumably code for the traits that are the basis of male competition and female choice. These traits are under strong selection owing to their role in reproductive success, manifesting in accelerated rates of protein change.
This is an attractive argument, as it provides a link between phenotypes and the genome. However, we do not yet know what evolutionary constraints operate on gene expression or the mix of selection pressures acting on these genes. Recent studies indicate that sex-biased genes share a suite of characters, and these characters have implications as to the role of positive selection, which includes sexual selection, in shaping observed patterns of molecular evolution. These characteristics suggest that genes with sex-biased expression patterns are more dispensable, and this dispensability in turn leads to relaxed evolutionary constraints that may be responsible for much of the increase in evolutionary divergence (Barker et al. 2005). More importantly, we hypothesize that dispensability may be an important predictor of sex-biased gene expression, and that indispensability may act as a brake on the resolution of sexually antagonistic selection and the evolution of sexual dimorphisms.
2. Dispensibility
Genes do not all share the same degree of importance in terms of an organism's ability to survive and reproduce. Definitions of dispensability vary, but genetic studies ranging from yeast to mice have identified those genes that are indispensable to survival or fertility (Hirsh & Fraser 2001; Giaever et al. 2002) and those at the other end of the continuum, which show no obvious knock-out phenotype (Barbaric et al. 2007; Liao & Zhang 2007). Therefore, some genes are required, others appear to be superfluous, and most are intermediate between these extremes. This degree of importance is often referred to as dispensability, and while it is difficult to connect the measures of dispensability from unicellular eukaryotes to metazoans, we employ the term loosely here, intending it as a gauge for the phenotypic effects of a gene.
Critical genes generally show lower rates of functional protein change when compared with dispensable genes (Hirsh & Fraser 2001; Jordan et al. 2002; Pal et al. 2003; Wall et al. 2005; Liao & Zhang 2006). This theoretically results from narrow fitness optima for critical genes, manifesting in strong purifying selection against functional mutations, the vast majority of which are deleterious. Genes that are less critical are subject to less purifying pressure, and evolve more rapidly simply through neutral processes.
Dispensable genes share key expression characteristics with sex-biased genes, suggesting that sex-biased genes themselves may be dispensable. Indeed, it is possible that genes with higher levels of dispensability may respond more quickly to sexually antagonistic selection, thereby evolving sex-biased expression, as different female and male transcription levels would be less likely to have deleterious effects for less critical genes.
3. Expression variance
Expression variance is not often discussed but has powerful implications. Both within- (Mank et al. 2008a) and among-population (Baker et al. 2007; Lawniczak et al. 2008) studies show higher variance for sex-biased genes. Similarly, critical genes have less variance among biological replicates (Fraser et al. 2004; Batada & Hurst 2007), and this may be due to the interactive network structure of the transcriptome, which requires precise titres of protein levels to initiate, regulate and buffer complex functions and pathways (Batada et al. 2006b). Perturbations in the protein components of critical pathways due to variance among individuals could theoretically have an amplifying effect downstream, resulting in maladaptive consequences. Thus, indispensable genes are tightly regulated, resulting in little variance in expression among individuals.
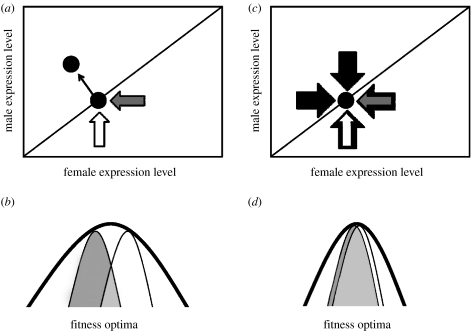
The fact that sex-biased genes have a higher variance in expression suggests that they are dispensable to some degree, though this may be confounded by differences in mutation rates. Variance in gene expression, at least part of which is genetic and heritable, provides the raw material for evolution, and may explain why sex bias can change rapidly among species (Zhang et al. 2007). Furthermore, dispensability may facilitate rapid changes in sex-biased gene expression without serious deleterious consequences to the organism. This also has implications for fitness optima (figure 1) of sex-biased versus unbiased genes. Variance in gene expression levels may be an indirect method to infer the width of fitness optima for individual genes, as higher variance may indicate wider optima.
Figure 1.
Sexually antagonistic selection on gene expression. (a) Sexual antagonism is resolved when sex-specific selection pressures (grey arrows for female specific, white for male specific) produce sex-biased gene expression. (b) For genes with broad overall fitness optima, sexual antagonism can produce clear male (white) and female (grey) distinctions as a result of sex-specific selection on any specific function. (c) For genes with pleiotropic effects, constraints from other functions, shown with black arrows prevent sex-specific selection from decoupling male and female expression levels, preventing the resolution of sexual antagonism. (d) These genes have narrow fitness optima, and therefore female and male profiles are largely overlapping.
4. Pleiotropy
Pleiotropy describes the phenomenon where one locus affects more than one trait, and can refer to various types of functions. Indispensable genes are more pleiotropic in terms of network connectivity than non-essential genes (Jeong et al. 2001; Hahn & Kern 2005). Genetic networks are highly redundant (Ulitsky & Shamir 2007; Wagner & Wright 2007), but redundancy decreases for proteins that have more interacting partners, and genes without redundant back-ups have a greater individual contribution to organismal fitness.
We do not yet have a full map of any metazoan genetic network; however, we do know that pleiotropy, measured by the expression breadth, or the number of tissues a transcript is found within, is lower for sex-biased genes (Mank et al. 2008b). Similarly, the regulatory elements controlling sex-biased genes indicate that they are involved in fewer functionalities (Lawniczak et al. 2008). If one accepts the analogy between pleiotropy in terms of expression breadth and functionality (Yanai et al. 2005; Liao & Zhang 2006) and pleiotropy in terms of genetic network connectivity (Batada et al. 2006a), sex-biased genes exhibit characteristics suggesting a large proportion are dispensable. This makes logical sense, as pleiotropic constraints would prevent the realization of sex-biased gene expression regardless of sexual antagonism.
5. Conclusions
The characteristics we described above suggest that sex-biased genes are dispensable. However, that is not to say that indispensable genes are not subject to sexual antagonism, rather that sexual antagonism for these genes is less likely to produce sex-biased gene expression. If sex-biased genes are less critical, they would be subject to less powerful purifying selection, and would show elevated rates of functional protein change through neutral processes alone. Additionally, this reduction in purifying selection allows for two additional processes to occur. First, the lack of purifying selection associated with some degree of dispensability allows for flexibility in the fitness landscape of sex-biased genes. This then allows positive selection, sexual or otherwise, to influence the evolution of sex-specific expression patterns when genes are sexually antagonistic. It may be that indispensable genes have sex-specific fitness optima in some of their functionalities; however, constraints acting from other pathways or functions prevent them from realizing independent male and female fitness peaks. Dispensability may therefore allow for sex-specific selection to produce sex-biased gene expression patterns, and this has implications to the evolution of sexually selected phenotypes. If the evolution of sex-biased expression is limited to those genes that are non-critical, dispensability of the underlying genes may act as a brake on the evolution of sexually dimorphic traits.
Testing these questions will require an integrated approach. Global dispensability is best measured in a systems biology framework, and systems biology data will be required to understand its role in metazoan evolution. Metazoan systems biology is a nascent field, and time is needed to develop network maps and functional genomics. In the meantime, we can focus on two persisting gaps. First, we need a better understanding of the actual loci under sexual selection in order to determine how sexual selection shapes gene expression patterns. Additionally, we need more extensive molecular data on divergence and standing polymorphisms in an array of animals in order to delineate the beacon of positive selection from relaxed constraint (McDonald & Kreitman 1991). These are crucial if we want to understand how selection shapes expression patterns.
Acknowledgements
Support was provided by the Swedish Research Council and the Wenner–Gren Foundation. We thank Göran Arnqvist for helpful discussions, and four anonymous reviewers for their comments.
Footnotes
One contribution of 11 to a Special Feature on ‘Whole organism perspectives on understanding molecular evolution’.
References
- Baker D.A., Meadows L.A., Wang J., Dow J.A., Russell S. Variable sexually dimorphic gene expression in laboratory strains of Drosophila melanogaster. BMC Genomics. 2007;8:454. doi: 10.1186/1471-2164-8-454. doi:10.1186/1471-2164-8-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric I., Miller G., Dear N. Appearances can be deceiving: phenotypes of knockout mice. Brief. Funct. Genomic. Proteomic. 2007;6:91–103. doi: 10.1093/bfgp/elm008. doi:10.1093/bfgp/elm008 [DOI] [PubMed] [Google Scholar]
- Barker M.S., Demuth J.P., Wade M.J. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet. 2005;1:527–530. doi: 10.1371/journal.pgen.0010057. doi:10.1371/journal.pgen.0010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batada N.N., Hurst L.D. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat. Genet. 2007;39:945–949. doi: 10.1038/ng2071. doi:10.1038/ng2071 [DOI] [PubMed] [Google Scholar]
- Batada N.N., Hurst L.D., Tyers M. Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2006a;2:e88. doi: 10.1371/journal.pcbi.0020088. doi:10.1371/journal.pcbi.0020088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batada N.N., Reguly T., Breitkreutz A., Boucher L., Breitkreutz B.J., Hurst L.D., Tyers M. Stratus not altocumulus: a new view of the yeast protein interaction network. PLoS Biol. 2006b;4:1720–1731. doi: 10.1371/journal.pbio.0040317. doi:10.1371/journal.pbio.0040317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D.J., Lindfors H.A. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 2005;22:2010–2021. doi: 10.1093/molbev/msi201. doi:10.1093/molbev/msi201 [DOI] [PubMed] [Google Scholar]
- Ellegren H., Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007;8:689–698. doi: 10.1038/nrg2167. doi:10.1038/nrg2167 [DOI] [PubMed] [Google Scholar]
- Fraser H.B., Hirsh A.E., Giaever G., Kumm J., Eisen M.B. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. doi:10.1371/journal.pbio.0020137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. doi:10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Hahn M.W., Kern A.D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. doi:10.1093/molbev/msi072 [DOI] [PubMed] [Google Scholar]
- Hirsh A.E., Fraser H.B. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. doi:10.1038/35082561 [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. doi:10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Rogozin I.B., Wolf Y.I., Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak M.K.N., Holloway A.K., Begun D.J., Jones C.D. Genomic analysis of the relationship between gene expression variation and DNA polymorphism in Drosophila simulans. Genome Biol. 2008;9:R125. doi: 10.1186/gb-2008-9-8-r125. doi:10.1186/gb-2008-9-8-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B.Y., Zhang J.Z. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol. Biol. Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. doi:10.1093/molbev/msj119 [DOI] [PubMed] [Google Scholar]
- Liao B.Y., Zhang J. Mouse duplicate genes are essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. doi:10.1016/j.tig.2007.05.006 [DOI] [PubMed] [Google Scholar]
- Mank J.E., Hultin-Rosenberg L., Axelsson E., Ellegren H. Rapid evolution of female-biased, but not male-biased, genes expressed in avian brain. Mol. Biol. Evol. 2007;24:2698–2706. doi: 10.1093/molbev/msm208. doi:10.1093/molbev/msm208 [DOI] [PubMed] [Google Scholar]
- Mank J.E., Hultin-Rosenberg L., Webster M.T., Ellegren H. The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics. 2008a;9:148. doi: 10.1186/1471-2164-9-148. doi:10.1186/1471-2164-9-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J.E., Hultin-Rosenberg L., Zwahlen M., Ellegren H. Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am. Nat. 2008b;171:35–43. doi: 10.1086/523954. doi:10.1086/523954 [DOI] [PubMed] [Google Scholar]
- McDonald J.H., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Pal C., Papp B., Hurst L.D. Rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. doi:10.1038/421496b [DOI] [PubMed] [Google Scholar]
- Pröschel M., Zhang Z., Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. doi:10.1534/genetics.106.058008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. doi:10.1126/science.1085881 [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shamir R. Pathway redundancy and protein essentiality revealed in the Saccharomyces cerevisiae interaction networks. Mol. Syst. Biol. 2007;3:104. doi: 10.1038/msb4100144. doi:10.1038/msb4100144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., Wright J. Alternative routes and mutational robustness in complex regulatory networks. Biosystems. 2007;88:163–172. doi: 10.1016/j.biosystems.2006.06.002. doi:10.1016/j.biosystems.2006.06.002 [DOI] [PubMed] [Google Scholar]
- Wall D.P., Hirsch A.E., Fraser H.B., Kumm J., Giaever G., Eisen M.B., Feldman M.W. Functional genomic analysis of the rates of protein evolution. Proc. Natl Acad. Sci. USA. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. doi:10.1073/pnas.0501761102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I., et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. doi:10.1093/bioinformatics/bti042 [DOI] [PubMed] [Google Scholar]
- Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. doi:10.1101/gr.5217506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hambuch T.M., Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. doi:10.1093/molbev/msh223 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sturgill D., Parisi M., Kumar S., Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–238. doi: 10.1038/nature06323. doi:10.1038/nature06323 [DOI] [PMC free article] [PubMed] [Google Scholar]