Abstract
Inbreeding causes increases in homozygosity and is commonly associated with reductions in fertility and embryogenesis. Although the mechanisms underlying such effects are unknown, recent work has suggested that inbred males may suffer impaired ejaculate quality, thus providing a functional explanation for reductions in reproductive function in inbred populations. However, the relationship between inbreeding and sperm quality remains controversial, particularly in wild populations where the level of inbreeding is typically estimated using neutral molecular markers. Such markers are thought to reflect genome-wide levels of heterozygosity only under restricted conditions, and rarely in outbred populations. Here we employ a comparative approach that takes account of these criticisms and evaluates the evidence linking inbreeding to reductions in sperm quality in 20 mammal species. We focus on sperm abnormalities and sperm motility, which are key determinants of male fertility in many species. We show that species with reduced mean heterozygosity have impaired ejaculated quality, although subsequent analyses revealed that these effects were confined to endangered populations. Our findings therefore support the notion that inbreeding can severely impair sperm quality while concomitantly addressing criticisms surrounding the use of heterozygosity estimates to estimate the level of inbreeding.
Keywords: heterozygosity fitness correlation, abnormal sperm, fertility, ejaculate, sperm competition
1. Introduction
Inbreeding depression is commonly associated with reductions in fertility and embryogenesis (Keller & Waller 2002). While reduced sperm quality has been implicated as a likely mechanistic explanation underlying these effects (Gage et al. 2006), the effect of inbreeding on sperm quality remains controversial. Following initial reports of inbreeding depression for sperm quality in endangered felids (e.g. Wildt et al. 1987), subsequent studies of captive and wild species revealed that inbreeding reduces sperm quality (Gomendio et al. 2000; Gage et al. 2006; van Eldik et al. 2006; Asa et al. 2007). However, studies examining captive-reared species with known pedigrees generally lack an appropriately controlled outbred comparison population (e.g. Caro & Laurenson 1994), and inbreeding depression for sperm quality is not universal (e.g. Gomendio et al. 2000). Furthermore, studies of wild populations, which have relied on molecular estimates of inbreeding (multilocus heterozygosity), have come under intense scrutiny (Balloux et al. 2004; Slate et al. 2004). Although inbred individuals are expected to have more homozygous genomes (Keller & Waller 2002), in generally outbred species there is a tenuous relationship between marker heterozygosity and inbreeding estimated from known pedigrees (Balloux et al. 2004; Slate et al. 2004). Therefore, researchers interested in understanding how inbreeding influences sperm quality face two key challenges: finding outbred populations for comparisons and accurately estimating the degree of inbreeding present in a species.
Here we employ a comparative approach to examine the association between inbreeding and sperm quality among 10 endangered and 10 non-endangered populations of mammal. We focused on two sperm parameters that are known to reflect male fertility and competitive fertilization performance: sperm abnormalities (e.g. Oettle 1993; Bartoov et al. 2002) and the proportion of motile sperm in the ejaculate (e.g. Denk et al. 2005). We examined the relationship between these traits and inbreeding using marker heterozygosity to estimate the level of inbreeding. The strength of the correlation between marker heterozygosity and genome-wide heterozygosity is dependent on the individual pedigree in question and the strength of historic inbreeding in the populations (Aparicio et al. 2007; Grueber et al. 2008). In endangered species, where reduced effective population sizes make inbreeding more common, marker heterozygosity should more accurately reflect genome-wide heterozygosity, and thus act as an appropriate measure of the degree of inbreeding (e.g. Hedrick et al. 2001; Aparicio et al. 2007; Grueber et al. 2008). Therefore, after first assessing the relationship between sperm characteristics and heterozygosity across all species, we performed separate analyses to evaluate the relationship between heterozygosity and sperm quality in endangered and non-endangered species. We predict that any relationship between marker heterozygosity and sperm quality will be confined to endangered species.
2. Material and methods
(a) Data collection
We obtained data from published sources for sperm characteristics and expected heterozygosity (He) for 20 mammalian species (mean number of males per species ±s.e. 16.3±4.6, range 4–97; see caption of figure 1 and the electronic supplementary material for species names and further details). Values for He were highly correlated with those for observed heterozygosity in the subset of studies (n=15) that reported both estimates (r=0.91, p<0.0001). Since the results were qualitatively similar irrespective of the measure used, we maximized statistical power by focusing on He in our subsequent analysis. For 18 species, data on the percentage of motile sperm were also available. We preferentially used sperm characteristics obtained at the peak of the reproductive season. Sperm abnormalities and motility were assessed using species-specific methodologies. However, generally sperm were classified as abnormal based on the structural defects of the acrosome, head, midpiece or flagellum, while sperm motility was estimated as the percentage of motile sperm within each sample (see electronic supplementary material).
For He, we used studies where the greatest numbers of microsatellite loci were examined (mean loci number ±s.e. 36.7±8.3, range 4–93; Balloux et al. 2004; Slate et al. 2004), matching the geographical locations where sperm characteristics and heterozygosity were measured as closely as possible. When data were available from multiple published sources, we used mean values.
Species conservation status was determined based on the International Union for Conservation of Nature's (IUCN) Red List (IUCN 2008). Species classified as ‘critically endangered’, ‘endangered’ or ‘vulnerable’ were considered endangered in our analyses, while those classified as ‘near threatened’ or ‘least concern’ were considered non-endangered. The only exception was the American bison (Bison bison), which we classified as endangered because this species is undergoing conservation programmes to maintain its near-threatened IUCN status.
Mating system, a proxy measure of sperm competition, did not affect the relationship between sperm parameters and heterozygosity when added as a covariate to the full model. Therefore, mating system was not considered in subsequent analyses.
(b) Phylogenetic analyses
To control for the effects of shared ancestry (Harvey & Pagel 1991), we determined the phylogenetic relationships among species using the phylogeny described in Bininda-Emonds et al. (2007). Because this phylogeny did not distinguish between lion subspecies, we placed lions (Panthera leo) from the Ngorongoro Crater and the Serengeti as sister subspecies and placed these African lions basal to the Asiatic lion (Panthera leo persica) (Yamaguchi et al. 2004). All branch lengths were set equal to one. Percentage data (abnormal and motile sperm) were angular transformed. All data were log transformed prior to phylogenetic analyses, which were performed using the R program (v. 2.6.1: R Foundation for Statistical Computing 2007).
Data were analysed using generalized least-squared (GLS) regressions while controlling for phylogenetic effects (Freckleton et al. 2002). We used likelihood ratio tests to assess the degree of phylogenetic dependence in the data by calculating the scaling parameter λ (Freckleton et al. 2002). To account for the independent examination of heterozygosity estimates on sperm characteristics, we calculated the effect size, r, from each test and used the non-central 95 per cent confidence interval (CI) for r to determine the statistical significance (Nakagawa & Cuthill 2007). Effect sizes were calculated from t values generated from the phylogenetic GLS models (Nakagawa & Cuthill 2007). See ESM for effect sizes, CIs and λ values for regressions.
3. Results
Although there were no significant differences in heterozygosity between endangered and non-endangered species (mean He; endangered: 0.45±0.07, non-endangered: 0.68±0.06, t=−1.36, p=0.19), endangered species had fewer motile sperm (t=−2.1, p=0.047) and more abnormal sperm (t=2.49, p=0.02) than non-endangered species (endangered species: percentage of motile sperm 66.7±5.1, percentage of abnormal sperm 47.8±8.28; non-endangered species: percentage of motile sperm 77.4±1.75, percentage of abnormal sperm 30.0±5.20).
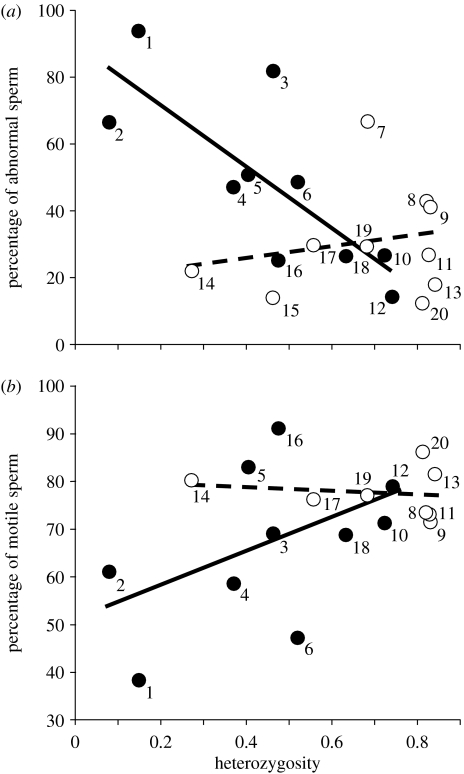
Across all species, heterozygosity was negatively associated with the percentage of abnormal sperm (, p=0.04; figure 1a) and positively associated with the percentage of motile sperm per ejaculate (, p=0.02; figure 1b). When we considered only endangered species, the relationships between heterozygosity and sperm quality remained significant (sperm abnormalities: , p=0.02; sperm motility: , p=0.02; figure 1). However, for non-endangered species, heterozygosity was not related with either of the two sperm traits (sperm abnormalities: , p=1.0; sperm motility: , p=0.68; figure 1).
Figure 1.
Relationship between the proportion of (a) abnormal and (b) motile sperm in endangered (filled circles, solid line) and non-endangered (open circles, broken line) mammals. Data in the figures are not controlled for phylogeny. Numbers indicate the species: (1) Florida panther, (2) Indian lion, (3) cheetah, (4) black-footed ferret, (5) Ngorongoro Crater lion, (6) black bear, (7) South American panther, (8) margays, (9) tigrinas, (10) red wolf, (11) jaguar, (12) giant panda, (13) ocelot, (14) brown bear, (15) European rabbit, (16) Serengeti lions, (17) howler monkey, (18) bison, (19) domestic cat, (20) coyotes. See electronic supplementary material for additional details.
4. Discussion
Our results demonstrate that extensive inbreeding leads to depressed sperm quality. However, while the negative relationship between species heterozygosity and sperm quality was evident when all species were examined, it is clear that this pattern was generated by the endangered species in the dataset. When we confined our analysis to endangered species, criteria that arguably most closely meet the conditions where heterozygosity reflects inbreeding (e.g. Slate et al. 2004; Aparicio et al. 2007; Grueber et al. 2008), more homozygous species had reduced sperm quality. There was no association between sperm quality and heterozygosity in non-endangered species. Thus, our results support the previously reported link between heterozygosity and sperm quality in European rabbits (Oryctolagus cuniculus; Gage et al. 2006) while bolstering the argument that estimates of heterozygosity may not accurately reflect the level of inbreeding unless certain restrictive conditions are met (Balloux et al. 2004; Slate et al. 2004; Grueber et al. 2008).
As argued elsewhere (Gage et al. 2006), spermatozoa may be particularly susceptible to inbreeding depression because spermatogenesis relies on highly regulated developmental genes, the disruption of which causes male infertility (e.g. Hargreave 2000). Consequently, exposure of deleterious alleles or losses of heterozygote advantage resulting from inbreeding will directly affect sperm quality. However, given the correlative nature of studies reporting relationships between heterozygosity and fitness, it is not clear whether heterozygosity leads to reduced sperm quality, or alternatively whether species with poor sperm quality suffer from greater rates of infertility, which in turn reduces heterozygosity (Slate & Pemberton 2006). The available evidence suggests, however, that sperm quality is directly influenced by inbreeding rather than vice versa. For example, experimentally inbred male domestic cats and dogs produce ejaculates containing fewer (Wildt et al. 1982) and more abnormal sperm (Pukazhenthi et al. 2006), and recent experimental work on guppies has suggested that highly inbred males produce ejaculates that are competitively inferior than their outbred counterparts (S. R. J. Zajitschek, A. K. Lindholm, J. P. Evans & R. C. Brooks 2008, unpublished data). Impaired sperm quality therefore appears to be a consequence of inbreeding.
Our analyses focused on mammals, as appropriate data were not available from other taxa. Yet inbreeding depression has been demonstrated in many non-mammalian species (Darwin 1876; Keller & Waller 2002) and we anticipate that similar reductions in sperm quality in response to inbreeding will be widespread. Regardless of the focal species, however, our finding that inbreeding is associated with potentially important declines in male reproductive function will have obvious implications for the management of endangered populations, where both genetic viability and reproductive potential are important considerations.
Acknowledgements
We thank Damian Dowling, Joe Tomkins, Leigh Simmons and two referees for their comments on the manuscript. This work was funded by the Natural Sciences and Engineering Research Council of Canada (J.L.F.) and the Australian Research Council (J.P.E.).
Supplementary Material
Electronic appendix
References
- Aparicio J.M., Ortego J., Cordero P.J. Can a simple algebraic analysis predict markers–genome heterozygosity correlations? J. Hered. 2007;98:93–96. doi: 10.1093/jhered/esl055. doi:10.1093/jhered/esl055 [DOI] [PubMed] [Google Scholar]
- Asa C., Miller P., Agnew M., Rebolledo J.A.R., Lindsey S.L., Callahan M., Bauman K. Relationship of inbreeding with sperm quality and reproductive success in Mexican gray wolves. Anim. Cons. 2007;10:326–331. doi:10.1111/j.1469-1795.2007.00116.x [Google Scholar]
- Balloux F., Amos W., Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. doi:10.1111/j.1365-294X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Bartoov B., Berkovitz A., Eltes F., Kogosowski A., Menezo Y., Barak Y. Real-time fine morphology of motile human sperm cells is associated with IVF-ICSI outcome. J. Androl. 2002;23:1–8. doi: 10.1002/j.1939-4640.2002.tb02595.x. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P., et al. The delayed rise of present day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Caro T.M., Laurenson M.K. Ecological and genetic factors in conservation: a cautionary tale. Science. 1994;263:485–486. doi: 10.1126/science.8290956. doi:10.1126/science.8290956 [DOI] [PubMed] [Google Scholar]
- Darwin C. John Murray; London, UK: 1876. The effects of cross and self fertilisation in the vegetable kingdom. [Google Scholar]
- Denk A.G., Holzmann A., Peters A., Vermeirssen E.L.M., Kempenaers B. Paternity in mallards: effects of sperm quality and female sperm selection for inbreeding avoidance. Behav. Ecol. 2005;16:825–833. doi:10.1093/beheco/ari065 [Google Scholar]
- Freckleton R.P., Harvey P.H., Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 2002;160:712–726. doi: 10.1086/343873. doi:10.1086/343873 [DOI] [PubMed] [Google Scholar]
- Gage M.J.G., Surridge A.K., Tomkins J.L., Green E., Wiskin L., Bell D.J., Hewitt G.M. Reduced heterozygosity depresses sperm quality in wild rabbits, Oryctolagus cuniculus. Curr. Biol. 2006;16:612–617. doi: 10.1016/j.cub.2006.02.059. doi:10.1016/j.cub.2006.02.059 [DOI] [PubMed] [Google Scholar]
- Gomendio M., Cassinello J., Roldan E.R.S. A comparative study of ejaculate traits in three endangered ungulates with different levels of inbreeding: fluctuating asymmetry as an indicator of reproductive and genetic stress. Proc. R. Soc. B. 2000;267:875–882. doi: 10.1098/rspb.2000.1084. doi:10.1098/rspb.2000.1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber C.E., Wallis G.P., Jamieson I.G. Heterozygosity-fitness correlations and their relevance to studies on inbreeding depression in threatened species. Mol. Ecol. 2008;17:3978–3984. doi: 10.1111/j.1365-294x.2008.03910.x. doi:10.1111/j.1365-294X.2008.03910.x [DOI] [PubMed] [Google Scholar]
- Hargreave T.B. Genetic basis of male fertility. Br. Med. Bull. 2000;56:650–671. doi: 10.1258/0007142001903454. doi:10.1258/0007142001903454 [DOI] [PubMed] [Google Scholar]
- Harvey P.H., Pagel M.D. Oxford University Press; Oxford, UK: 1991. The comparative method in evolutionary biology. [Google Scholar]
- Hedrick P., Fredrickson R., Ellegren H. Evaluation of d2, a microsatellite measure of inbreeding and outbreedin, in wolves with a known pedigree. Evolution. 2001;55:1256–1260. doi: 10.1111/j.0014-3820.2001.tb00646.x. [DOI] [PubMed] [Google Scholar]
- International Union for Conservation of Nature (IUCN) 2008 IUCN red list of threatened species. International Union for conservation of Nature, available at: www.iucnredlist.org (accessed 18 November 2008).
- Keller L.F., Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Nakagawa S., Cuthill I.C. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. doi:10.1111/j.1469-185X.2007.00027.x [DOI] [PubMed] [Google Scholar]
- Oettle E.E. Sperm morphology and fertility in the dog. J. Reprod. Fertil. 1993:257–260. [PubMed] [Google Scholar]
- Pukazhenthi B.S., Neubauer K., Jewgenow K., Howard J., Wildt D.E. The impact and potential etiology of teratospermia in the domestic cat and its wild relatives. Theriogenology. 2006;66:112–121. doi: 10.1016/j.theriogenology.2006.03.020. doi:10.1016/j.theriogenology.2006.03.020 [DOI] [PubMed] [Google Scholar]
- Slate J., Pemberton J. Does reduced heterozygosity depress sperm quality in wild rabbits (Oryctolagus cuniculus)? Curr. Biol. 2006;16:R790–R791. doi: 10.1016/j.cub.2006.08.047. doi:10.1016/j.cub.2006.08.047 [DOI] [PubMed] [Google Scholar]
- Slate J., David P., Dodds K.G., Veenvliet B.A., Glass B.C., Broad T.E., McEwan J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity. 2004;93:255–265. doi: 10.1038/sj.hdy.6800485. doi:10.1038/sj.hdy.6800485 [DOI] [PubMed] [Google Scholar]
- van Eldik P., van der Waaij E.H., Ducro B., Kooper A.W., Stout T.A.E., Colenbrander B. Possible negative effects of inbreeding on semen quality in Shetland pony stallions. Theriogenology. 2006;65:1159–1170. doi: 10.1016/j.theriogenology.2005.08.001. doi:10.1016/j.theriogenology.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Wildt D.E., Baas E.J., Chakraborty P.K., Wolfle T.L., Stewart A.P. Influence of inbreeding on reproductive performance, ejaculate quality and testicular volume in the dog. Theriogenology. 1982;17:445–452. doi: 10.1016/0093-691x(82)90026-7. doi:10.1016/0093-691X(82)90026-7 [DOI] [PubMed] [Google Scholar]
- Wildt D.E., Bush M., Goodrowe K.L., Packer C., Pusey A.E., Brown J.L., Joslin P., Obrien S.J. Reproductive and genetic consequences of founding isolated lion populations. Nature. 1987;329:328–331. doi:10.1038/329328a0 [Google Scholar]
- Yamaguchi N., Cooper A., Werdelin L., Macdonald D.W. Evolution of the mane and group-living in the lion (Panthera leo): a review. J. Zool. 2004;263:329–342. doi:10.1017/S0952836904005242 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic appendix