Summary
Intramembrane proteases, which cleave TM helices, participate in numerous biological processes encompassing all branches of life. Several crystallographic structures of Escherichia coli GlpG rhomboid protease have been determined. In order to understand GlpG dynamics and lipid interactions in a native-like environment, we have examined the molecular dynamics of wild-type and mutant GlpG in different membrane environments. The irregular shape and small hydrophobic thickness of the protein cause significant bilayer deformations that may be important for substrate entry into the active site. Hydrogen-bond interactions with lipids are paramount in protein orientation and dynamics. Mutations in the unusual L1 loop cause changes in protein dynamics and protein orientation that are relayed to the His-Ser catalytic dyad. Similarly, mutations in TM5 change the dynamics and structure of the L1 loop. These results imply that the L1 loop has an important regulatory role in proteolysis.
Keywords: lipid bilayers, lipid-protein interactions, intramembrane protease, molecular dynamics simulations, phosphatidylcholine, phosphatidylethanolamine
Introduction
Intramembrane proteases are membrane proteins that cleave TM helices (Wolfe and Kopan, 2004) to liberate proteins that participate in important processes, such as cell signalling and gene regulation (Brown and Goldstein, 1997; McQuibban et al., 2003; Struhl and Greenwald, 1999; Urban, 2001; Weihofen et al., 2002; Wolfe and Kopan, 2004). The first rhomboid serine protease was identified in Drosophila Melanogaster where it was found to cleave the membrane-anchored Spitz growth factor that stimulates the epidermal growth factor receptor (Bier et al., 1990; Freeman et al., 1992; Urban, 2001). Rhomboid proteases were subsequently found to be present in almost all organisms, including Escherichia coli (Koonin et al., 2003; Wasserman et al., 2000). The three-dimensional structures of the E. coli rhomboid (GlpG) solved recently to atomic resolution provided important insights into the possible mechanism of substrate cleavage (Ben-Shem et al., 2007; Wang et al., 2007; Wang et al., 2006; Wu et al., 2006). Albeit invaluable, these are frozen snapshots of GlpG in a crystal environment, which provide little information on lipid interactions and dynamics.
Knowledge of how GlpG interacts with lipids is important for beginning to understand how GlpG interacts with substrates and how the composition of the lipid membrane could influence GlpG catalysis. Lipids can affect the activity of membrane proteins (Lee, 2004), and may be particularly important for membrane-embedded proteases (Kalvodova et al., 2005; Narayanan et al., 2007; Urban and Wolfe, 2005). For example, GlpG is active when reconstituted with phosphatidylethanolamine (PE) lipids, but is inactive in phosphatidylcholine (PC) (Urban and Wolfe, 2005). The highly irregular shape of GlpG revealed by the crystal structures makes the dynamics of the lipid interactions very intriguing. The role of L1 in rhomboid protease function is also poorly understood; although it extends away from the active site (Figure 1B,C), its deletion inactivates the protein (Wang et al., 2007). The observation that mutation of specific L1 amino acids not directly connected to the catalytic site drastically reduces the activity (Baker et al., 2007) was interpreted to suggest that L1 has an important structural role (Baker et al., 2007). The effect of the L1 mutations implies that perturbation of L1 is relayed as a change in the microenvironment of the catalytic groups. To begin to understand at the atomic level the dynamics and the lipid interactions of the GlpG intramembrane protease, we have performed extensive all-atom molecular dynamics (MD) simulations of wild-type and two mutant E. coli GlpG rhomboids embedded in two different lipid membrane environments.
Figure 1.
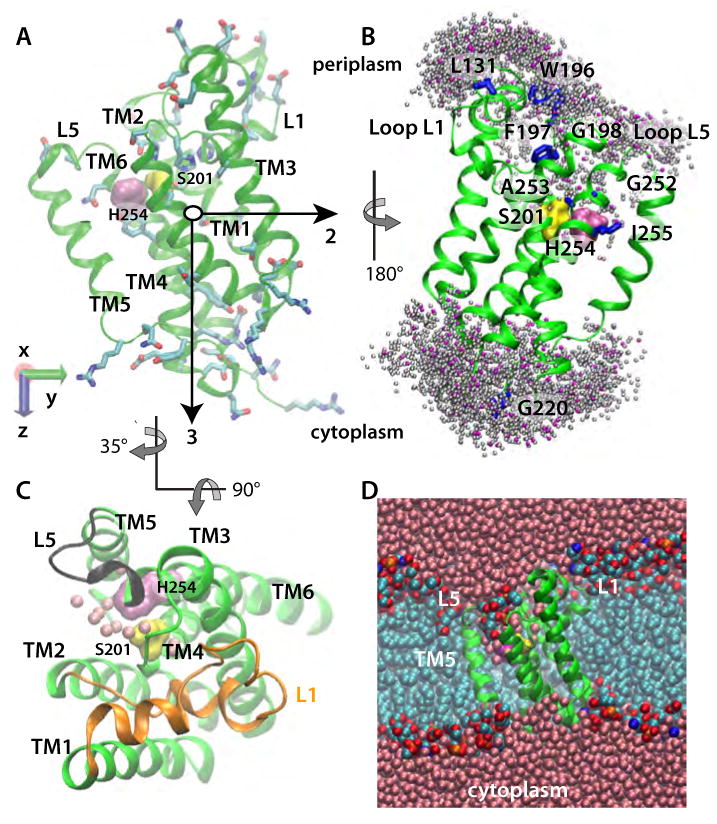
E. Coli GlpG in a POPE lipid bilayer (Sim2). (A) Starting geometry and principal axes (black arrows) of GlpG. S201 and H254 are shown as yellow and mauve surfaces, respectively; amino acid sidechains with H-bonding capability are depicted as bonds. (B) Dynamic water distribution in the active site. Locations of the water-molecule oxygen atoms located within 5 Å of the protein in a snapshot from Sim2 are shown as pink spheres. Shown in grey are water locations sampled during ten equally spaced snapshots during a subsequent 1ns segment of Sim2. Amino acids known from experiments to be accessible to bulk water (Maegawa et al., 2007) are shown as blue bonds, or as surfaces (S201 and H254). (C) View from the luminal side of GlpG from Sim2, depicting water molecules within 5Å of S201 and H254 as pink spheres, L1 in orange, and L5 in black. (D) Cut away view of GlpG in a POPE lipid bilayer, with the protein shown in green, bulk-water oxygen atoms in pink, lipid alkyl carbon cyan, lipid phosphorus orange, lipid oxygen red, and lipid nitrogen atoms blue.
GlpG consists of a catalytic TM domain and a cytosolic N-terminal domain that is not essential for catalysis (Wu et al., 2006). The recent crystal structures of the TM domain of GlpG (Ben-Shem et al., 2007; Wang and Ha, 2007; Wang et al., 2006; Wu et al., 2006) reveal six TM helices (TM1-TM6) with very different tilt angles, and the unusual periplasmic L1 loop accommodated between TM1 and TM3 (Figure 1). Alignment of the several structures shows that the greatest structural variability occurs in TM5 and loop L5 (White, 2006). The catalytic residues (S201 and H254 on TM4 and TM6, respectively) are located within the bilayer boundaries (Ben-Shem et al., 2007; Urban, 2001; Wang et al., 2006; Wu et al., 2006) (Figure 1D). The presence in one of the crystal structures of a phosphoglycerol lipid headgroup close to the catalytic dyad suggests exposure of the active-site to the lipid membrane (Ben-Shem et al., 2007).
The structural events along the reaction pathway of GlpG are not known and there is no structural information about the complex between GlpG and its substrate(s). Even though GlpG can cleave the Drosophila Spitz peptide (Urban et al., 2002) and the TM2 helix of LacY permease (Maegawa et al., 2005), and may participate in quorum sensing (Clemmer, 2006), the physiological substrate of GlpG remains unclear. Two different scenarios have been proposed to explain how structural rearrangements of GlpG may assist the formation of the enzyme-substrate complex. In one scenario, TM5 acts as a lateral gate to control substrate access to the active site (Baker et al., 2007; Wu et al., 2006). This scenario is supported by the significant range of TM5 tilt angles (∼30°) observed in the crystal structures (Ben-Shem et al., 2007; Wang et al., 2006; Wu et al., 2006) and by the effect of site-directed mutations on catalytic efficiency (Baker et al., 2007). In the second scenario, the cap loop L5 (Figure 1C) opens to allow docking of the substrate to the active site (Wang et al., 2007), consistent with the conformational variability of L5 in the crystal structures. Of course, L5 and TM5 are connected, implying that the reaction path may involve both L5 and TM5.
To derive a dynamic view of GlpG in a fluid lipid bilayer, we examined the lipid interactions of GlpG embedded in 1-palmytoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPE) and (1-palmytoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine) POPC lipid bilayers. The roles of the L1 loop and the TM5 helix were examined by modeling two different mutant GlpG phenotypes in which activity is significantly reduced (Y138S/F139S/L143S) or enhanced (L229V/F232V/W236V) (Baker et al., 2007). Our analysis of these mutants suggests that TM5 and L1 are dynamically coupled so that changes in the dynamics of one are relayed to the other.
Results
We performed four all-atom MD simulations (Sims, Table 1) on systems comprised of one protein molecule, approximately 500 lipids, and 30,000 waters, for a total of about 160,000 atoms (see Methods). Two independent simulations of wild-type GlpG were carried out in POPC (Sim1) and POPE (Sim2). In Sim3 and Sim4, we explored the dynamics of the mutant phenotypes Y138S/F139S/L143S (the triple-Ser mutant; Sim3) and L229V/F232V/W236V (the triple-Val mutant; Sim4) in POPE bilayers. Because there are no crystal structures of these mutants, starting coordinates were prepared from the wild-type crystal structure (Ben-Shem et al., 2007). Consequently, the limited timescales of our simulations may not capture the full extent of the protein conformational changes induced by the mutations. All simulations were run at least 10 nanoseconds (ns) beyond the time point at which the dimension of the unit cell reached plateau values. All histograms and average values were computed based on 10000 sets of coordinates saved every 1 ps during the last 10 ns of each simulation run. The time-dependence plots of distances and root-mean-squared deviation (rmsd) values are for the unconstrained simulation runs.
Table 1.
Summary of MD simulations of the GlpG protease in different membrane environments.
| Simulation | Protein | Lipida | Active-site lipid | Length (ns)b |
|---|---|---|---|---|
| Sim 1 | Wild-type | POPC | PGV | 34.7 |
| Sim 2 | Wild-type | POPE | POPE | 34.4 |
| Sim 3 | triple-Ser mutantc | POPE | POPE | 20.4 |
| Sim 4 | triple-Val mutantd | POPE | POPE | 26.9 |
Lipids: POPC, palmitoyloleoylphosphatidylcholine; POPE, palmitoyloleoylphosphatidylethanolamine; PGV, palmitoylvaccenoylphosphatidylglycerol.
Unconstrained simulation time in nanoseconds
Triple Ser mutant: Y138S/F139S/L143S
Triple Val mutant: L229V/F232V/W236V
Rhomboid protease orientation and lipid bilayer thinning
Defining protein orientation as the angle between the bilayer normal and the third principal axis of the protein (Figure 1A), we found average orientation angles of 11.7±1.8° and 15.9±2.3° for POPE and POPC bilayers, respectively (Figure 2A). The difference between these angles appears to be a result of protein-bilayer interactions that optimize H-bonding between protein surface groups and lipid headgroups (below). Optimization is achieved mainly by the surrounding lipid molecules adjusting their geometry (Figures 2B,C,D and 3A,B) to accommodate the relatively stable TM part of the protein (Figure 4A,B).
Figure 2.
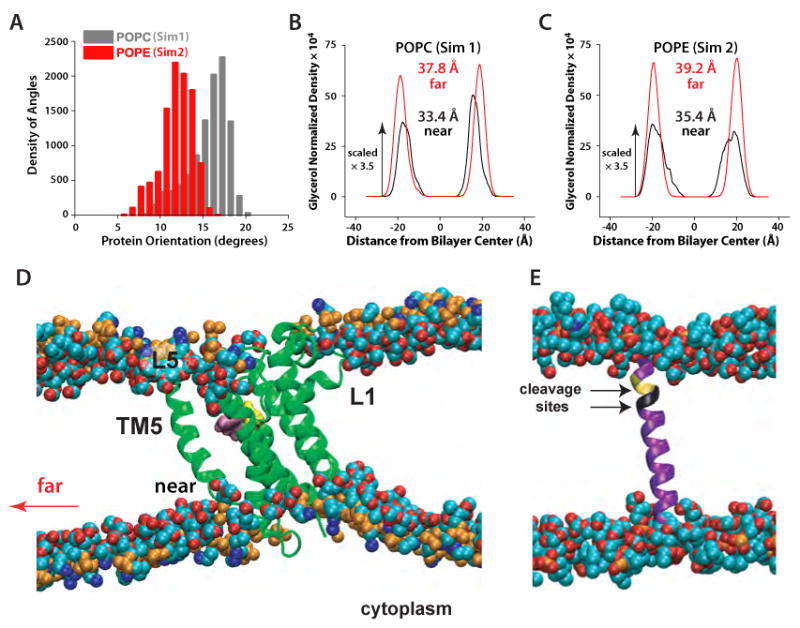
Local perturbations of lipid bilayers caused by GlpG rhomboid protease. (A) Distribution of orientational angles of GlpG relative to the membrane normal in Sim1 (gray) and Sim2 (red). (B) Hydrocarbon thickness of the POPC bilayers (Sim1) and (C) POPE bilayers (Sim2) close to the protein (black curves; computed from ∼90 lipids within the first 2-3 shells of lipids around the protein) and far from the protein in the unperturbed region of the bilayer (red curves). The hydrocarbon thickness was estimated as the distance between the peaks of the distribution for the glycerol groups of the two lipid leaflets (Wiener and White, 1992) taken along the membrane normal and normalized by the volume of the simulation cell. In the case of the distorted density peaks for POPE lipids close to the protein (z ∼20Å), we used the value of z at the center of the distribution. (D) Thinning of the lipid bilayer close to the protein; only a ∼38 Å-wide section of the bilayer containing the protein is depicted. (E) Snapshot of the Spitz substrate in a POPC lipid bilayer. The two substrate cleavage sites (Baker et al., 2007) are shown in yellow (Ala-Ser) and black (Gly-Ala). Little perturbation of the bilayer in the vicinity of Spitz was observed.
Figure 3.
Lipid and water interactions of GlpG in POPC (Sim1) and POPE (Sim2). Pink spheres indicate the location of the bulk water oxygen atoms within 5 Å of the protein amino acid residues shown as bonds. Phospholipid headgroups are also shown as bonds. (A) Luminal interactions of GlpG residues with POPE headgroups. (B) Cytoplasmic interactions of GlpG residues with POPE headgroups. (C) Active site interactions of GlpG in POPE membranes. (D) Active-site interactions of GlpG in a POPC membrane, with a PGV lipid close to the active site. (E) Loop L1 interactions with POPE headgroups. (F,G) Loop L1 interactions with POPC headgroups during a segment of ∼20ns of Sim1 (F) and late in the simulation (G).
Figure 4.
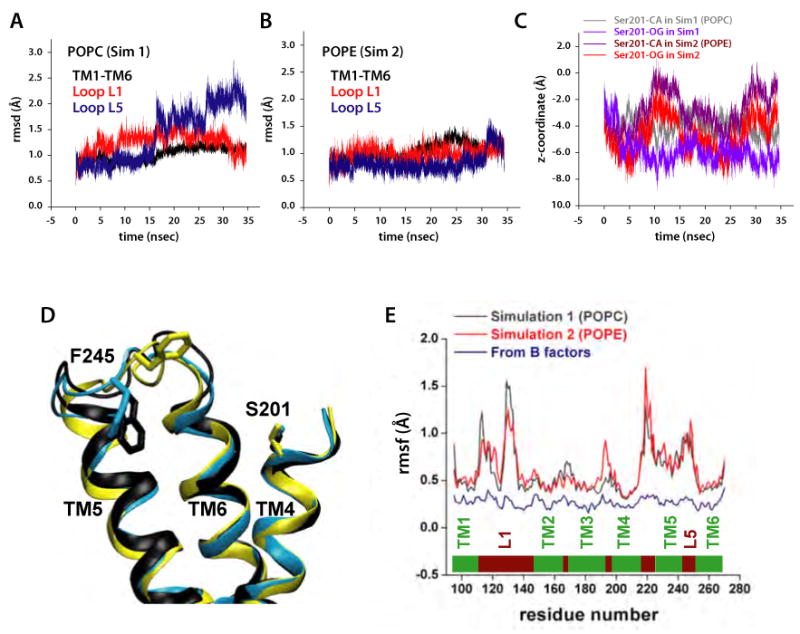
Dynamics of GlpG in POPC (Sim1) and POPE (Sim2) bilayers. (A) Dynamics of loops L1 (red), L5 (blue), and of helices TM1-TM6 (black) in POPC and (B) POPE bilayers measured as the rmsd (Å) relative to the starting crystal structure coordinates. (C) Position of the catalytic S201 in POPC (Sim1) and POPE (Sim2) bilayers. (D) Conformations sampled by L5 in POPC (Sim1) during the time periods 0-15ns (yellow), 15-25ns (cyan), and 25-34.5ns (black). (E) Rmsf (Å) of GlpG residues in Sim1 (black, POPC) and Sim2 (red, POPE) are dramatically greater than those computed from the crystal structure B-factors, shown in blue (Ben-Shem et al., 2007).
The rearrangement of the lipid molecules led to non-uniform thinning of the membrane in the vicinity of the protein (Figure 2D). Although the average glycerol-to-glycerol thickness of the bilayer in the first 2-3 shells of lipids around the protein is only about 4 Å smaller than at remote distances (Figure 2B,C), the thickness varies considerably around the protein perimeter (black curves). The largest lipid rearrangements and bilayer thinning occurred on the cytoplasmic side, underneath the L1 loop (Figure 2D). The average thicknesses of the unperturbed, distant regions of the POPE and POPC lipid bilayers (∼39 Å and ∼38 Å, respectively) are compatible with earlier bilayer simulations (Jensen and Mouritsen, 2004). Bilayer thinning close to the protein was also proposed by Wang et al. (2007). However, direct comparison with the thinning we observe is difficult because of the complex nature of the thinning close to GlpG (Figure 2B-D) and the wide distribution of the protein's orientation angles in the fluid bilayer (Figure 2A), which could not have been anticipated from the X-ray structure alone.
The catalytic serine is located deep within the lipid bilayer (Figure 4C). The positions of S201 oxygen fluctuate around the mean values of ≈ 6 Å (POPC) and ≈ 5 Å (POPE) above the bilayer midplane in the periplasmic leaflet. Relative to the midplane of the 20 Å-thick detergent belt in their x-ray structure, Wang et al. (2007) estimated a distance of ≈ 7 Å. What distinguishes our result from that of Wang et al. (2007) is that the location of S201 is dynamic, and is influenced by the composition of the lipid bilayer (Figure 4C). Even though S201 is relatively close to the bilayer midplane, it is nevertheless in a hydrophilic environment comprised of water and lipid headgroups (Figures 1 and 3) due to bilayer thinning in the vicinity of the protein (Figure 2D). An aqueous environment at the active site was predicted from experiments (Maegawa et al., 2005; Maegawa et al., 2007), and is required for proteolysis.
Membrane hydrogen bonding of GlpG depends on the lipid environment
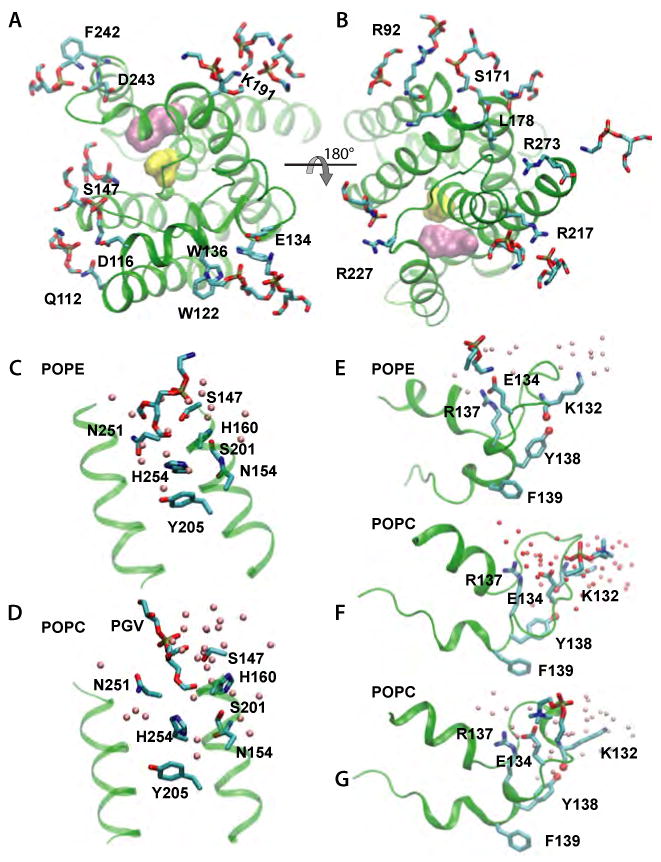
The charged protein residues that can readily form hydrogen bonds (H-bonds) with the lipid headgroups are located remote from the active site, at the cytoplasmic termini of the TM segments, and on L1 (Figure 1A). Importantly, some of the lipid-protein H-bonds depend on the type of lipid headgroup. The positively charged protein groups located at the membrane interface (Figure 1A) H-bond to the both POPE and POPC (Figure 3A,B). A detailed analysis of the protein-lipid H-bonds indicates that some interactions are remarkably stable, whereas others are dynamic, breaking and re-forming during the simulations. For example, the lipid H-bonding of K191 (Figure 3A), is very stable (donor-acceptor distances of ∼2.7Å for the last ∼20 ns of the both Sim1 and Sim2). On the other hand, the lipid interactions of R92, R227, or R168 break and re-form during the simulations. The lipid molecule close to the catalytic site in the crystal structure (Ben-Shem et al., 2007) displaces somewhat towards the bulk and is part of a H-bonded network that comprises the lipid molecule, water, the catalytic S201 and H254, and other polar amino acids (Figure 3C,D).
The different H-bonding capabilities of the —N+H3 (POPE) and the bulkier —N+(CH3)3 (POPC) groups lead to significant differences in L1:membrane interactions (Figure 3E,F,G). In POPE, strong H-bonds between E134, R137, and a POPE amine form a structural barrier impenetrable to water molecules (Figure 3E). In contrast, during most of Sim1, E134 forms a water-mediated H-bond with a POPC phosphate, which allows water molecules to penetrate deeper into the lipid bilayer (Figure 3F). Structural rearrangements of POPC towards the end of Sim1 lead to the formation of an H-bond between E134 and the POPC choline (Figure 3G). The dependence of L1-membrane interactions on lipid type may influence its local structure and intra-protein H-bonding: In POPE the H-bond between Y138 and the K132 carbonyl group is stable during Sim2 at ∼2.8Å, whereas in Sim1 the presence of the Y138:K132 H-bond correlates with the lipid interactions of E134 (Figures 3F,G, and 5D). A correlation between the L1 H-bonding and hydration can also be observed in the structural analysis of the W136A mutant, in which the space initially occupied by W136 became filled with water molecules upon mutation (Wu et al., 2006).
Figure 5.
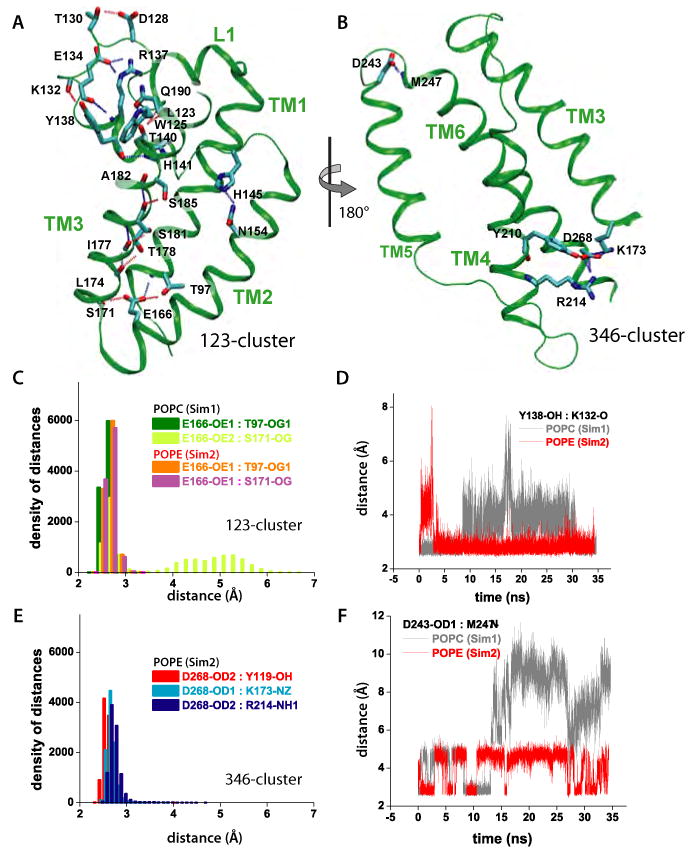
Intra-protein H-bonding of GlpG. (A) H-bonding within L1 and TM1/TM2/TM3 (123-cluster). (B) H-bonding within TM3/TM4/TM6 (346-cluster). For simplicity, only the backbone carbonyl or amide groups are depicted for non-polar residues. (C) Histograms of H-bonding distances in the 123-cluster for POPC and POPE. The E166-S171 H-bond breaks and reforms in POPC. (D) Effect of lipid-type on the dynamics of the Y138-K132 H-bonding in the 123-cluster. (E) Histograms of H-bonding distances within the 346 cluster in POPE. The histograms for the 346-cluster are very similar for POPE and POPC (data not shown). (F) Dynamics of H-bonds in the lipid interface of L5 depend upon lipid type.
Consistent with the stronger L1:lipid H-bonding in POPE vs. POPC, the root-mean-squared fluctuation (rmsf) values of the L1 atoms are somewhat smaller in POPE (Figure 4E). H-bonding to lipid and intra-protein H-bonding (see below) explain the structural stability of L1, indicated by small rmsd values comparable to those of the TM region (Figure 4A,B). Structural stability of L1 is also suggested by the structure of L1 being similar in the wild-type and in the W136A mutant in vitro (Wang et al., 2007).
The emergent pattern is that protein sidechains anchor the protein to the lipid interface by H-bonding. To assess further the importance of the H-bonds, we performed a sequence analysis (Supplementary Information) for three data sets extracted from the PFAM database version 23.0 (Finn et al., 2008): the family of Enterobacteriaceae (46 sequences), the PFAM seed (65 hand-curated sequences of rhomboids from Archea, Bacteria, Plants, and Eukaryotes), and in the complete PFAM rhomboid domain (1582 sequences).
Amino acids with H-bonding capability are present in the sequences of other rhomboids at the sites where GlpG H-bonds to the lipid bilayer. For example, half of the rhomboid sequences from the PFAM database seed and 870 sequences from the full PFAM alignment have residues with H-bonding capability corresponding to R217; 55 and 836 sequences of the seed and, respectively, of the full PFAM alignments, have a residue with H-bonding capability corresponding to E134 (Figure 3A,B; Tables S2-S3). R227 is conserved in all except for two Enterobacteriacea sequences, but not in other organisms (Tables S1-S3). H-bonding at the periplasmic K191 (Figure 3A) is not significantly conserved - 45% of the Enterobacteriacea sequences, 27% of the PFAM seed, and 18% of the full PFAM alignment (Tables S1-S3). Differences in the nature of residues located at the interface with lipid headgroups could lead to a different response to lipids. For example, GlpG and the P. stuartii rhomboid respond differently to PC lipids (Koonin et al., 2003; Urban and Wolfe, 2005). Although further studies are needed to understand how lipids affect rhomboid proteases, we note that differences in the L1 sequences of these two rhomboids could contribute to their different response to lipids. For example, both GlpG and the P. stuartii rhomboid have E or D at position 134, R or K at 217, R or Y at 227 (the P. stuartii rhomboid also has Arg at position 226), but they differ in the 125 position (Koonin et al., 2003; Urban et al., 2002): P. stuartii has Asn, whereas GlpG has Trp, which is located in a sensitive region of GlpG where its H-bonding dynamics change upon perturbation of L1 and TM5 (Figures 7E and 9D).
Figure 7.
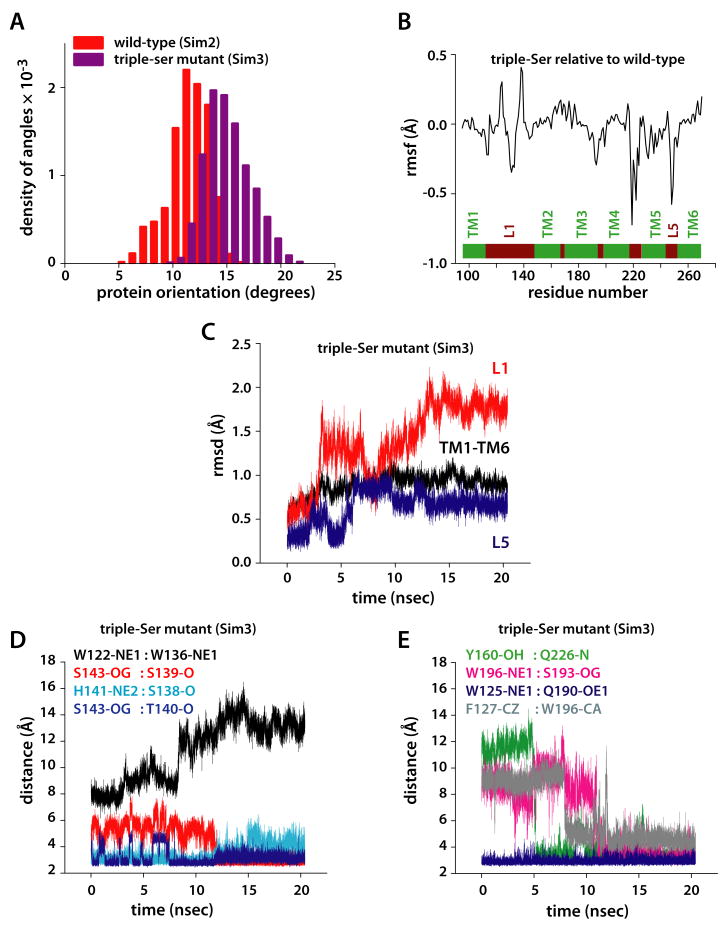
Dynamics of the triple-Ser mutant (Y138S/F139S/L143S) in POPE (Sim3). (A) Histograms of the orientation of the protein relative to the membrane normal (measured in degrees) in the wild-type (red) and the triple-Ser mutant (wine). (B) Rmsf (in Å) of the amino acid residues in the triple-Ser mutant relative to the wild-type protein. (C) Rmsd (in Å) of the triple-Ser mutant relative to the equilibrated structure of wild-type GlpG in POPE (Sim2). (D) Dynamics of selected distances in the triple-Ser mutant involving amino acids from L1. (E) Dynamics of selected distances in the triple-Ser mutant involving amino acids not in the L1 loop.
Figure 9.
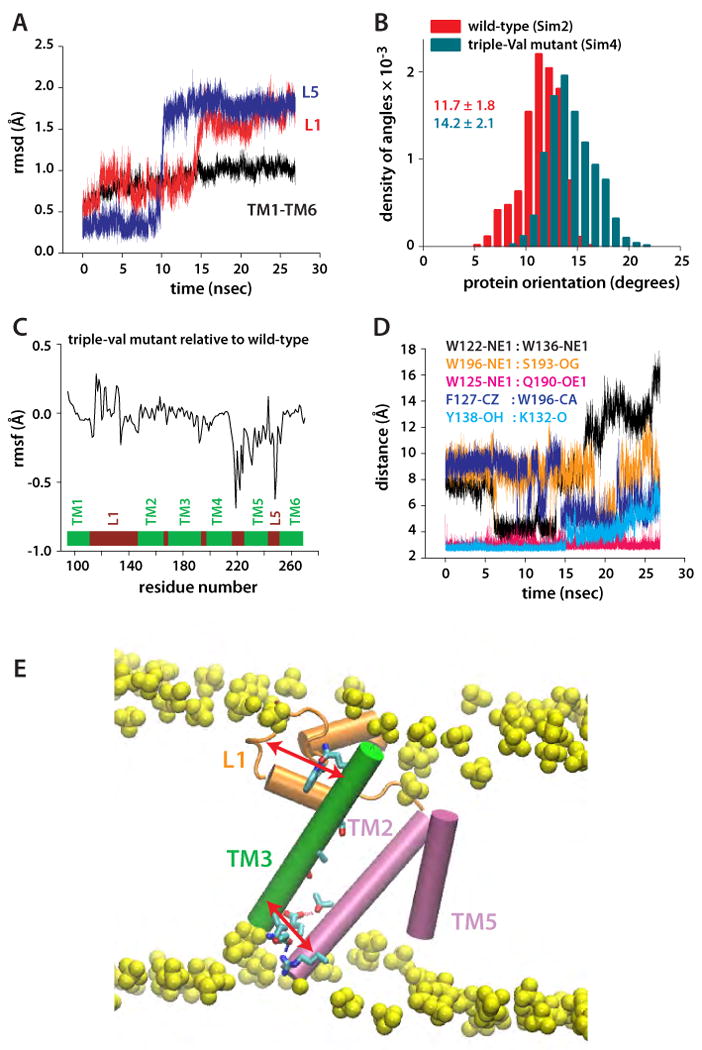
Protein orientation and dynamics of the triple-Val mutant (W236V/F232V/L229V). (A) Rmsd of TM1-TM6 and of loops L1 and L5, relative to the starting coordinates. The spontaneous and nearly simultaneous switching of L1 and L5 suggest conformational coupling between TM2/TM5 and L1. (B) Orientation of triple-Val GlpG (compare with Figure 7A). (C) Changes in the rmsf of triple-Val GlpG relative to the wild-type are observed not only in the region of TM5, but also in L1. (D) Dynamics of selected intra-protein interactions in Sim4 indicating that perturbation of TM5 is transmitted to L1. (E) Suggested relay mechanism in GlpG. Due to the array of H-bonds along TM3 (green), this structural element can act as a rigid arm that relays perturbations between L1 (orange) and the TM2/TM5 helices (orange) through intra-helical and L1:TM3 H-bonding.
Dynamics of intra-protein H-bond interactions
Multiple intra-protein H-bonds are observed between amino acid residues with polar or charged side-chains distributed asymmetrically (Figure 5A,B). The polar side-chains are located preferentially at the cytoplasmic ends of the TM helices, on L1, and along TM3. TM5 is not connected through H-bonds to other helices (Figure 5B), which could explain its rmsf values being larger than for the other TM helices (Figure 4E). In contrast, the cytoplasmic termini of the remaining five helices are interconnected by two clusters of H-bonds that bridge TM1/TM2/TM3 (123-cluster, Figure 5A) and TM3/TM4/TM6 (346-cluster, Figure 5B). Both clusters involve a highly-conserved amino acid residue: TM2-E166 in the 123-cluster and TM3-K173 in the 346-cluster are present as E and as R/K in 83% and respectively 73% of the sequences from the PFAM seed (Table S2); 69% and respectively 58% of the 1582 sequences from the PFAM full alignment have E166 and R/K173 (Table S3). TM3, the only TM segment involved in both H-bonding clusters, also connects to L1 via the W125:Q190 H-bond (Figure 5A), and has an array of intra-helical H-bonds involving T178, S181, and S185. S181 is present as S/T in 15% of the PFAM seed sequences, and 44% sequences from the full alignment. T178 and S185 are less conserved than S181 (Table S3).
The cytoplasmic clusters of H-bonds are present regardless of the membrane composition. The interactions between the various groups are stable during the simulations with characteristic distances of 2.6-2.8Å (Figure 5C,E). The only exception is the H-bond between TM2-E166 and TM3-S171, which is stable at 2.6±0.1 Å in POPE, whereas in POPC it breaks and reforms rapidly such that a H-bond (donor-acceptor distance of 2.7±0.1Å) is present half of the time (Figure 5C).
The presence of the stable H-bond clusters at the cytoplasmic site likely contributes to the structural rigidity of the TM region indicated by the small rmsd values (Figure 4A,B). The largest number of H-bonds is observed for TM3, which H-bonds to TM1, TM2, TM4, TM6, and L1; there is also an array of H-bonds along TM3 (Figures 5A,B). Because TM3 is a H-bonding partner in both H-bond clusters and for L1, we suggest that TM3 could act as messenger of structural and dynamical perturbations throughout the protein. Although a systematic experimental study of the effects of mutating H-bonding amino acids has not been performed yet, we note that the TM3-K173A mutant has a reduced proteolytic activity (Urban, 2001), which might perturb both H-bonding clusters. The seemingly normal activity of the TM2-E166A mutant in D. melanogaster rhomboid expressed in mammalian COS cells (Urban, 2001) might be due to a lipid headgroup substituting for the Glu sidechain.
The E134:R137 and Y138:K132 H-bonds (Figure 5A) may be particularly important for the local geometry of L1, and could contribute to the rmsd values of L1 being similar to those of the TM region (Figures 4 A,B). R137 is part of the highly conserved WR motif (Koonin et al., 2003; Urban, 2001), which is present in more than 80% of the both seed and full PFAM alignments (see Supplementary Information). The presence of the Y138-K132 H-bond depends somewhat on the interactions of L1 with the lipid bilayer (Figures 3E-G, 5D). Site-directed mutagenesis suggests a possible role of R137 and Y138 in GlpG function: The R137A mutant has reduced proteolytic activity in cells, and is inactive in detergent (Wang et al., 2007); the Y138F mutant has a reduced activity in detergent (Baker et al., 2007). Many rhomboid sequences have amino acid residues with H-bonding capability at positions 134 and 137 (Tables S1-S3; see also (Koonin et al., 2003)). Y138 is conserved only in Enterobacteriaceae (∼52%) (Table S1).
Conformational flexibility of the cap loop L5
A key question about rhomboid protease function is how substrate access to the catalytic site is controlled. One scenario assigns a central role to the L5 cap loop (Figure 1C), which presumably opens upon docking of the substrate to the protease (Wang and Ha, 2007). On the timescale of our simulations, we observed two conformational transitions of L5 in Sim1 after ∼15ns and ∼27ns of unconstrained dynamics (Figure 4A). In Sim2, a conformational transition of L5 was observed after ∼30ns of unconstrained dynamics (Figure 4B). The conformational transitions of L5 occurred without any significant change of the distances between TM5 and TM6 (Figure 4D). That motions of L5 do not necessarily have to be accompanied by changes in the TM region is also suggested by the x-ray crystallographic study of (Wang and Ha, 2007). The differences in the dynamics of L5 in POPC and POPE lipids can be explained by L5 being located within the interfacial region (Figure 2D) of the membrane (Wang et al., 2007), where its dynamics can be affected by the lipid headgroups. Indeed, the dynamics of the D243:M247 H-bonding interactions (Figure 5B) depend significantly on the lipid environment (Figure 5F).
The observation of fast structural rearrangements of L5 is consistent with its relatively large rmsf values (Figure 4E), and with the conformational variability of L5 in the crystal structures. Due to the limited length of our simulations, we cannot exclude the possibility that the conformational transitions of L5 are reversible, or that further structural rearrangements of the loop might occur on a longer timescale.
Loop L1 is a key determinant of the protein orientation within the bilayer
The drastically perturbed proteolytic activity of some L1 mutants (Baker et al., 2007) and the lack of activity in the absence of L1 (Wang et al., 2007) indicate an important role for L1 in GlpG function. To gain insights into its mechanistic role, we performed an MD simulation (Sim3) of the L1 triple-Ser mutant Y138S/F139S/L143S (Figure 6A,B), which is inactive in proteolysis (Baker et al., 2007). The triple-Ser mutation affects the orientation of the protein within the lipid bilayer and the location of the catalytic Ser. On the time-scale of the present simulations, the average tilt of the protein relative to the membrane normal is 14.9 ± 2.1°, as compared to 11.7 ± 1.8° in the wild-type (Figure 7A), and the catalytic Ser is buried ∼2 Å deeper into the membrane. These results suggest that L1 participates in controlling the orientation of the protein within the lipid bilayer, which is important for the productive interaction with the substrate. A role for loop L1 in maintaining and adjusting the orientation of GlpG has also been suggested by Wang et al. (2007).
Figure 6.
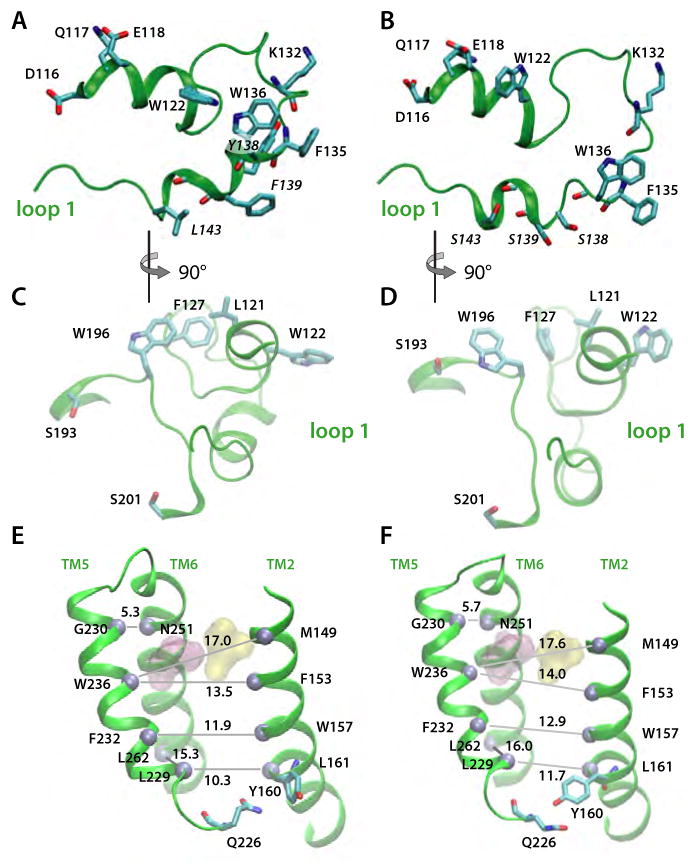
Simulation snapshots illustrating relay of Loop L1 perturbations to the substrate access site in the triple-Ser mutant (Y138S/F139S/L143S). Snapshots of the wild-type and mutant conformations are shown in the left-hand (A,C,E) and right-hand (B,D,F) panels, respectively. (A) Periplasmic view of L1 in the wild-type protein. All distances are in Å. (B) Periplasmic view of L1 in the triple-Ser mutant. (C). View of wild-type L1 viewed from the active site. site in the wild-type protein (D) View of triple-ser L1 from the active site. (E) Mean spacing between selected Cα atoms (gray spheres) in TM2/TM5/TM6 for the wild-type protein. (F) Mean Cα spacings for triple-Ser mutant. For panels E and F, the standard deviations of the distances are ≤ 0.6Å.
Perturbation of L1 is transmitted to the active site and TM2/TM5
The triple-Ser mutation of loop L1 causes structural rearrangements of L1 and, unexpectedly, the perturbation is transmitted to the active site and the TM2/TM5 helices (Figures 6 and 7). Optimization of the H-bonding of the three substituted Ser residues causes small structural rearrangements of L1 (Figure 6A,B) that in part minimize the energy penalty for burying polar hydroxyl groups within the membrane hydrophobic region. The lipid-mediated interaction between the L1 residues W122 and W136 weakens (Figure 7D), and W122 flips with its Nε1-H group towards the bulk water (Figure 6A,B). Perturbation of the hydrophobic cluster formed by L121, L127, and W196 and the repositioning of F127 relative to W196 (Figure 7E), allow W196 to reorient towards the substrate access-site where it H-bonds to TM3-S193 (Figure 6C-D, 7E). That is, perturbation of L1 leads in our simulation to the formation of a new H-bond seven residues downstream from the catalytic S201. Moreover, the mutation induces the formation of a H-bond between TM2-Y160 and TM5-Q225 (Figures 6E and 7E), perturbs the relative distance between TM2 and TM5 (Figure 6E-F), and affects the fluctuations of amino acids as far away as the TM5-L5 region (Figure 7B). Thus, the changes in L1 cause of a cascade of collectively important small-scale structural events that reach the substrate access site and TM2/TM5.
The changes in the structure, dynamics, and orientation of GlpG in the L1 triple-Ser mutant suggest a coupling between L1, the active site, and TM2/TM5. Because L1 interacts with TM3 (Figure 5A), which in turn is part of the two H-bonding clusters connecting the cytoplasmic termini of the helices (Figure 5A,B), structural and dynamical perturbations of L1 can be transmitted by TM3 to TM1, TM2, TM4, and TM6. The hydrophobic contacts between residues on the cytoplasmic sides of TM2 and TM5 (Figure 6E) (Ben-Shem et al., 2007; Wang et al., 2006; Wu et al., 2006) can further propagate the perturbation to TM5. Indeed, even in the early stages of Sim3, changes in the dynamics of interactions between L1 atoms (Figure 7D) were accompanied by changes in the interactions between L1 and groups from other structural elements (e.g., F127:W196, Figure 7E), and between protein groups located at distances remote from L1 (Y160:Q226, Figure 7E).
Perturbation of TM5 influences protein dynamics
The conformational variability of the TM5 helix observed in the crystal structures (Ben-Shem et al., 2007; Wang et al., 2006; White, 2006; Wu et al., 2006) and experiments on mutant phenotypes (Baker et al., 2007) were interpreted to suggest that TM5 functions as a dynamic lateral gate that controls substrate access to the active site. If TM5 is indeed the lateral access gate, it is unknown how its conformation and dynamics change upon interaction with substrates and whether this interaction leads to further changes in the rest of the protein. To begin to understand the effect of TM5 perturbations, we performed simulations (Sim4) of the TM5 triple-Val mutant (W236V/F232V/L229V) (Figure 8A,B). This mutant is of special interest, because it only involves amino acids of TM5, causes a 4-fold increase in activity relative to the wild-type (Baker et al., 2007), and has the potential to increase significantly the space between TM2 and TM5 (Figure 6E).
Figure 8.
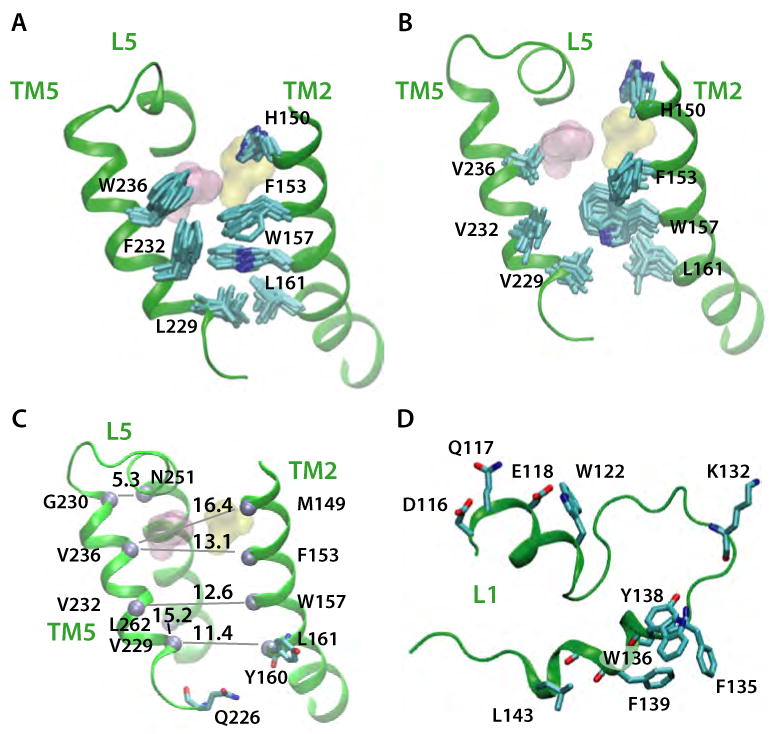
Simulation snapshots showing structural changes in TM5 of the triple-Val mutant in POPE (Sim4). Snapshots of wild-type and mutant conformations are shown in the left-hand (A,C) and right-hand (B,D) panels, respectively. (A) Loop L5, TM2, and TM5 in the wild-type protein (B) Same features as in for the triple-Val mutant (B). (C) Effect of the triple-Val mutant on the average distances between TM5, TM2, and TM6 Cα atoms (gray spheres). Compare with the distances in Figure 6E. All distances are average values (given in Å), with standard deviations ≤ 0.6 Å. (D) Snapshot of loop L1 at the end of Sim4. Compare with Figure 6A.
The structures of the TM segments did not change significantly in the triple-Val mutant (Figure 9A) but, due to the smaller size of the Val relative to the wild-type residues (Trp, Phe, and Leu), the accessibility of the catalytic Ser from the lateral side increased (Figure 8A,B). This change alone helps explain the enhanced activity of the triple-Val mutant. However, the dynamical changes in the protein go beyond just increasing access to the catalytic dyad. Relative to wild-type (Figure 8A), the mutation shifted the distributions of the protein orientation angles toward larger values (Figure 9B), perturbed the fluctuations of residues throughout the protein (Figure 9C), and led to structural rearrangements of L1 and L5 (Figure 9A,D). Importantly, the dynamics of intra-protein interactions that are affected by the triple-ser mutation of L1 are also perturbed in the triple-val mutant (e.g., F127-W196, W122-W136; Figure 7D,E and Figure 9D). This observation supports our suggestion of a communication between remote regions of GlpG (Figure 9E).
The long-distance effects of mutating L1 or TM5 demonstrated here suggest long-range coupling in GlpG, and help explain why GlpG's enzymatic activity is drastically perturbed when residues remote from the active are mutated. Long-range coupling has also been proposed, for example, for V. cholerae RTX cysteine protease (Lupardus et al., 2008), and for the E. coli dihydrofolate reductase (Rod et al., 2003). Further computations and experiments are necessary to understand fully why catalytic activity is significantly reduced in triple-Ser, but increased in triple-Val. One potentially important difference between the two mutants is that, although in both the interactions between W193, S193, and F127 are perturbed (Figures 7E, 9D), only the (inactive) triple-Ser mutant appears to be locked in a conformation that strengthens the interactions between these groups.
Systematic computer simulation studies of additional GlpG mutants are necessary to understand why perturbing certain interactions affects the catalytic activity, and which steps in GlpG's mechanism of action are affected by specific classes of mutations. For example, it is unclear why the double mutation W236A/F153A results in a 10-fold increase of the activity, while the triple-Val mutant has a more modest 4-fold increase of activity relative to the wild type (Baker et al., 2007).
Discussion
We have carried out MD simulations of the wild-type E. coli GlpG rhomboid protease in POPC and POPE and two GlpG mutants in POPE. The structure of the TM helices of GlpG in bilayers is close to the crystallographic structures (Figure 4A,B). The TM5 helix and the L5 loop, which have the greatest structural variability among the three GlpG structures (White, 2006), are mobile regions of GlpG in lipid bilayers (Figures 4A,B,E). The S201/H254 catalytic dyad is positioned deep in the lipid bilayer, where it nevertheless remains well hydrated at all times (Figures 1B, 4C). Differences in the H-bonding of the lipid headgroups with the protein affect the orientation (Figure 2A) and dynamics (Figures 4A,B,E) of the protein. The orientation of GlpG within the lipid bilayer may be an essential determinant of enzyme efficiency, because orientation can influence the likelihood of a productive interaction between the catalytic dyad and the substrate cleavage site. Our simulations of the triple-Ser and triple-Val mutations reveal subtle, but important changes in protein conformation and dynamics that are relayed between L1 and TM5 (Figures 6-9).
Optimization of the interactions of the protein with the lipid bilayer is mainly achieved by structural rearrangement of lipid molecules. These rearrangements lead to changes in the hydrophobic thickness of the bilayer in the vicinity of the protein due to H-bonding between surface residues and lipid headgroups (Figure 2 B,C,D). This localized bilayer thinning close to the protein may influence the conformation (Wang et al., 2007) or orientation of the substrate, which by itself does not perturb of the lipid bilayer (Figure 2E). Based on their estimate of 30% bilayer thinning, Wang et al. (2007) suggested that hydrophobic mismatch between the substrate and the bilayer around GlpG could force an unwinding of the cleavage site. When measured as the distances between the peaks in the average distributions of glycerol moieties in the two leaflets, the 4-5 Å thinning of the lipid bilayer indicated by our simulations (Figures 2B,C) could be interpreted to suggest that only the top 3 residues of the substrate would unwind. However, the lipid bilayer might undergo further perturbations when the substrate and GlpG are within a close distance, and the thinning of the lipid bilayer around GlpG might induce tilting of the substrate in addition to unwinding.
Due to the different H-bonding capabilities of the POPC and POPE headgroups, the details of the protein-lipid interactions naturally depend on the lipid headgroup. This effect of the headgroup H-bonding capability is most prominent in the case of the L1 loop, where H-bond bridging between E134, R137, and a lipid molecule forms a barrier impenetrable to water molecules in POPE, but not in POPC (Figures 3E-G). The dependence of the conformation of L1 on protein interactions with the lipid bilayer could allow L1 to act as a sensor of the GlpG environment. This sensitivity, together with the perturbing effect of L1 mutations on the protein orientation and dynamics, likely contributes to the effect of lipids on GlpG's proteolytic activity (Urban and Wolfe, 2005), and helps rationalize why the effect of certain L1 mutations (W136A, R137A) depends on the whether the mutant is assessed in detergent or in various lipid environments (Lemberg et al., 2005; Maegawa et al., 2005; Urban, 2001; Wang et al., 2007). Further studies are required for a detailed understanding of how intramembrane proteases are affected by their environment. The functional role of H-bonding between lipids and surface protein groups has been proved recently for the secondary multidrug transporter LmrP, whose function requires H-bonding between a protein aspartate residue and the lipid membrane (Hakizimana et al., 2008).
Conformational transitions of L5 were observed both in the wild-type (Figure 4A-B) and in the TM5 triple-Val mutant (Figure 9A). Our observation of a conformational change of L5 in the absence of substrate (using the coordinates of GlpG in its closed conformation (Ben-Shem et al., 2007)), suggest that opening and closing of the loop is an intrinsic protein motion. Although the timescale of our simulations is too short to assess whether the cap loop closes back and then re-opens, or whether further conformational changes of L5 occur on longer timescales, the significant flexibility of the cap loop indicated by experiments and our simulations supports such motions. If the optimal docking of the substrate in the active site indeed requires an opening of L5 (Wang et al., 2007), the intrinsic flexibility of L5 may allow sampling of conformations compatible with substrate docking, even in the absence of substrate. This suggestion is inspired by the observation of the equilibrium, in the absence of the substrate, between the open and closed states of the lid domain controlling the access to the adenylate kinase active (Hanson et al., 2007). However, the GlpG structure used here, which has a lipid headgroup bound to the active site (Ben-Shem et al., 2007), is of a closed-state enzyme. The dynamics of L5 and TM5 in the more open conformation of GlpG (Wu et al., 2006) may be different than observed here for the closed state (Ben-Shem et al., 2007).
Perturbations of L1 induced by mutation or changes in the interaction with the lipid membrane are transmitted to the catalytic site, and can influence protein orientation and the location of the catalytic site (Figures 6 and 7). Furthermore, perturbation of TM5 affects the local structure and fluctuations of L1 (Figures 8C-D, 9A,C,D) as well as the orientation of the protein (Figure 9B). Clusters of H-bonds involving amino acids with significant conservation (TM2-E166 and TM3-K173) may be essential for the structure and dynamics of the protein, and may assist the propagation of perturbations throughout the protein. This transmission of structural and dynamic changes between remote regions of the protein suggests an important role of L1 in regulating GlpG. It is possible that coupling between remote structural elements is essential for GlpG enzymatic activity (Figure 9E).
Experimental procedures
We used molecule B of the Ben-Shem et al. (2007) structure (PDB code 2IRV) for the starting coordinates. Amino-acid residues R92, A272, and R273 were constructed with the CHARMM software (Brooks et al., 1983). Missing hydrogen atoms were constructed using the HBUILD facility of CHARMM. All titratable amino-acid residues were considered in their standard protonation states. The tautomeric state of the four His residues in the TM domain can be influenced by their local electrostatic environment (Baran et al., 23412). We tested the effect of the tautomeric state of H145 and H254 on GlpG dynamics and lipid interactions by comparing ∼10ns long test simulations with the two His residues in the either NE1 or in the ND1 tautomeric state. No significant effect of the tautomeric state could be observed. In the simulations discussed here, H145 and H254 are in the ND1 tautomeric state, and H141 and H150 are NE1. The distance between N154-ND2 and H145-NE2 is 3.2 Å in the crystal structure, and 3.3 ± 0.3 Å, 3.2 ± 0.3 Å, 3.1 ± 0.3 Å, and 3.3 ± 0.4 Å in Sim1, Sim2, Sim3 and Sim4, respectively.
The protein was aligned with its three principal axes along the x, y, z directions using the VMD software (Humphrey et al., 1996) and embedded in a solvated lipid bilayer so that its center of mass coincided with center of the bilayer coordinates. Two independent sets of MD simulations of wild-type GlpG were performed with GlpG embedded in bilayers of 511 POPC (Sim1) and 531 POPE lipid molecules (Sim2), respectively. The coordinates of the phosphatidylglycerol headgroup of 1-palmitoyl-2-vaccenoyl-sn-glycerol-3-phosphatidylglycerol (PGV) close to the active-site in the crystal structure (Ben-Shem et al., 2007) were used to construct a PGV lipid molecule in Sim1, and POPE in Sim2. We included 30077 and 30983 water molecules in Sim1 and Sim2, respectively. Chloride ions (1 in Sim1; 2 in Sim2) were added for charge neutralization. Non-bonded and electrostatic interactions were calculated every 2 and 4 time-steps, respectively, with a cut-off of 11 Å. The SHAKE algorithm (Ryckaert et al., 1977) was used to constrain the lengths of bonds involving hydrogen atoms. Coulomb interactions were computed using the particle mesh Ewald summation (Darden et al., 1993). Temperature was kept constant using Langevin dynamics; a Nose-Hoover Langevin piston (Feller et al., 1995; Martyna et al., 1994) was employed for pressure control (NPT, 1 bar and 300K). MD simulations were performed using NAMD (Kalé et al., 1999; Phillips et al., 2005) with the CHARMM22 protein force field (MacKerell et al., 1998) and CHARMM27 lipid force field (Feller and MacKerell, 2000). Waters were modeled using TIP3P (Jorgensen et al., 1983). An integration step of 1 fs was used during the first ∼2 ns of the simulation. After that, we used the reversible multiple time-step algorithm (Tuckerman and Berne, 1992) to integrate the equations of motion with time-steps of 1 fs for the bonded forces, 2 fs for the short-range non-bonded forces, and 4 fs for the long-range electrostatic forces.
During minimization, heating, and the first ∼1 ns of equilibration, we used harmonic constraints of 5 kcal mol-1 for the protein atoms and 2 kcal mol-1 for ions, solvent water molecules, and lipid molecules further than 15 Å from protein atoms. During the subsequent ∼2 ns of equilibration, we used constraints of 2 kcal mol-1 for all groups. All harmonic constraints were then switched off.
The triple-Ser (Y138S/F139S/L143S) and triple-Val (L229V/F232V/W236V) mutants were modeled using a set of coordinates of wild-type GlpG equilibrated in POPE (Sim2) for about 10 ns. Sim3 and Sim4 were then performed according to the protocol described above for the wild type. A test ∼22ns simulation on the Spitz GlpG substrate (Sim5) was performed by embedding the 23-residue TM region of Spitz in the center of a solvated lipid bilayer consisting of 280 POPC lipid molecules and 12140 solvent water molecules. Sim1 and the Spitz-containing POPC bilayer were equilibrated at 300K; Sim2, Sim3, and Sim4 at 310K.
Supplementary Material
Acknowledgments
We thank Prof. Sin Urban, Dr. J. Alfredo Freites and Prof. Douglas J. Tobias for valuable discussions; Profs. Charlie Zender, Steve Frank, and Robin Bush for their generous contribution of time on their clusters; and Mr. Joseph Farran of the UC Irvine MPC Cluster for excellent technical support. Research supported in part by the National Institute of General Medical Sciences (GM-74637 to SHW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci USA. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran KL, Chimenti MS, Schlessman JL, Fitch CA, Herbst KJ, Garcia-Moreno BE. Electrostatic effects in a network of polar and ionizable groups in Staphylococcal nuclease. J Mol Biol. 2008;379:1045–1062. doi: 10.1016/j.jmb.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN. rhomboid, a gene required for dosoventral axis establishment and peripheral nervous system develompent in Drosophila melanogaster. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics. J Comput Chem. 1983;4:187–217. [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Clemmer KM, Sturgill GM, Veenstra A, Rather PN. Functional characterization of Escherichia Coli GlpG and additional rhomboid proteins using an aarA mutant of Providencia stuartii. J Bacteriol. 2006;188:3415–3419. doi: 10.1128/JB.188.9.3415-3419.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M, Kimmel BE, Rubin GM. Identifying targets of the rough homeobox gene of Drosophila: Evidence that rhomboid functions in eye development. Development. 1992;116:335–346. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- Hakizimana P, Masureel M, Gbaguidi B, Ruysschaert JM, Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP a multidrug transporter. J Biol Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- Hanson JA, Duderstadt K, Watkins LP, Bhattacharyya S, Brokaw J, Chu J, Yang H. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc Natl Acad Sci USA. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jensen MØ, Mouritsen OG. Lipids do influence protein function: The hydrophobic matching hypothesis revisited. Biochim Biophys Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K. Lipds as modulators of proteolytic activity of BACE. J Biol Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: A nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Ito K, Akiyama Y. Proteolytic action of G1pG, a rhomboid protease in the escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol Microbiol. 2007;64:435–447. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun B, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rod TH, Radkiewicz JL, Brooks CL. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc Natl Acad Sci USA. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access to notch in drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Tuckerman M, Berne BJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–183. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ha Y. Open-cap conformation of intramembrane protease GIpG. Proc Natl Acad Sci USA. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomoboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104–1113. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to active EGF receptor signaling. Genes Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- White SH. Rhomboid intramembrane protease structures galore! Nature Struct Mol Biol. 2006;13:1049–1051. doi: 10.1038/nsmb1206-1049. [DOI] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R. Intramembrane proteolysis: Theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nature Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.