Abstract
Clostridium difficile is the leading cause of infectious diarrhea in hospitals worldwide, because of its virulence, spore-forming ability and persistence1,2. C. difficile-associated diseases (CDAD) are induced by antibiotic treatment or disruption of the normal gastrointestinal flora3,4. Recently, morbidity and mortality resulting from CDAD have increased significantly due to changes in the virulence of the causative strains and antibiotic usage patterns1,2,5,6. Since 2002, epidemic toxinotype III NAP1/027 strains1,2, which produce high levels of the major virulence factors, toxin A and toxin B, have emerged. These toxins have 63% amino acid sequence similarity7 and are members of the large clostridial glucosylating toxin family, which are monoglucosyltransferases that are proinflammatory, cytotoxic and enterotoxic in the human colon8–10. Inside host cells, both toxins catalyze the transfer of glucose onto the Rho family of GTPases, leading to cell death8, 11. However, the role of these toxins in the context of a C. difficile infection is unknown. Here we describe the construction of isogenic tcdA and tcdB mutants of a virulent C. difficile strain and their use in the hamster disease model to show that toxin B is a key virulence determinant. Previous studies showed that purified toxin A alone can induce most of the pathology observed following infection of hamsters with C. difficile8,9, 12 and that toxin B is not toxic in animals unless it is co-administered with toxin A, suggesting that the toxins act synergistically12. Our work provides evidence that toxin B, not toxin A, is essential for virulence, which represents a major paradigm shift. Furthermore, it is clear that the importance of these toxins in the context of infection cannot be predicted exclusively from studies using purified toxins, reinforcing the importance of using the natural infection process to dissect the role of toxins in disease.
C. difficile is difficult to genetically manipulate; reproducible methods for the construction of chromosomal mutants have only recently become available13,14. We have developed a method that involves a plasmid, designated the recombination vector, which is unstable in C. difficile; this plasmid carries DNA homologous to part of the genome and becomes inserted at that genomic location, resulting in a specific gene disruption. Using this process, we have successfully mutated two regulatory genes13 and others have generated mutations in additional genes15. This methodology has now made possible studies aimed at dissecting the role of toxin A and toxin B in CDAD by constructing and analyzing isogenic strains of this bacterium.
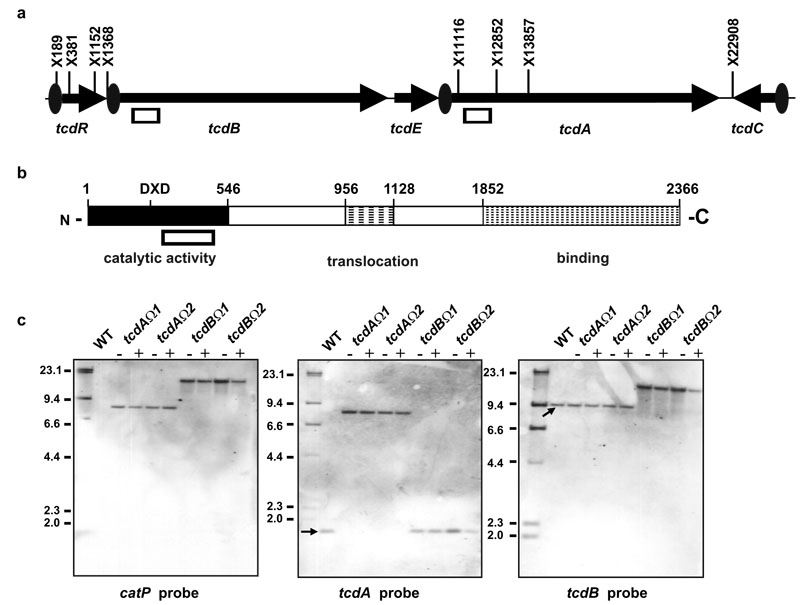
Toxin A and toxin B are encoded by the tcdA and tcdB genes, respectively, located within a 19.6kb pathogenicity locus, the PaLoc9 (Fig. 1a), which encodes three other genes involved in toxin production16,17. To construct the plasmids required for the targeted inactivation of tcdA and tcdB, 566 bp tcdA- or tcdB-specific PCR products were cloned into pJIR1456. These PCR products corresponded to an essential region of the N-terminal domain between the DXD catalytic motif and amino acid 546 (Fig. 1b)18. Insertion of these plasmids into the corresponding chromosomal gene region by homologous recombination should lead to disruption of the gene, resulting in a truncation of the toxins (at leucine residues 492 and 493 in toxins A and B, respectively), thereby attenuating toxin function.
Fig. 1.
Genetic organization of the C. difficile PaLoc and analysis of toxin mutants. (a) PaLoc genetic organization. XbaI sites (X) are shown, with the PaLoc co-ordinate indicated. Boxes indicate regions used in the recombination vectors and the tcdB- and tcdA-specific probes. Ovals show the gene-specific promoters and arrows the direction of transcription. (b) Structural organization of toxin B (toxin A is similar). Co-ordinates indicate the amino acid number. Relevant domains are shown18. The box indicates the region in the recombination vectors. (c) Southern hybridization of mutants. Genomic DNA was XbaI-digested and blots probed as shown. λ-HindIII markers are indicated (kb). WT represents strain JIR8094, the mutant lanes are indicated. DNA was purified from cultures grown in the absence (−) or presence (+) of thiamphenicol selection.
The tcdA- and tcdB-specific recombination vectors were transferred to JIR809413, a derivative of C. difficile strain 630, by RP4-mediated conjugative transfer from Escherichia coli. Two putative independently derived toxin A (tcdAΩ1 and tcdAΩ2) and five putative independently derived toxin B (including tcdBΩ1 and tcdBΩ2) mutants were isolated. Note that since each of these mutants was derived from a separate conjugation experiment, the mutants are truly independent. Their genotypes were confirmed by PCR analysis (data not shown) and Southern blotting (Fig. 1c). The insertion of the recombination vectors was shown to be specific and to have disrupted the respective tcdA and tcdB genes, with the wild-type bands replaced by bands corresponding to insertion of the recombination vectors. For each specific mutant the other toxin gene had a profile identical to wild-type. Even though the insertions were generated by single crossovers, and were therefore reversible13, the resultant chromosomal mutants were very stable; no evidence of deletion of the inserted plasmids was detected in the Southern blots (Fig. 1c). Autonomously replicating plasmids could not be extracted from these strains, suggesting that once chromosomal insertion had occurred, the plasmid was maintained in an inserted state, as previously found13.
Western blots using toxin A-specific antibodies showed that the tcdA mutants no longer produced toxin A and had a similar profile to an avirulent strain that does not carry the PaLoc (Fig. 2a). By comparison, the tcdB mutants produced toxin A, although they reproducibly appeared to make more toxin A than the wild-type. This finding was confirmed by quantitative real time reverse transcriptase PCR (QRT-PCR), which showed that the tcdB mutants expressed tcdA at 2.2–3.3-fold higher levels than the wild-type. The basis for this increased expression of toxin A is not known. QRT-PCR showed that no differences in expression of the other PaLoc genes (tcdR, tcdE, and tcdC) were observed in any of the mutants (data not shown).
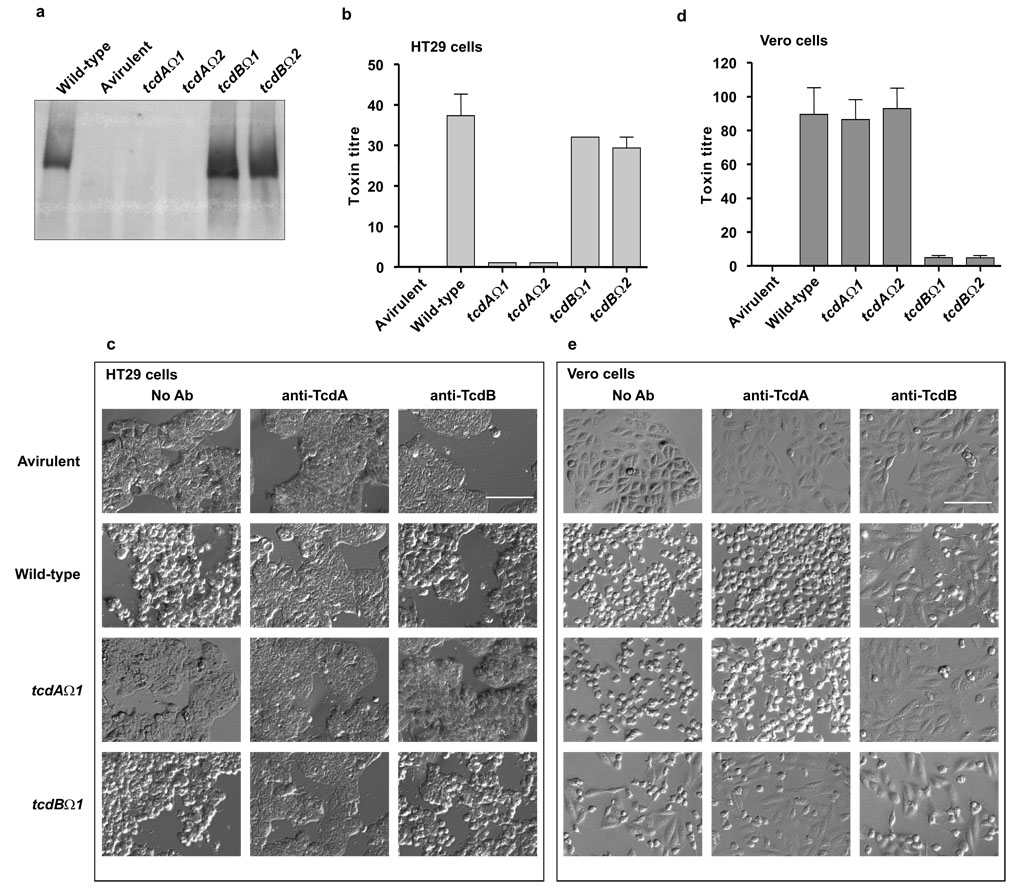
Fig. 2.
Comparative analysis of toxin production by wild-type and mutants. The wild-type is JIR8094 and the avirulent strain is CD37, which does not produce toxins29. (a) Western blot using toxin A-specific antibodies. (b) and (d) Toxin A and toxin B cytotoxicity assays using HT-29 and Vero cells, respectively. Data represent the mean±SEM. n=3. (c) and (e) Neutralization of toxicity. Culture supernatants were added to the cells and morphological changes observed after 24 h (No Ab) or pretreated with toxin A (anti-TcdA) and toxin B (anti-TcdB) neutralizing antibodies prior to their addition to the cells. Representative images are shown. No neutralization was observed when supernatants were pretreated with anti-ErmB methylase antibodies (data not shown). Bar indicates 1 mm.
Cytotoxicity assays using HT29 cells, which are more sensitive to toxin A than other cell lines19,20 showed that supernatants from the toxin A mutants were significantly less toxic (p=0.021) than supernatants from the wild-type (Fig. 2b). These assays also showed that toxin A produced by the wild-type strain and by the tcdB mutants was cytotoxic and therefore functional (Fig. 2b). The latter activity was neutralised by toxin A-specific antibodies (Fig. 2c), verifying that the observed toxicity resulted from the action of toxin A. Vero cell cytotoxicity assays were also carried out on these supernatants, the cytotoxic activity observed is predominantly toxin B-mediated8,19. Consequently, the tcdA mutants exhibited the same cytotoxic phenotype as the wild-type strain (Fig. 2d). These cytopathic effects were neutralized by toxin B-specific antibodies, confirming that toxin B was responsible for the observed Vero cell toxicity (Fig. 2e). By contrast, supernatants from the tcdB mutants were significantly less cytotoxic (p=0.0058) for Vero cells than wild-type supernatants. These mutants exhibited a similar phenotype to the avirulent strain although they did have low levels of toxicity (Fig. 2d), consistent with the relatively lower cytopathic effects of toxin A on Vero cells. This low level of cytotoxicity was not neutralized by preincubation with toxin B antibodies (Fig. 2e).
All of the genetic and phenotypic assays used to analyze the mutant strains therefore showed that each pair of mutants, although isolated independently, behaved identically. Attempts were made to complement these mutations with the wild-type genes, but these experiments were not successful. The intact genes are very difficult to clone into shuttle vectors because they are very large (8.1- and 7.1-kb for tcdA and tcdB, respectively) and contain repeat regions21,22. For this reason, in subsequent virulence experiments we used two independently isolated mutants for each gene, to rule out the possibility that any observed phenotypic effects were the result of other mutations.
The hamster disease model parallels most of the recognized features of human CDAD, especially with respect to the susceptibility of the animal to C. difficile infection following the administration of antimicrobial agents. Since it is currently the best animal model available23 it was used to compare the virulence of the C. difficile mutants.
Upon challenge with the wild-type, all ten infected hamsters were colonized approximately two days after inoculation and 9/10 animals died approximately five days after colonization (Fig. 3). The two independent toxin A mutants, which produced wild-type levels of toxin B in vitro, yielded similar results, with 9/10 and 8/10 hamsters being colonized following challenge with the tcdAΩ1 and tcdAΩ2 mutants, respectively. The mean number of days (+SD) to colonization was similar following inoculation with wild-type (2.1+1.2), tcdAΩ1 (1.9+0.3) and tcdAΩ2 (1.9+1.1) strains. These results matched in vitro studies that investigated the adherence of these strains to polarized Caco-2 human intestinal epithelial cells under anaerobic conditions, where no significant differences in adherence were observed (data not shown).
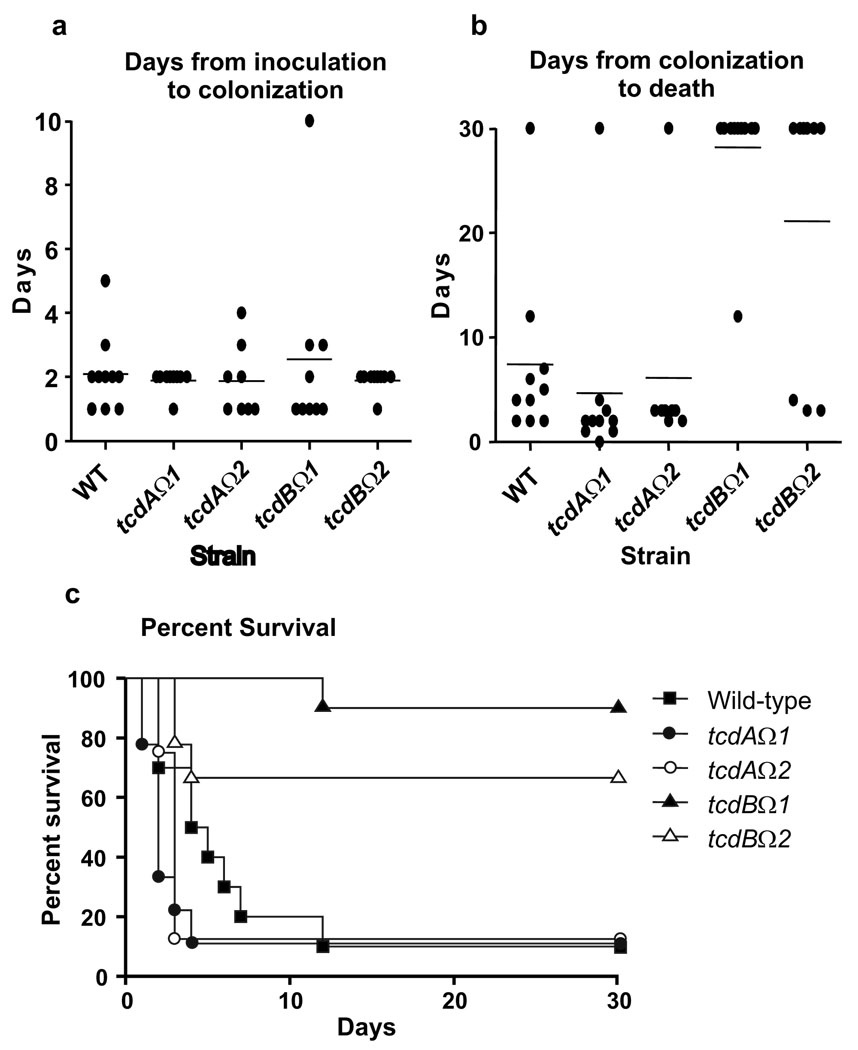
Fig. 3.
Virulence of C. difficile wild-type and mutant strains in hamsters. (a) Days from inoculation of C. difficile to colonization of clindamycin treated hamsters. The rate of colonization was 10/10 for WT, 9/10 for tcdAΩ1, 8/10 for tcdAΩ2, 9/10 for tcdBΩ1 and 9/10 for tcdBΩ2. n=10. (b) Days from colonization to death or sacrifice. All hamsters that survived were sacrificed at day 30. n=10. (c) Kaplan-Meier survival estimate demonstrating days from colonization with C. difficile strains to death. n=10.
The mean time from colonization to death was similar for hamsters colonized with the wild-type (9/10) and the (tcdAΩ1 (9/9) and tcdAΩ2 (7/8) mutants (Fig.3). Fecal pellets from hamsters infected with the tcdAΩ2 mutant were analyzed for plasmid stability, toxin analysis was also performed on these specimens. All of the colonies (n=800) recovered from the eight tcdAΩ2 colonized hamsters were thiamphenicol resistant, confirming that the inserted recombination vector was retained in these isolates. Seven of the eight colonized hamsters died (Fig. 3c), the one surviving animal lost colonization after only two days. Confirmatory PCRs, with primers specific for the gene disruption, were performed on a minimum of four post-colonization isolates (from different hamsters) for each group of animals; the results showed that they all carried an insertionally inactivated tcdA gene.
All of the fecal pellets isolated from tcdAΩ2-infected hamsters were strongly cytotoxic (using a cytotoxin assay that primarily measures toxin B), indicating the presence of active toxin B, and were negative for toxin A in an enzyme immunoassay, as expected. Overall, 16/17 (94%) of the hamsters colonized with the toxin A mutants died, providing clear evidence that the tcdA mutants were still virulent even though they no longer produced toxin A, either in vitro or in vivo. These data provide clear evidence that toxin A is not essential for virulence in the hamster model, at least for a derivative of strain 630.
By contrast, the virulence trials involving the toxin B mutants demonstrated that toxin B was required for virulence (Fig. 3). For each of the tcdB mutants tested, 9/10 hamsters were colonized after challenge. The time from challenge to colonization (tcdBΩ1 (2.6+2.9); tcdBΩ2 (1.9+0.3)) was similar to that observed for the wild-type and the toxin A mutants. However, only 4/19 of the hamsters (21%) colonized with the toxin B mutants died (Fig. 3c), which was statistically different (P=0.001) from the wild-type strain (10/10 deaths). The hamster that died following challenge with the tcdBΩ1 mutant died 12 days after colonization although the interval from colonization to death in the three hamsters that died following challenge with the tcdBΩ2 mutant was similar to that of the wild-type. Plasmid stability analysis and toxin testing of fecal pellets from the tcdBΩ2 trial explained these results. All of the colonies (n=600) recovered from the six surviving hamsters were thiamphenicol resistant, indicating that the integrated recombination vector was carried by these strains. Fecal pellets from 5/6 surviving hamsters were EIA positive for toxin A and were either cytotoxin negative (n=2) or weakly cytotoxic (n=4), consistent with the absence of toxin B and the relatively lower cytotoxicity of toxin A. These results provided evidence that the isolates recovered from the surviving hamsters were no longer virulent, even though they continued to produce toxin A in vivo. Of the three hamsters that died, colonies from two animals (n=200) were all thiamphenicol resistant. However, only 74% of the isolates from the third hamster were thiamphenicol resistant. Restriction endonuclease analysis (REA) typing of two of the thiamphenicol sensitive isolates confirmed that they were derived from the wild-type strain, suggesting that they represented revertants that had lost the integrated plasmid and therefore displayed a wild-type (toxin A+B+) phenotype. In addition, fecal pellets from all three hamsters that died were strongly cytotoxic (toxin B positive) and toxin A positive by EIA. We conclude that the thiamphenicol sensitive isolates were revertants and were able to produce toxin B, reinforcing the requirement for toxin B in the virulence of C. difficile. We suggest that the two hamsters that carried thiamphenicol resistant strains, but whose fecal isolates were toxin A and B positive, had mixed infections of the mutant strain and a revertant strain capable of producing wild-type toxin B, at a level that was not detected by our analysis.
In summary, hamsters challenged with the toxin A mutants were as likely to die as those infected with the wild-type strain, even though these strains produce more toxin A than the wild-type (Fig. 2a), and animals challenged with the toxin B mutants were much more likely to survive (Fig. 3c). Among the few hamsters that died following challenge with the toxin B mutants, our analysis suggested that a proportion of the C. difficile population infecting those animals produced toxin B. These results support the conclusion that the production of toxin B, not toxin A, correlates with a virulent C. difficile phenotype.
In conclusion, in this study, four independently derived toxin A or toxin B mutants of C. difficile were constructed and characterized in vitro and in vivo using the hamster CDAD model. The results demonstrated that toxin B is an essential virulence factor since disruption of the tcdB gene led to a significantly attenuated virulence phenotype. Isolates that produced toxin B but not toxin A retained a wild-type virulence phenotype, which provides further confirmation of the paramount role of toxin B in disease pathogenesis. Unexpectedly, the presence of toxin A in the absence of toxin B, even at levels higher than in the wild-type strain, was not lethal in the hamster model. Although this finding ostensibly contradicts a study which showed that antibodies to toxin A protect toxoid A-immunised hamsters from disease24, vaccination studies do not provide direct evidence of the role of a bacterial component in disease. Therefore, it is concluded that toxin A is not an essential virulence determinant in a derivative of the virulent C. difficile strain 630, although it may still play a non-essential role in the disease process.
Some previous studies have suggested that toxin A may not be the major virulence factor of C. difficile and that toxin B might not be dependent on toxin A for function23,25, however, a lack of genetic tools did not allow these hypotheses to be tested. Other studies suggested that toxin B was unable to initiate disease unless toxin A was present9,12. The data generated from our study do not support the hypothesis that both toxin A and toxin B are required to cause disease. Clinical variants expressing toxin B but not toxin A are being isolated with increasing frequency26 and disease resulting from these strains is similar to that following infection with strains producing both toxins27. The observation that clinical isolates that lack toxin A are still capable of causing severe disease reinforces the proposition that toxin A is not essential for virulence and that toxin B does not require the presence of toxin A for its in vivo activity9,23,26,27. Note that naturally occurring toxin A-positive, toxin B-negative isolates of C. difficile have not been reported. Earlier studies on toxin A and toxin B function and their putative cooperativity involved purified toxins or culture supernatants9,12. The disparity between these studies and those described here accentuates the importance of using the natural infection process to dissect the role of toxins in disease pathogenesis and reinforces the critical need for the use of isogenic strains in such studies.
The clinical diagnosis of C. difficile predominantly involved immunoassays for the detection of toxin A until the discovery of clinical disease caused by toxin A−B+ isolates that were not detected by these assays. It is now evident from our data that in the absence of culturing for the organism, the diagnostic emphasis should focus on the detection of both toxins by EIA and/or a standard cytotoxin assay that measures primarily toxin B26, especially with toxin A-deficient strains being isolated with increasing frequency. Rapid and accurate diagnosis is of critical importance because of the changing epidemiology and increased virulence of C. difficile strains1,2, and because this organism is now causing disease in individuals previously thought not to be at risk, such as children and pregnant women28. In summary, it is clear that our findings represents a paradigm shift in our understanding of the pathogenesis of C. difficile-mediated disease and will have major implications for the diagnosis, control and treatment of these syndromes.
METHODS SUMMARY
The toxin A- and toxin B-specific recombination vectors pJIR3051 and pJIR3050 were constructed by cloning a 566 bp tcdA (accession M30307) PCR product (nt 1071–1637) and the equivalent tcdB (accession X53138) region, respectively, into the shuttle vector pJIR1456 (accession U90554). These plasmids were transferred by conjugation to C. difficile strain JIR8094 and the resultant chromosomal mutants analyzed by PCR, Southern blotting and plasmid rescue13.
Filtered supernatants of 2TY broth cultures (72 h) were used for Western blots, cytotoxicity and toxin neutralization assays. Serial two-fold dilutions of these supernatants were used in Vero and HT-29 cell cytotoxicity assays, with the end-point scored as the last dilution yielding complete CPE. Toxins were neutralized with affinity purified polyclonal antibodies specific for toxin A and toxin B; neutralization was carried out at the last dilution giving complete CPE. Partially purified ammonium sulphate-precipitated toxin proteins were Western blotted using toxin A-specific affinity purified polyclonal goat antibodies.
Syrian hamsters (n=10) were infected with C. difficile spores for virulence testing23. Clindamycin was administered orogastrically to render the hamsters susceptible to infection (day zero). On day five hamsters received 1000 cfu of spores by gastric inoculation. Fecal pellets were collected daily for 12 days, then weekly until day 30 and inoculated onto selective TCCFA plates to monitor colonization. Colonies were typed by REA to confirm the identity of the infecting isolate. Fecal pellets were analyzed for the presence of toxin A using a toxin A-specific ELISA and for the presence of toxin B using a human foreskin fibroblast cytotoxicity assay.
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.
Acknowledgements
Research at Monash University was supported by Program Grant 284214 from the Australian National Health and Medical Research Council, funding from the ARC Centre of Excellence in Structural and Functional Microbial Genomics and grant AI057637 from the United States National Institute of Allergy and Infectious Diseases. S.J., D.N.G. and G.V. were supported by Merit Review Grants from the United States Department of Veterans Affairs Research Service. We thank D. Lyerly (TechLab), K. Aktories and C. von-Eichel Streiber for kindly providing toxin A-specific and toxin B-specific antibodies, K. Nagaro and A. Cheknis for assistance with the hamster experiments, V. K. Viswanathan for providing intestinal epithelial cell lines, E. Hartland for providing the HT29 cell line and M. Merrigan for adherence assays.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.McDonald LC, et al. An epidemic, toxin gene–variant strain of Clostridium difficile. N. Engl. J. Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 2.Warny M, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett JG, et al. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology. 1978;75:778–782. [PubMed] [Google Scholar]
- 4.Bartlett JG. Antibiotic-associated diarrhea. N. Engl. J. Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 5.Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg. Infect. Dis. 2007;13:1417–1419. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O′Brien JA, Lahue BJ, Caro JJ, Davidson DM. The emerging infectious challenge of Clostridium difficile–associated disease in Massachusetts hospitals: clinical and economic consequences. Infect. Control Hosp. Epidemiol. 2007;28:1219–1227. doi: 10.1086/522676. [DOI] [PubMed] [Google Scholar]
- 7.von Eichel-Streiber C, et al. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol. Gen. Genet. 1992;233:260–268. doi: 10.1007/BF00587587. [DOI] [PubMed] [Google Scholar]
- 8.Just I, Gerhard R. Large clostridial cytotoxins. Rev. Physiol. Biochem. Pharmacol. 2004;152:23–47. doi: 10.1007/s10254-004-0033-5. [DOI] [PubMed] [Google Scholar]
- 9.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 2005;18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 2008;16:222–229. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Just I, et al. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 12.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect. Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O′Connor JR, et al. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 2006;61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 14.Heap JT, et al. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods. 2007;70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Dineen S, Villapakkam AC, Nordman JT, Sonenshein AL. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 2007;66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 16.Mani N, et al. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 2002;184:5971–5978. doi: 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 2007;64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 18.Schirmer J, Aktories K. Large clostridial cytotoxins: cellular biology of Rho/Ras-glucosylating toxins. Biochim. Biophys. Acta. 2004;1673:66–74. doi: 10.1016/j.bbagen.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Torres J, Camorlinga-Ponce M, Munoz O. Sensitivity in culture of epithelial cells from rhesus monkey kidney and human colon carcinoma to toxins A and B from Clostridium difficile. Toxicon. 1992;30:419–426. doi: 10.1016/0041-0101(92)90538-g. [DOI] [PubMed] [Google Scholar]
- 20.Giesemann T, et al. Cholesterol-dependent pore formation of Clostridium difficile toxin A. J. Biol. Chem. 2006;281:10808–10815. doi: 10.1074/jbc.M512720200. [DOI] [PubMed] [Google Scholar]
- 21.Burger S, et al. Expression of recombinant Clostridium difficile toxin A using the Bacillus megaterium system. Biochem. Biophys. Res. Commun. 2003;307:584–588. doi: 10.1016/s0006-291x(03)01234-8. [DOI] [PubMed] [Google Scholar]
- 22.Tang-Feldman YJ, et al. One-step cloning and expression of Clostridium difficile toxin B gene (tcdB) Mol. Cell. Probes. 2002;16:179–183. doi: 10.1006/mcpr.2002.0409. [DOI] [PubMed] [Google Scholar]
- 23.Sambol SP, et al. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J. Infect. Dis. 2001;183:1760–1766. doi: 10.1086/320736. [DOI] [PubMed] [Google Scholar]
- 24.Kim PH, Iaconis JP, Rolfe RD. Immunization of adult hamsters against Clostridium difficile-associated ileocecitis and transfer of protection to infant hamsters. Infect. Immun. 1987;55:2984–2992. doi: 10.1128/iai.55.12.2984-2992.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du T, Alfa MJ. Translocation of Clostridium difficile toxin B across polarized Caco-2 cell monolayers is enhanced by toxin A. Can. J. Infect. Dis. 2004;15:83–88. doi: 10.1155/2004/292580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. Int. J. Infect. Dis. 2007;11:5–10. doi: 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Johnson S, et al. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann. Intern. Med. 2001;135:434–438. doi: 10.7326/0003-4819-135-6-200109180-00012. [DOI] [PubMed] [Google Scholar]
- 28.Benson L, Song X, Campos J, Singh N. Changing epidemiology of Clostridium difficile-associated disease in children. Infect. Control Hosp. Epidemiol. 2007;28:1233–1235. doi: 10.1086/520732. [DOI] [PubMed] [Google Scholar]
- 29.Smith CJ, Markowitz SM, Macrina FL. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 1981;19:997–1003. doi: 10.1128/aac.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–790. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.