Abstract
Expression of the tumor suppressor deleted in liver cancer-1 (DLC-1) is lost in non-small cell lung (NSCLC) and other human carcinomas, and ectopic DLC-1 expression dramatically reduces proliferation and tumorigenicity. DLC-1 is a multidomain protein that includes a Rho GTPase Activating Protein (RhoGAP) domain which has been hypothesized to be the basis of its tumor suppressive actions. To address the importance of the RhoGAP function of DLC-1 in tumor suppression, we performed biochemical and biological studies evaluating DLC-1 in NSCLC. Full length DLC-1 exhibited strong GAP activity for RhoA as well as RhoB and RhoC, but only very limited activity for Cdc42 in vitro. In contrast, the isolated RhoGAP domain showed 5- to 20-fold enhanced activity for RhoA, RhoB, RhoC and Cdc42. DLC-1 protein expression was absent in six of nine NSCLC cell lines. Restoration of DLC-1 expression in DLC-1-deficient NSCLC cell lines reduced RhoA activity, and experiments with a RhoA biosensor demonstrated that DLC-1 dramatically reduces RhoA activity at the leading edge of cellular protrusions. Furthermore, DLC-1 expression in NSCLC cell lines impaired both anchorage-dependent and -independent growth, as well as invasion in vitro. Surprisingly, we found that the anti-tumor activity of DLC-1 was due to both RhoGAP-dependent and -independent activities. Unlike the rat homologue p122RhoGAP, DLC-1 was not capable of activating the phospholipid hydrolysis activity of phospholipase C-δ1. Combined, these studies provide information on the mechanism of DLC-1 function and regulation, and further support the role of DLC-1 tumor suppression in NSCLC.
Keywords: GTPase activating protein, Rho GTPases, tumor suppressor
INTRODUCTION
DLC-1 (deleted in liver cancer-1) was identified originally as a gene whose expression was lost in hepatocellular carcinomas (HCC). Subsequent studies found loss of DLC-1 gene expression in liver, breast, colon, gastric, prostate, cervical, esophageal and other cancers [1-5]. The downregulation of DLC-1 expression is particularly dramatic in non-small cell lung carcinomas (NSCLC); DLC-1 transcription was significantly reduced or lost in 95% (20/21) of primary tumors and 58% (11/19) of NSCLC cell lines [6]. Ectopic restoration of DLC-1 expression in DLC-1-deficient NSCLC, HCC, breast and esophageal cancer cell lines resulted in reduced growth in vitro and tumorigenicity and metastasis in vivo, supporting the role of DLC-1 as a tumor suppressor [2,6-10].
DLC-1 is a 122 kDa multidomain protein that consists of an N-terminal SAM domain, a C-terminal START domain, and an internal RhoGAP domain (Fig. 1A). The RhoGAP domain is a conserved catalytic domain found in GTPase activating proteins for members of the Rho family of small GTPases (RhoGAPs) [11]. SAM domains (~70 amino acids) are putative protein interaction modules [12] and START domains (~120 amino acids) are lipid-binding domains found in proteins that transfer lipids between organelles [13]. The roles of the SAM and START domains in DLC-1 function have not been determined.
Figure 1.
DLC-1 is a RhoGAP. (A) Schematic of the structure of wild type and mutant human DLC-1 proteins used in this study. (B) DLC-1 is a RhoGAP for RhoA, (C) RhoB, and (D) RhoC. The GTP hydrolysis rate of recombinant GST fusion proteins of RhoA, RhoB, and RhoC was measured in the presence and absence of 0.30 μM DLC-1. GAP activity of a missense mutant of DLC-1, with substitution of a conserved arginine residue in the RhoGAP catalytic domain (DLC-1(R718E)), was also determined. The small GTPases were preloaded with GTP, and incubated with a fluorescently-labeled phosphate binding protein (PBP) which undergoes a dramatic increase in fluorescence upon binding inorganic phosphate released from GTP hydrolysis. Real time GTP hydrolysis was monitored by measuring the increases in fluorescence, which directly correlated with GTP hydrolysis. Data shown are representative of two independent experiments.
Rho GTPases comprise a major branch of the Ras superfamily of small GTPases, with RhoA, Rac1 and Cdc42 the most intensely studied [14,15]. Like Ras, Rho GTPases function as binary switches that cycle between active, GTP-bound and inactive, GDP-bound forms. In normal cells, the Rho GDP/GTP cycle is tightly regulated. Extracellular stimulus-mediated activation of Rho GTPases is facilitated by guanine nucleotide exchange factors (RhoGEFs) that promote formation of Rho-GTP. Rho-GTP preferentially binds and regulates multiple downstream effectors that in turn regulate cytoplasmic signaling networks that control actin cytoskeletal organization, cell cycle progression, gene expression, cell movement and polarity, and intracellular vesicular transport [16]. RhoGAPs negatively regulate GTPase function by accelerating the weak intrinsic GTP hydrolysis activity of the GTPase, promoting formation of the inactive, GDP-bound form. Whereas a variety of mechanisms that regulate RhoGEFs have been identified [17], the regulation of RhoGAPs remains poorly understood.
In vitro analyses of rat (p122-RhoGAP) and human DLC-1 demonstrated that DLC-1 exhibits robust GAP activity for RhoA [18,19]. Similarly, in vivo analyses showed that ectopic expression of rat DLC-1 caused cell rounding and disruption of actin stress fibers, activities consistent with inhibition of RhoA [20]. While DLC-1 [19] and the related DLC-2 protein [21,22] have been reported to have weak or no in vitro activity for Cdc42 and Rac1, it has not been determined whether DLC-1 functions as a GAP for the structurally and biochemically-related, yet biologically-divergent, RhoB and RhoC GTPases [23,24].
Like Ras, the aberrant activation of Rho family GTPases has also been associated with cancer progression and growth [25]. However, whereas mutational activation of Ras is prevalent in human cancers, mutation of Rho GTPases has not been observed in human cancers. Instead, aberrant Rho GTPase activity has been found in human cancers as a result of the altered expression and/or function of Rho GTPases or their regulators. Of the diversity of mechanisms that have been found, perhaps the most frequent and widespread involves the loss of DLC-1 expression in a variety of human cancers. By analogy to the loss of expression of the NF1 and TSC2 tumor suppressors, that function as GAPs for Ras and the Ras-like GTPase Rheb [26], the tumor suppressive functions of DLC-1 have been hypothesized to be due to its RhoGAP activity. However, the significance of the RhoGAP activity to DLC-1-mediated tumor suppression remains unclear. Since DLC-1 has been described to exhibit other functions, such as the activation of phospholipase C delta (PLCδ) by the rat DLC-1 homologue, p122RhoGAP [18], DLC-1 anti-tumor activity may be due to non-RhoGAP activities.
Because the importance of the RhoGAP function of DLC-1 in tumor suppression has not been determined, we initiated studies to evaluate the biochemical and biological activity of DLC-1 in NSCLC. Our observations expand the Rho GTPase targets of DLC-1 and suggest that intramolecular regulation may be limiting the activity of the full length protein. Furthermore, we demonstrate that, while DLC-1 expression reduces RhoA activity, the anti-tumor activity of DLC-1 is reliant on both RhoGAP-dependent and - independent activities. Combined, our observations expand our understanding of DLC-1 function and regulation, and further support the role of DLC-1 as a tumor suppressor in NSCLC.
MATERIALS AND METHODS
DLC-1 Expression Plasmids
The cDNA sequence encoding human DLC-1 (nucleotides 321-3596 of accession # NM_006094) was amplified through PCR of human embryonic kidney 293-T cDNA and cloned into the pEGFP-N1 (Clontech) vector. Site directed mutagenesis was utilized to change nucleotides 2472-2473 from GA to CG, creating the cDNA sequence encoding the DLC-1(R718E) missense mutant. The following primer sequences were used: primer-1, 5'-CATGCTGAAGCAGTATTTTGAAGATCTTCCTGAGCCACTA-3' and primer-2, 5'-TAGTGGCTCAGGAAGATCTTCAAAATACTGCTTCAGCATG-3'. For NSCLC expression experiments, the wild type and mutant DLC-1 cDNA sequences were subcloned into the BamHI site of the pBabe-puro retroviral expression vector. Oligonucleotide-directed PCR mutagenesis was utilized to restore the stop codon in the last triplet and add a 3' SalI site. Parallel manipulations were performed with a pEGFP-N1 vector encoding DLC-1(R718E) to generate the pBabe-puro DLC-1(R718E) expression construct. For transient assays, the open reading frame of DLC-1 was excised from pBabe-puro DLC-1 with BamHI and Sal1, and subcloned into the BamHI-Sal1 sites of pcDNA 3.1+ (Invitrogen). The pcDNA3.1+ vectors for PLC-β2, Rac3 G12V, and PLC-δ1 were obtained from the Guthrie cDNA Resource Center. For the RhoA biosensor experiments, the cDNA sequence encoding the mCherry fluorescent protein tag was PCR-amplified and subcloned into the Kpn1-BamHI sites of pcDNA DLC-1 to create the coding sequence for the mCherry DLC-1 fusion protein.
For expression of recombinant protein, cDNA sequences for DLC-1, DLC-1(R718E), and the isolated DLC-1 RhoGAP domain were subcloned into the pGEX-5X-3 (GE Healthcare) bacterial expression vector for expression of glutathione S-transferase (GST)-DLC-1 fusion proteins. All plasmid cDNA coding sequences were sequence verified before use. pGEX plasmids for expression of GST fusions of wild type human RhoA, RhoB, RhoC, Rac1 and Cdc42 have been described previously [27,28].
Bacterial Protein Expression and Purification
Plasmids encoding GST-fusion proteins of DLC-1 and Rho GTPases were transformed into the BL-21 E. coli strain. GST-DLC-1 (full length DLC-1, DLC-1(R718E), and DLC-1 GAP expression was induced with 100 μM isopropyl-B-D-1-thiogalactopyranoside (IPTG) for 16 h at room temperature. GST fusion proteins of human RhoA, RhoB, and RhoC were induced with 250 μM IPTG for 16 h at room temperature. Rac1 and Cdc42 were induced with 1 mM IPTG for 3 h at 37°C. GST-fusion proteins were purified by glutathione Sepharose 4B chromatography, and the purity of soluble proteins was assessed by Coomassie staining. Purified proteins were quantified by the method of Bradford [29].
Single Turnover GTP Hydrolysis Assay
The in vitro GAP activity of DLC-1 was measured with a fluorescence-based technique [30]. This assay utilizes a fluorophore-conjugated phosphate binding protein (MDCC-PBP) that dramatically increases in fluorescence upon binding phosphate. Purified glutathione-agarose bead bound-Rho GTPases were preloaded with GTP in an exchange buffer (20 mM Tris, 50 mM NaCl, 200 mM ammonium sulfate, 10 mM EDTA, and 0.5 mM GTP) for 1 min at 37°C. Following elution, single turnover hydrolysis assays were performed with 15 μM MDCC-PBP and 2 μM GTPase in a buffer containing 20 mM Tris, 50 mM NaCl, and 1 mM MgCl2. Assays were initiated with addition of 0.30 μM DLC-1, and a SpectraMAX Gemini (Molecular Devices) spectrofluorimeter was used to measure increases in fluorescence (λex = 425 nm and λem = 465 nm), reflecting increases in Pi production from GTP hydrolysis.
Cell Culture and Generation of Stable Cell Lines
293-T and Cos-7 cells were maintained in Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum (FBS; Hyclone). NSCLC cells were obtained from the ATCC (Rockville, MD) and grown in RPMI-1640 supplemented with 10% FBS. To generate NSCLC cell lines stably-expressing ectopic DLC-1 proteins, infectious virus of the pBabe-puro vectors encoding DLC-1 or DLC-1(R718E) was generated and used to infect the NSCLC cell lines. After selection in growth medium containing 1.0 μg/mL puromycin, multiple drug resistant colonies were pooled together to establish mass populations stably expressing each DLC-1 protein. Multiple, independently established stable cell lines (<3 passages) were used for all analyses, to evaluate reproducibility and to minimize secondary consequences of prolonged maintenance of cell lines ectopically expressing growth suppressing proteins.
RhoA Activity Assay
A GST-fusion of the Rho-GTP binding domain (RBD) of Rhotekin (amino acids 7-89), an effector of RhoA, RhoB, and RhoC, was used in pull down assays to detect expression of activated RhoA-GTP as we have described previously [31]. Total and GST-RBD-precipitated lysate samples were subjected to SDS-PAGE and analyzed by western blot analysis using anti-RhoA antibody (Santa Cruz Biotechnology). Exogenous DLC-1 expression was confirmed by probing the total lysate samples with the monoclonal anti-DLC-1 antibody (612020, BD Biosciences).
RhoA Biosensor Experiments
The genetically encoded RhoA biosensor based on CFP/YFP intramolecular fluorescence resonance energy transfer (FRET) was described previously [32]. Briefly, the sensor is comprised of the Rhotekin-RBD which specifically binds to RhoA-GTP, followed by cyan fluorescent protein, an unstructured linker of optimized length, a pH-insensitive variant of yellow fluorescent protein Citrine YFP, and the full length RhoA. Upon GTP loading and activation of RhoA, the binding domain within the biosensor binds to the activated RhoA and changes the relative orientation of CFP and Citrine YFP to affect FRET. By monitoring the ratio of donor and acceptor fluorescence emission, spatio-temporal dynamics of RhoA activation can be studied.
MEF/3T3 cells stably expressing RhoA biosensor under tetracycline-off regulator [32] were induced to express the RhoA biosensor by removal of doxycyclin and replating at 4 × 104 cells in a 10 cm diameter tissue culture dish. Twenty four h after induction, cells were detached and replated at 1 × 105 cells in a 10 cm tissue culture dish, and transfected with 1 μg of mCherry-DLC-1 DNA using Fugene 6 (Roche Diagnostics, Indianapolis, IN). Twenty four h post transfection, cells were replated on glass coverslips coated with fibronectin (10 μg/ml) (Sigma, St. Louis, MO) at 4 × 104 cells per coverslip and allowed to adhere and spread for 5 h prior to imaging experiments.
Cells were imaged in Ham's F-12K medium without phenol red (Biosource), with 2% FBS, 10 mM HEPES buffer and 10 μg/ml Oxy-Fluor reagent supplemented with 5mM dl-lactate (Oxyrase Inc.), in a heated Atto-fluor chamber (Molecular Probes). Images were obtained using a Zeiss 40× 1.3 NA EC-Plan NeoFluar DIC lens on a Zeiss Axiovert 100TV microscope, a CoolsnapES charged coupled device camera (Roper Scientific), and Metamorph software (Universal Imaging). For ratio imaging of the RhoA biosensor, the following filters were used (Chroma Technology): CFP: D436/20, D470/40; FRET: D436/20, HQ535/30; YFP: HQ500/20, HQ535/30. mCherry-DLC-1 was imaged using HQ580/30, HQ630/40. A “Custom Quad” dichroic mirror was custom manufactured by Chroma Company for compatibility with all of these filters. Filterwheels were used to change the filters in the excitation and emission light paths (Ludl Electronic Products, Hawthorne, NY). Cells were illuminated with a 200 W Hg/Xe arc lamp (Optiquip, Highland Mills, NY) through a 36% neutral density filter. At each time point, autofocus was performed in DIC followed by DIC image acquisition, the three fluorescence images were recorded with the following exposure times: CFP (800ms), FRET (400ms) and mCherry-DLC-1 (500ms) at binning 2×2.
Metamorph version 7.1.2 (Universal Imaging) was used to perform the image processing and data analysis. All images were flat-field corrected and background subtracted. FRET and CFP images were then carefully aligned either manually or automatically [33] to ascertain correct pixel-to-pixel registration. The FRET image, because it had the largest signal-to-noise ratio and therefore provided the best distinction between the cell and the background, was intensity thresholded to generate a binary mask with a value of zero outside the cell and a value of one inside the cell. FRET and CFP images were multiplied by this binary mask to set areas outside the cell uniformly to zero to minimize noise and other artefacts. The masked FRET image was divided by the masked CFP image to yield a ratio reflecting RhoA activation throughout the cell. A linear pseudocolor lookup table was applied and the ratio values were normalized to the lower scale value. In every data set, CFP and FRET images were carefully inspected to verify that all portions used to create the ratio image had a high enough signal/noise ratio. This was especially important in thin parts of the cell where fluorescence was low. In time-lapse experiments, CFP and YFP bleached at different rates. The ratio was corrected for bleaching using a published method [34]. Because ruffles and cell edges can move rapidly, it was important to exclude motion artefacts; we routinely reversed the order of FRET and CFP acquisition to ascertain that motion artefacts were negligible, or used fixed cells (data not shown).
Growth Transformation and Invasion Assays
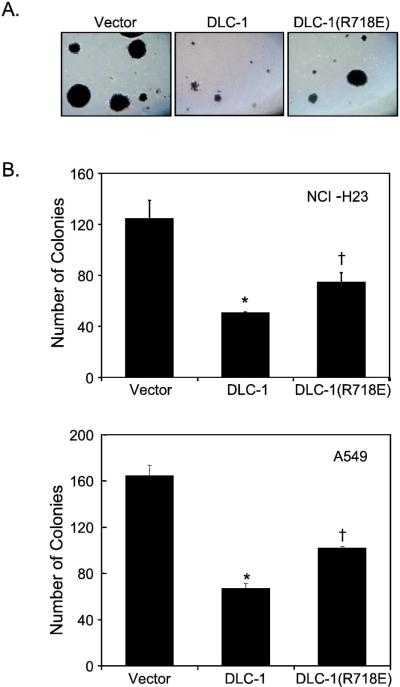
Mass populations of drug-resistant NCI-H23 and A549 cells stably-infected with the empty pBabe-puro vector, or encoding DLC-1 proteins, were analyzed in soft agar assays as we have described previously [35,36]. In triplicate, 2 × 104 NCI-H23 or 104 A549 cells were trypsinized and resuspended in growth medium containing 0.4% agar. Cultures were maintained at 37°C for 21 days, at which point viable colonies were stained with the MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) viability stain and photographed. The number of proliferating colonies (>10 cells) was quantified by counting the number of colonies in five representative fields of view within each plate.
Invasion in vitro was measured by using the Matrigel transwell assay with 8.0 μm pore Growth Factor Reduced Matrigel Invasion Chambers (BD Biosciences) according to the manufacturer's protocol. NCI-H23 cells were dissociated with TrypLE Express (Invitrogen), and 7.5 × 103 cells resuspended in serum-free RPMI-1640 containing 1% bovine serum albumen were seeded into the upper chamber. RPMI-1640 containing 3% FBS (chemoattractant) was added to the well beneath the chamber. After 22 h at 37°C, non-invaders were removed, and invading cells were fixed, stained, photographed, and quantified. Five fields were counted for each chamber, and the total number of cells counted per chamber was used for calculating the average number of invading cells. The relative number of invading cells was determined by arbitrarily assigning the cells transfected with empty vector a value of 1.0. The two-tailed Student's t test was used to quantify statistical differences between experimental groups for the soft agar and Matrigel assays. Results are expressed as mean ± SD.
PLC Activity Assays
The ability of DLC-1 to promote phoshoinositide hydrolysis activity of PLC-δ1 was quantified by measuring the accumulation of 3[H]inositol phosphates as previously described [37]. Briefly, Cos-7 cells were plated in 12-well culture dishes transfected with the indicated DNA expression vector using FuGENE 6 transfection reagent (Roche) according to the manufacturer's protocol. Empty-vector DNA was used as necessary to maintain a constant total amount of DNA per well. Twenty-four h after transfection, the medium was replaced with inositol- and serum-free DMEM containing 1 uCi/well [myo-3H]inositol (American Radiolabeled Chemicals, St. Louis, MO). Phospholipase activity was quantified 12 h after labeling by incubation in inositol-free DMEM containing 10 mM LiCl. After approximately 60 min, ice-cold 50 mM formic acid was added to stop the reaction. 150 mM ammonium hydroxide was used to neutralize the reaction, and the accumulation of [3H]inositol phosphates was quantified by Dowex chromatography as described previously [37].
RESULTS
DLC-1 Stimulates RhoA, RhoB, and RhoC GTP Hydrolysis Activity In Vitro
Previous studies demonstrated clearly that DLC-1 has potent GAP activity for RhoA in vitro. Whether DLC-1 is also a GAP for the biochemically-related, yet biologically-distinct, RhoB and RhoC proteins is not known. In contrast to RhoA, conflicting observations have been made for the ability of DLC-1 to serve as a GAP for Cdc42 and Rac1. Therefore, we evaluated the ability of DLC-1 to accelerate the GTP hydrolysis of RhoA, RhoB, RhoC, Cdc42, and Rac1.
For these analyses we purified bacterially-expressed DLC-1 (Fig. 1A) and GST fusion proteins of human RhoA, RhoB, RhoC, Cdc42 and Rac1. Using a fluorescence-based technique that measures single turnover GTP hydrolysis [30], we measured the ability of full length DLC-1 to stimulate the intrinsic GTP hydrolysis activity of these GTPases. In agreement with previous studies, we found that DLC-1 had robust activity on RhoA in vitro. Furthermore, DLC-1 also had strong GAP activity on both RhoB and RhoC (Fig. 1B - D). To aid our studies on the contribution of the RhoGAP function to DLC-1 tumor suppression, we introduced a missense mutation in the conserved arginine-718 residue critical for RhoGAP catalytic activity [38]. We found that DLC-1(R718E) was completely impaired in stimulating the intrinsic hydrolysis activity of GTP-loaded RhoA, RhoB, and RhoC in vitro (Fig. 1B - D).
Intramolecular Regulation of DLC-1 Impairs its GAP Activity
Our previous observations with another RhoGAP (p190RhoGAP), which is also a GAP for RhoA, RhoB, and RhoC, found that ectopic overexpression of p190RhoGAP caused significant cell rounding [39]. In contrast, our initial observations found that ectopic overexpression of DLC-1 did not cause significant cell rounding (data not shown). Therefore, we speculated that full length DLC-1 may not be fully active. The RhoGAP domain of DLC-1 comprises only 146 residues (amino acids 655-801) of the full length, 1,091 residue protein. The presence of significant N- and C-terminal flanking regions suggests that DLC-1 could be highly regulated, possibly by intramolecular interactions with flanking sequences and domains. Therefore, we expressed the isolated RhoGAP domain of DLC-1 (residues 609-878; designated DLC-1 GAP) and compared the activity with full length DLC-1.
We determined that DLC-1 GAP was extremely robust on RhoA in vitro, accelerating the GTP hydrolysis nearly 70-fold, or approximately 20-fold more than the activity seen with the full length protein (Fig. 2A and B). DLC-1 GAP was also particularly robust on RhoB and RhoC, accelerating the GTP hydrolysis 5- to 10-fold more than full-length DLC-1 (Fig. 2C - F). For Cdc42, acceleration of intrinsic GTP hydrolysis increased from a modest 1.4-fold activity for full length DLC-1 to 10-fold stimulation with DLC-1 GAP, corresponding to a 7-fold increase in RhoGAP activity (Fig. 2G and H). We also observed similar activities in cells, where transient expression of DLC-1 RhoGAP, but not full length DLC-1, caused considerable cell rounding (data not shown). Additionally, introduction of the R718E mutation into DLC-1 RhoGAP completely abolished this rounding activity, demonstrating that this mutation abolished RhoGAP activity in vivo.
Figure 2.

The isolated RhoGAP domain shows greatly enhanced catalytic activity for RhoA, RhoB, RhoC and Cdc42. Recombinant full length and an isolated RhoGAP catalytic domain fragment (amino acids 609-878) of DLC-1 were expressed and purified for analysis of in vitro GAP activity. Real time GTP hydrolysis was measured with a fluorescently labeled PBP, as described in Fig. 1, for GST fusion proteins of wild type (A, B) RhoA, (C, D) RhoB, (E, F) RhoC, (G, H) Cdc42, and (I, J) Rac1. Data shown are representative of three independent experiments.
Interestingly, neither DLC-1 nor DLC-1 GAP displayed any activity towards Rac1 (Fig. 2I and J). Our results contrast with observations made with the related DLC-2 protein [21]. The isolated RhoGAP fragment of DLC-2 (80% identical) had altered substrate specificity, as it was active on Cdc42 and Rac1, but not RhoA, whereas the full length DLC-2 was reported to be robust towards RhoA and Cdc42, with mild activity on Rac1. Unlike DLC-2, the flanking regions of the DLC-1 GAP domain do not affect Rho GTPase substrate specificity.
Dlc-1 Protein Expression is Lost in Non-Small Cell Lung Cancers
Yuan et al. employed RT-PCR analyses and demonstrated that the DLC-1 transcript is lost in both primary tumors (20/21, 95%) and NSCLC cell lines (11/19, 58%) [6]. We utilized a DLC-1-specific monoclonal antibody to evaluate DLC-1 protein expression in a panel of NSCLC cell lines, and determined that expression of DLC-1 was lost in 6 of 9 (67%) NSCLC cell lines (Fig. 3). This pattern of DLC-1 protein expression correlated with previous analyses of DLC-1 transcription for 5 of the 6 lines that were evaluated in both studies [6]. The only exception was the NCI-H2228 cell line, which was reported to be positive for DLC-1 mRNA but we determined was negative for DLC-1 protein expression. However, we utilized RT-PCR analyses and determined that our strain of NCI-H2228 cells was deficient for DLC-1 mRNA expression (data not shown), suggesting that the discrepancy is due to variations in cell line history. Hence, loss of DLC-1 protein expression correlated absolutely with the absence of DLC-1 gene transcription. Three NSCLC cell lines (A549, NCI-H23, and NCI-358) that we found to be DLC-1 negative for both the transcript and protein analysis were chosen for further biological studies.
Figure 3.
DLC-1 protein expression is lost in NSCLC cell lines and associated with increased RhoA-GTP formation. (A) DLC-1 protein expression is lost in some NSCLCs. Lysates from nine NSCLC cell lines were resolved by SDS-PAGE and transferred to a PVDF membrane. A monoclonal antibody was used for western blot analysis of DLC-1 expression (612020; BD Biosciences). A parallel blot for β-actin was done to ensure equivalent loading of total cell lysate protein. (B) Ectopic expression of wild type but not mutant DLC-1 is associated with a reduction of RhoA-GTP activity in NSCLCs. Mass populations of two NSCLC cell lines, deficient in endogenous DLC-1 protein expression, were established via infection with the pBabe-puro retrovirus vector encoding wild type full length DLC-1 or the GAP-dead mutant, DLC-1(R718E). RhoA-GTP levels were assessed by incubating lysates with GST-Rhotekin-RBD that was precoupled to glutathione-sepharose beads. Precipitated and total lysates (4% loading control) were analyzed by western blotting with anti-RhoA and -DLC-1 antibodies. Data shown are representative of two independent experiments.
DLC-1 Expression Modulates Rho GTPase Activity In Vivo
Whether DLC-1 expression can modulate RhoA activity in vivo has not been demonstrated. To address this question, we ectopically restored DLC-1 expression in two DLC-1-deficient NSCLC cell lines (NCI-H23 and NCI-H358). For all of the ectopic DLC-1 expression studies, the level of expression ranged from comparable to ~3-fold greater than endogenous DLC-1 (NCI-H1703 cells). Restoration of wild type, but not R718E, DLC-1 expression significantly reduced the level of RhoA-GTP in both NCI-H23 and NCI-H358 cell lines (Fig. 3B). These results suggest that loss of DLC-1 will cause increased Rho GTPase activity.
DLC-1 Expression Reduces Rho GTPase Activity at the Edge of Cellular Protrusions
In addition to using pull down analyses to demonstrate that DLC-1 can reduce total cellular RhoA-GTP levels, we utilized a RhoA biosensor to quantify spatially distinct changes in RhoA activity. MEFs stably expressing a RhoA biosensor were transfected with an mCherry DLC-1 vector, and fluorescence resonance energy transfer (FRET) analysis was performed to quantify RhoA activity, as previously described [32]. Interestingly, DLC-1 expression resulted in a preferential reduction of RhoA activity at the leading edge of cellular protrusions (Fig. 4). To verify that the DLC-1 expression was not limited to this region in the cell, we analyzed mCherry-DLC-1 expression and, consistent with published reports [40,41], DLC-1 was localized to focal adhesions throughout the cells (Fig. 4C). These results indicate that there are some DLC-1 localizations for which RhoGAP activity is more activated, and that the RhoGAP activity of DLC-1 plays a role in leading edge dynamics.
Figure 4.
DLC-1 expression reduces RhoA activity at the leading edge of cellular protrusions. MEFs stably expressing a RhoA biosensor were transiently-transfected with mCherry-tagged DLC-1 and plated on fibronectin. (A) Time-lapse images of RhoA activity during cellular protrusion. Regions of intense RhoA activity are shown in red. Scale bar, 10 μm. (B) Quantification of RhoA activity at varying distances from the edge of the cell. RhoA activity was quantified along 120 line-scans drawn perpendicular to the edge of 8 mCherry control cells and 103 line-scans perpendicular to the edge of 8 mCherry DLC-1 cells. Data points represent the average ± SD (C) mCherry DLC-1 is expressed at focal adhesions in the MEFs. Expression of the mCherry fluorescence is shown for representative control and DLC-1 cells.
DLC-1 Displays both GAP-Dependent and -Independent Tumor Suppression
To assess the importance of GAP activity to DLC-1 mediated tumor suppression, we utilized mass populations of NCI-H23 and A549 cells stably expressing ectopic wild type DLC-1 or DLC-1(R718E). Interestingly, using a clonogenic growth assay to monitor the proliferation of retrovirus-infected cells, we observed that cells infected with either the wild type or the R718E mutant showed a much reduced frequency of drug-resistant colonies than was seen with the empty pBabe-puro retrovirus-infected cells (data not shown). The DLC-1-mediated reduction in clonogenic growth was similar to what had been reported previously [6], while the DLC-1(R718E)-mediated reduction provided an initial suggestion that DLC-1 possesses RhoGAP-independent growth inhibitory activity.
To extend previous biological studies, we analyzed the impact of DLC-1 expression on the anchorage-independent growth of two DLC-1-deficient NSCLC cell lines that grow well in soft agar. We observed that DLC-1 caused an approximately 60% reduction in colony forming efficiency, as well as colony size, in soft agar for both NCI-H23 and A549 NSCLC cells (Fig. 5). Surprisingly, expression of the GAP-dead DLC-1(R718E) also significantly inhibited anchorage-independent growth, though to a lesser extent (~40% colony reduction) than seen with the wild type protein.
Figure 5.
Ectopic re-expression of DLC-1 impairs NSCLC anchorage-independent growth. NCI-H23 and A549 NSCLC cells stably-infected with the pBabepuro vector encoding DLC-1 or DLC-1(R718E) were evaluated for colony formation in soft agar and the number of proliferating viable colonies was quantitated after 21 days. (A) Viable colonies were stained with MTT and photographed. (B) The total number of colonies (> 10 cells) within five representative fields of view was quantified. Data shown are the average ± SD of triplicate wells and are representative of three independent experiments. *, significant at P ≤ 0.005 vs. vector; †, significant at P < 0.01 vs. vector and vs. wild type DLC-1.
We also examined the impact of DLC-1 expression upon NSCLC cell migration and invasion. In NCI-H23 and A549 NSCLC cells, expression of DLC-1 did not alter cellular motility in wound healing assays (data not shown). However, DLC-1 expression did reduce invasion in Matrigel assays by 50% in NCI-H23 cells (Fig. 6). The GAP-dead DLC-1(R718E) mutant reduced invasion by approximately 25%, though this decrease was not statistically significant (P = 0.18). Likewise, the DLC-1-mediated reduction in A549 invasiveness (~30%) was not statistically significant (data not shown). These results, together with the anchorage-independence assays, suggest that DLC-1 uses both GAP-dependent and -independent mechanisms to suppress NSCLC growth and invasion.
Figure 6.
DLC-1 reduces NSCLC invasion in vitro. NCI-H23 cells stably-infected with the pBabe-puro vector encoding DLC-1 or DLC-1(R718E) were assessed for invasion through Matrigel. Cells were dissociated, resuspended in serum-free growth media containing 1% BSA, and incubated for 22 h at 37°C in a Matrigel invasion chamber; the lower well contained growth medium supplemented with 3% FCS. Non-invaders were removed, and the chambers were fixed and stained. (A) Invading cells were photographed under 10X magnification. (B) Quantification of invading cells. Data shown are the mean ± SD of triplicate wells and are representative of two independent experiments. *, significant at P ≤; 0.01 vs. vector.
DLC-1 does not stimulate the phospholipids hydrolysis activity of PLC-δ1
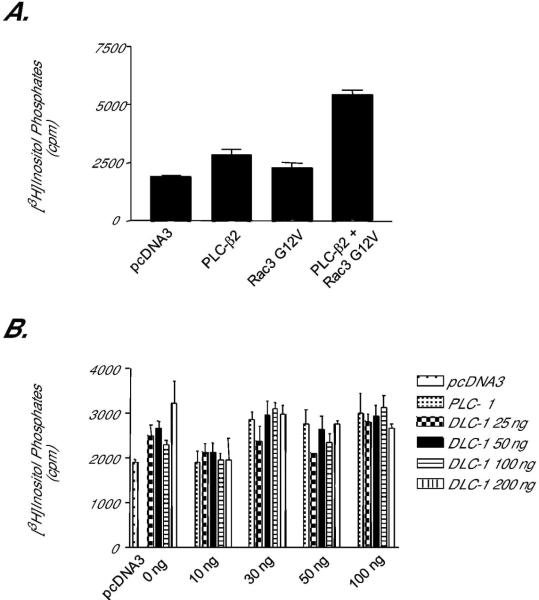
To investigate a possible mechanism of GAP-independent activity, we investigated the ability of DLC-1 to activate PLC-δ1, as has been reported for the rat homologue p122RhoGAP. Activation of PLC was quantified by measuring the accumulation of [3H]inositol phosphates, as we have described previously [37]. Co-transfection of an activated Rac3 mutant (Rac3 G12V) along with the PLC-β2 isozyme resulted in a dramatic increase in [3H]inositol phosphates (Fig. 7A), demonstrating the sensitivity of the cell-based system for detecting changes in PLC activity. Surprisingly, co-transfection of DLC-1 with PLC-δ1 did not result in an increase in [3H]inositol phosphate accumulation (Fig. 7B), suggesting that, unlike rat DLC-1, human DLC-1 does not activate the phospholipid hydrolysis activity of PLC-δ1.
Figure 7.
DLC-1 does not stimulate phosphoinositide hydrolysis activity of PLC-δ1. Cos-7 cells were transiently transfected with vectors encoding various PLC isoforms and known or putative activators. Intracellular inositol phosphate accumulation was quantified as described in Methods. (A) PLC-β2 phospholipid hydrolysis activity is activated by constitutively activated Rac3. To verify the assay conditions, we coexpressed PLC-β2 with Rac3 G12V, an established activator of PLC-β2, and quantified [3H]inositol phosphate accumulation. (B) PLC-δ1 activity is not stimulated by DLC-1. Zero toδ100 ng of expression vector encoding PLC-δ1 was co-transfected with 0, 25, 50, 100, or 200 ng of DNA encoding human DLC-1, and the accumulation of [3H]inositol phosphates was quantified. Data shown are the mean ± SD of triplicate determinations and are representative of results obtained in two independent experiments.
DISCUSSION
Expression DLC-1, a member of the RhoGAP family of negative regulators of Rho GTPases, is lost in NSCLC and other human cancers [6]. Ectopic restoration of DLC-1 expression in DLC-1-deficient tumor cell lines reduced growth in vitro and tumorigenicity and metastatic growth in vivo, supporting a tumor suppressor function for DLC-1 [7-9]. By analogy to the NF1 and TSC2 tumor suppressors, which function as GAPs for Ras and Rheb small GTPases [26], respectively, the loss of DLC-1 is hypothesized to promote oncogenesis by causing hyperactivation of Rho GTPases. However, the mechanistic basis of DLC-1's tumor suppression has remained largely unknown. Unexpectedly, we found that DLC-1 growth suppression may be mediated by both RhoGAP-dependent and -independent functions.
Previous studies demonstrated that DLC-1 is a potent GAP for RhoA in vitro [19]. We have verified and extended these observations, and showed that DLC-1 is a potent GAP for the structurally and biochemically-related RhoB and RhoC proteins. Though RhoA, RhoB and RhoC form a closely related subgroup (~90% amino acid identity) of the Rho GTPase family, they have strikingly different biological functions [23,24]. For example, Hynes and colleagues found that increased expression and activity of RhoC but not RhoA was important for melanoma invasion and metastasis [42]. Mercurio and colleagues found that RhoA impeded but RhoC stimulated breast cancer cell invasion [43]. However, unlike RhoA and RhoC, RhoB is commonly under expressed in cancers, and this GTPase suppresses various aspects of cancer progression [44]. Thus, it is not clear how the loss of DLC-1 GAP activity on this negative regulator of cancer progression could contribute to tumor suppression. A recent report demonstrated that RhoB is frequently lost in NSCLCs [45], and thus it is possible that the potential effects of DLC-1 acting as a GAP on RhoB are limited by the reduced expression of RhoB in DLC-1 positive tumors. Nonetheless, our finding of DLC-1 GAP activity on RhoB and RhoC expands the known targets of DLC-1, increasing the potential mechanisms by which DLC-1 functions.
The isolated DLC-1 GAP domain was 5- to 20-fold more active than the full length protein in vitro, responsible for limiting its activity. We also found that the isolated RhoGAP domain, but not full length DLC-1, caused potent cell rounding, indicating that the isolated RhoGAP fragment is a constitutively activated DLC-1 in vivo. Our further analyses of N-terminally-deleted variants of DLC-1 determined that deletion of the SAM domain alone increased GAP activity in vitro and was sufficient for constitutive RhoGAP activity in vivo (manuscript in preparation). Thus, our present studies are focused on determining a possible mechanism where the SAM domain functions as an intramolecular negative regulator of DLC-1 RhoGAP catalytic activity.
The impact of DLC-1 on Rho-GTP levels in vivo had not been addressed directly. Microinjection of rat DLC-1 caused a reduction in actin stress fibers and inhibited lysophosphatidic acid-stimulated stress fiber formation, phenotypes suggestive of reduced RhoA-GTP levels [20]. Likewise, Wong et al. recently demonstrated that DLC-1 inhibited stress fiber formation in HCC cells [46]. We addressed this directly and showed that ectopic restoration of wild type, but not RhoGAP-deficient, DLC-1 expression in DLC-1-deficient NSCLC cells reduced RhoA-GTP levels, demonstrating that DLC-1 functions as a GAP for RhoA in vivo. Thus, loss of DLC-1 expression is expected to result in persistent activation of Rho GTPases in NSCLCs. Our future studies will determine if elevated RhoA-GTP levels are associated with DLC-1-deficiency in NSCLC cell lines and patient tumors.
We also utilized a RhoA biosensor to demonstrate that DLC-1 expression significantly reduces RhoA activity, specifically at the edge of cellular protrusions. MEFs expressing DLC-1 had reduced motility and formed fewer protrusions (data not shown). It is likely that the ability of DLC-1 to reduce RhoA activity at the edge of cells contributes to the reduced migratory ability of DLC-1 expressing cells, and may contribute to DLC-1's ability to suppress invasion and metastasis [7].
Our studies determined that DLC-1 growth suppression is mediated by both RhoGAP-dependent and -independent functions. We found that exogenous expression of DLC-1 in the DLC-1 deficient NCI-H23 and A549 NSCLC cells resulted in an approximately 60% decrease in anchorage-independent growth. Surprisingly, the GAP-deficient DLC-1(R718E) mutant still suppressed anchorage-independent growth very significantly (~40% reduction). Similar trends were observed in Matrigel invasion assays, in which DLC-1 suppressed invasion of H23 cells ~50%, whereas DLC-1(R718E) reduced invasion by about 25%. These results suggest that the RhoGAP activity of DLC-1 is only partially responsible for its anti-tumor activity. Our results contrast with observations from a previous study investigating the significance of GAP activity to DLC-1-mediated growth inhibition. In their clonogenic growth analyses, ectopic DLC-1 expression reduced the number of HCC colonies grown on culture dishes following transfection and puromycin selection, whereas their putative GAP-deficient variant of DLC-1 (K714E) had no effect on colony formation [46]. Our different observations are not entirely surprising since Rho GTPases have been implicated in many facets of the tumorigenic and malignant phenotype, but with cell type differences.
The ability of the rat homologue of DLC-1, p122RhoGAP, to activate PLC-δ is the only other known catalytic activity associated with DLC-1. To investigate the RhoGAP-independent anti-tumor functions of DLC-1, we conducted experiments to quantify the ability of DLC-1 to activate PLC-δ1. Surprisingly, we found that DLC-1 did not activate the phospholipid hydrolysis activity of PLC-δ1, suggesting that the protein has distinct functions from rat DLC-1, p122RhoGAP. DLC-1 interaction with caveolin-1 [47] or tensin2 [41] remains interesting possibilities for GAP-independent functions for tumor suppression, since there is evidence for the function of both of these proteins in oncogenesis.
In summary, our studies provide information on the mechanisms by which DLC-1 functions as a tumor suppressor in NSCLC. In particular, we determined that DLC-1 expression will alter the level of Rho GTPase activity in NSCLCs and that DLC-1 growth suppression involves both Rho-dependent and Rho-independent mechanisms. Lastly, our studies suggest that DLC-1 will be regulated by posttranslational mechanisms. Identifying the interacting partners of DLC-1 is essential to developing a more comprehensive understanding of the mechanism of regulation of DLC-1 function.
ACKNOWLEDGEMENTS
We thank Misha Rand for assistance in figure preparation.
Our research was supported by grants from the National Institutes of Health to R.L.J. (PO1HL4500), to K.M.H (GM64346 and GM57464), to T.K.H (GM57391) and to C.J.D. (CA063071 and CA67771). K.D.H. was supported by a fellowship from the American Cancer Society and A. S. by a fellowship from the Susan G. Komen Breast Cancer Foundation.
Abbreviations
- DLC-1
deleted in liver cancer-1
- HCC
hepatocellular carcinomas
- NSCLC
non-small cell lung cancer
- RhoGAP
Rho GTPase activating protein
- RhoGEF
Rho guanine nucleotide exchange factors
- PLC
phospholipase C
- IPTG
isopropyl-B-D-1-thiogalactopyranoside
- GST
glutathione S-transferase
- PBP
phosphate binding protein
- RBD
Rho binding domain
- MEFs
mouse embryo fibroblasts
- FRET
fluorescence resonance energy transfer
- SD
standard deviation
REFERENCES
- 1.Kim TY, Jong HS, Song SH, et al. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22:3943–3951. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 2.Seng TJ, Low JS, Li H, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 3.Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int J Oncol. 2006;29:1127–1132. [PubMed] [Google Scholar]
- 4.Yuan BZ, Durkin ME, Popescu NC. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet. 2003;140:113–117. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- 5.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- 6.Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 7.Goodison S, Yuan J, Sloan D, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng IO, Liang ZD, Cao L, Lee TK. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. [PubMed] [Google Scholar]
- 9.Yuan BZ, Zhou X, Durkin ME, et al. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 11.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 12.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 13.Strauss JF, 3rd, Kishida T, Christenson LK, Fujimoto T, Hiroi H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol Cell Endocrinol. 2003;202:59–65. doi: 10.1016/s0303-7207(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 14.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 15.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 17.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 18.Homma Y, Emori Y. A dual functional signal mediator showing RhoGAP and phospholipase C-delta stimulating activities. Embo J. 1995;14:286–291. doi: 10.1002/j.1460-2075.1995.tb07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- 20.Sekimata M, Kabuyama Y, Emori Y, Homma Y. Morphological changes and detachment of adherent cells induced by p122, a GTPase-activating protein for Rho. J Biol Chem. 1999;274:17757–17762. doi: 10.1074/jbc.274.25.17757. [DOI] [PubMed] [Google Scholar]
- 21.Ching YP, Wong CM, Chan SF, et al. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 22.Nagaraja GM, Kandpal RP. Chromosome 13q12 encoded Rho GTPase activating protein suppresses growth of breast carcinoma cells, and yeast two-hybrid screen shows its interaction with several proteins. Biochem Biophys Res Commun. 2004;313:654–665. doi: 10.1016/j.bbrc.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Wennerberg K, Der CJ. Rho-family GTPases: it's not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler AP, Ridley AJ. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 26.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Abe K, Rossman KL, Liu B, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275:10141–10149. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- 28.Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem. 2002;277:42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Shutes A, Der CJ. Real-Time In Vitro Measurement of Intrinsic and Ras GAP-Mediated GTP Hydrolysis. Methods Enzymol. 2005;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
- 31.Reuther GW, Lambert QT, Booden MA, et al. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J Biol Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- 32.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 33.Shen F, Hodgson L, Hahn K. Digital autofocus methods for automated microscopy. Methods Enzymol. 2006;414:620–632. doi: 10.1016/S0076-6879(06)14032-X. [DOI] [PubMed] [Google Scholar]
- 34.Hodgson L, Nalbant P, Shen F, Hahn K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 2006;406:140–156. doi: 10.1016/S0076-6879(06)06012-5. [DOI] [PubMed] [Google Scholar]
- 35.Cox AD, Der CJ. Biological assays for cellular transformation. Methods Enzymol. 1994;238:277–294. doi: 10.1016/0076-6879(94)38026-0. [DOI] [PubMed] [Google Scholar]
- 36.Solski PA, Abe K, Der CJ. Analyses of transforming activity of Rho family activators. Methods Enzymol. 2000;325:425–441. doi: 10.1016/s0076-6879(00)25463-3. [DOI] [PubMed] [Google Scholar]
- 37.Wing MR, Houston D, Kelley GG, Der CJ, Siderovski DP, Harden TK. Activation of phospholipase C-epsilon by heterotrimeric G protein betagamma-subunits. J Biol Chem. 2001;276:48257–48261. doi: 10.1074/jbc.C100574200. [DOI] [PubMed] [Google Scholar]
- 38.Fidyk NJ, Cerione RA. Understanding the catalytic mechanism of GTPase-activating proteins: demonstration of the importance of switch domain stabilization in the stimulation of GTP hydrolysis. Biochemistry. 2002;41:15644–15653. doi: 10.1021/bi026413p. [DOI] [PubMed] [Google Scholar]
- 39.Wennerberg K, Forget MA, Ellerbroek SM, et al. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol. 2003;13:1106–1115. doi: 10.1016/s0960-9822(03)00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao YC, Si L, deVere White RW, Lo SH. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- 42.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 43.Simpson KJ, Dugan AS, Mercurio AM. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res. 2004;64:8694–8701. doi: 10.1158/0008-5472.CAN-04-2247. [DOI] [PubMed] [Google Scholar]
- 44.Huang M, Prendergast GC. RhoB in cancer suppression. Histol Histopathol. 2006;21:213–218. doi: 10.14670/HH-21.213. [DOI] [PubMed] [Google Scholar]
- 45.Sato N, Fukui T, Taniguchi T, et al. RhoB is frequently downregulated in non-small-cell lung cancer and resides in the 2p24 homozygous deletion region of a lung cancer cell line. Int J Cancer. 2007;120:543–551. doi: 10.1002/ijc.22328. [DOI] [PubMed] [Google Scholar]
- 46.Wong CM, Yam JW, Ching YP, et al. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 47.Yamaga M, Sekimata M, Fujii M, et al. A PLCdelta1-binding protein, p122/RhoGAP, is localized in caveolin-enriched membrane domains and regulates caveolin internalization. Genes Cells. 2004;9:25–37. doi: 10.1111/j.1356-9597.2004.00698.x. [DOI] [PubMed] [Google Scholar]