Abstract
The poor inhibitory activity of circulating antithrombin (AT) is critical to the formation of blood clots at sites of vascular damage. AT becomes an efficient inhibitor of the coagulation proteases only after binding to a specific heparin pentasaccharide, which alters the conformation of the reactive center loop (RCL). The molecular basis of this activation event lies at the heart of the regulation of hemostasis and accounts for the anti-coagulant properties of the low molecular weight heparins. Although several structures of AT have been solved, the conformation of the RCL in native AT remains unknown because of the obligate crystal contact between the RCL of native AT and its latent counterpart. Here we report the crystallographic structure of a variant of AT in its monomeric native state. The RCL shifted ~20Å , and a salt bridge was observed between the P1 residue (Arg-393) and Glu-237. This contact explains the effect of mutations at the P1 position on the affinity of AT for heparin and also the properties of AT-Truro (E237K). The relevance of the observed conformation was verified through mutagenesis studies and by solving structures of the same variant in different crystal forms. We conclude that the poor inhibitory activity of the circulating form of AT is partially conferred by intramolecular contacts that restrain the RCL, orient the P1 residue away from attacking proteases, and additionally block the exosite utilized in protease recognition.
Antithrombin (AT) 2 is activated by heparin through two distinct mechanisms depending on the target protease and the length of the heparin chain. Thrombin inhibition is accelerated by a simple template mechanism where heparin serves as a bridge to improve diffusion and to stabilize the Michaelis complex, whereas heparin stimulation of factor IXa and Xa inhibition depends on an AT conformational change involving the reactive center loop (RCL) (for reviews see Refs. 1 and 2). Thus, low molecular weight heparin and synthetic pentasaccharides, representing the minimal high-affinity binding sequence, are capable of stimulating the inhibition of factors IXa and Xa by two orders of magnitude but accelerate thrombin inhibition by only ~2-fold. The slow reactivity of the circulating native form of AT is crucial in preventing bleeding, which occurs with the overdosing of therapeutic heparin, whereas the localized activation of AT is critical in preventing venous thrombosis. The molecular basis of the activation of AT is thus a subject of great interest.
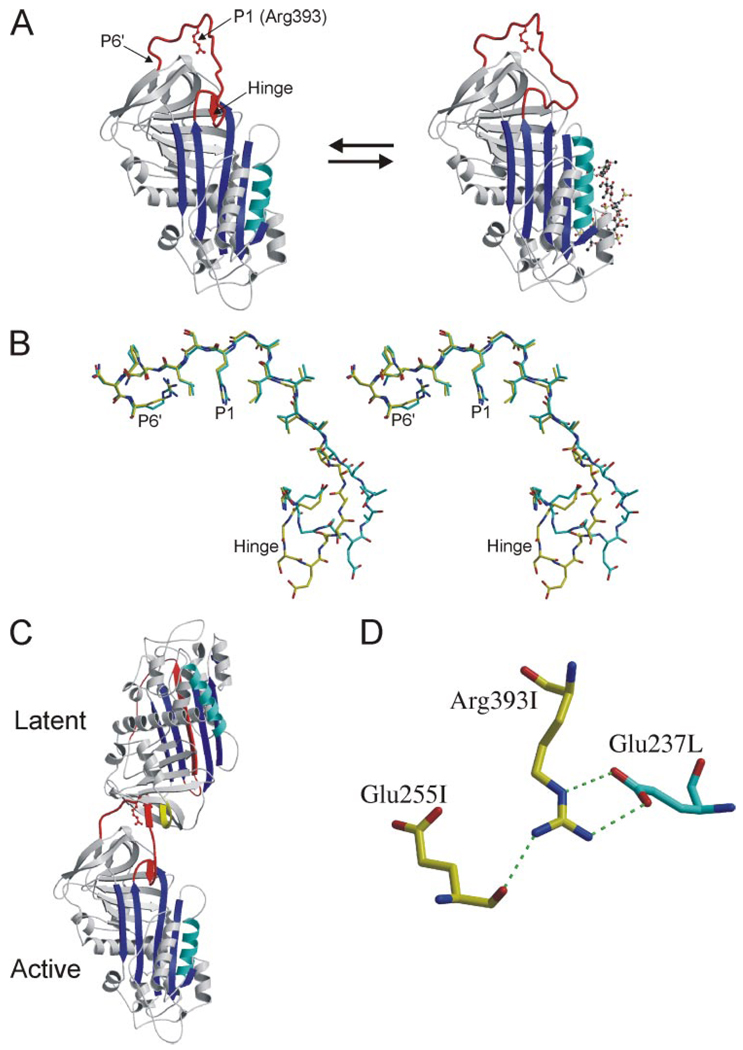
It is natural when considering the activation of AT to focus on the conformation of the RCL. The crystal structure of native AT revealed the partial incorporation of the very N terminus of the RCL (the hinge region) intoβ-sheet A (3,4). Both biochemical (5, 6) and structural (7) studies later demonstrated that pentasaccharide binding induced the expulsion of the hinge region from β-sheet A (Fig. 1A). Although it was assumed that the expulsion of the hinge region would induce a conformational change in the rest of the RCL, thereby increasing its availability for proteolytic attack, the crystal structure revealed an RCL conformation identical to that of native AT (Fig. 1B). This was clearly because of a crystal contact between the active AT component and its latent counterpart, which together formed a heterodimer comprising the asymmetric unit (Fig. 1C). The stable dimerization contact involved the incorporation of RCL residues P7–P3 3 of the active monomer into the vacated s1C position of the latent monomer.
FIGURE 1. RCL conformation and contacts from crystals of the AT heterodimer.
A, The structures of native (left) and pentasaccharide-bound AT (right), solved in the context of the heterodimer with a latent AT counterpart, are shown as ribbon diagrams with the RCL in red, β-sheet A in blue, and heparin-binding helix D in cyan (the pentasaccharide is depicted as a ball-and-stick, and P1 Arg-393 is shown as a red ball-and-stick). The hinge region of the RCL (indicated) is partially incorporated as strand 4 of β-sheet A in the native state and is expelled upon heparin binding. Although the conformation of the hinge region was different in the two structures, the rest of the RCL, constrained by contacts with latent AT, did not differ. B, stereo diagram of the RCL of native (yellow) and pentasaccharide-bound (cyan) ATs (from P17 in the hinge region to the C-terminal end of the RCL at P6′) illustrates the identical RCL conformations from P8 to P6′. C, ribbon diagram of the AT heterodimer between active AT (bottom) and the latent counterpart (top) shows the intimate interaction involving the RCL of the active monomer with s2C (yellow) of the latent monomer. D, one of the principal contacts between the active (I, yellow) and latent (L, cyan) monomers involves the P1 residue (Arg-393) of active AT with Glu-237 from latent AT. The only intramolecular contact involving the side chain of the P1 residue is a hydrogen bond with the main chain of Glu-255.
Native AT converts slowly to the hyperstable latent form through the incorporation of the entire RCL as the fourth strand in β-sheet A. This conformational change is possible only through the extraction of strand 1 from β-sheet C. The inevitable conversion of active AT to the latent form, coupled with the propensity of the active-latent heterodimer to crystallize, meant that crystal trials of active AT alone always resulted in crystallization of the heterodimer. The contacts between the RCL of the active monomer and the latent counterpart of the dimer were not limited to the β-sheet interaction and included residues from P8 to P3′. One such contact is a salt bridge between the P1 Arg of the active monomer to Glu-237 of the latent monomer (Fig. 1D). Although this interaction is an artifact of crystallization, there was strong evidence that the P1 Arg residue was involved in an intramolecular salt bridge in native AT (8). Recombinant and natural AT mutations at the P1 position showed a consistent perturbation in heparin binding affinity, consistent with a loss of a single salt bridge interaction (8 –10). The structures of AT did reveal an intramolecular contact between Arg-393 (P1) and Glu-255; however, the interaction was a hydrogen bond between the side chain of Arg-393 and the main chain oxygen of Glu-255 (Fig. 1D), not the predicted salt bridge.
It was thus unlikely that the conformation of the RCL observed in the crystal structure of the active-latent AT heterodimer represented that of the native circulating form. Despite many efforts by members of this laboratory and others to obtain crystals of the native counterpart alone, all attempts have led only to crystals of the heterodimer because of the slow conversion of a fraction of the material to the latent form. We report here the crystal structure of a variant of AT that is fully resistant to the latent transition and thus represents the first structure of native AT in a monomeric state. Although the overall structure was similar to that observed previously, the RCL shifted ~20 Å to make intimate intramolecular contacts with the body of AT. Included in these interactions is a salt bridge between Arg-393 and Glu-237. The relevance of the observed RCL conformation and interactions was tested by mutagenesis and by solving the structure of the mutant in other crystal forms. We conclude that the structure presented here more closely represents the main circulating conformation of AT than previous structures and explains, in molecular detail, its poor inhibitory activity.
EXPERIMENTAL PROCEDURES
Materials
Human α-thrombin was obtained from Hematologic Technologies (Essex Junction, VT), and human factor Xa was from Enzyme Research Laboratories (Swansea, UK). The physiological pentasaccharide (fondaparinux) and the hexadecasaccharide SR123781 were kindly provided by Maurice Petitou (previously of Sanofi-Synthelabo, Toulouse, France). Latent AT was produced from expired fresh-frozen plasma as described previously (11). S195A thrombin was kindly provided by Charles T. Esmon (Oklahoma Medical Research Foundation).
AT Expression and Purification
AT variants V317C/T401C, E237A, and E237K were constructed on the recombinant β-glycoform S137A to reduce glycosylation heterogeneity, as described previously (12). The S137A/V317C/T401C, S137A/E237A, and S137A/E237K ATs are described in the text as V317C/T401C, E237A, and E237K, respectively. S137A AT was used throughout as the control protein. Mutagenesis, expression, and purification were conducted as described previously (13). All variants exhibited the same two-peak elution profile from heparin-Sepharose, and for each variant the late eluting peak was used in all experiments (14). Purified material was stored at−80 °C. The concentration of AT was determined by absorbance at 280 nm and verified by stoichiometric titrations of 1 µM AT against a known concentration of pentasac-charide. Active concentrations corresponded to those determined by absorbance for all variants studied. The absence of significant amounts of inactive material was confirmed by an ability to form a stoichiometric complex with thrombin by SDS-PAGE.
Rate of Latency Transition
Control and V317C/T401C ATs were incubated at 50 °C in 50 mm NaPO4, 40% glycerol, pH 6.0 (conditions that promote transition to the latent form but limit polymer formation), and at the indicated times, samples were removed, snap-frozen, and stored at −80 °C. Remaining thrombin inhibitory activity was assessed for all time points in a 96-well plate as described previously (13).
Crystallization and Data Collection
Crystals of V317C/T401C AT were grown in hanging drops containing 2 µl of AT (7.4 mg/ml in 20 mm Tris pH 7.4) and 1 µl of precipitant solution (0.13 m NaI, 13% PEG 3350). After dehydration in increasing concentrations of PEG 3350 from 13 to 25% a single crystal was cryoprotected in 0.13 m NaI, 25% PEG 3350, 10% PEG 400 prior to mounting in a cryoloop and cooling in a vapor nitrogen stream to 100 K. The crystal was then annealed by blocking the nitrogen stream for 10 s. Data were collected from a single crystal at SRS Daresbury Station 9.6 (λ=0.87Å) on an ADSC Quantum 4 detector (Area Detector Systems Corp.). V317C/T401C was also crystallized as a dimer with latent AT. Hanging drops were set up containing 2µl of AT (10 mg/ml V317C/T401C, 10 mg/ml latent AT in 20 mm Tris, pH 7.4) and 2 µl of precipitant (17.5% PEG 4000, 50mm Na/KPO4 pH 6.7). A single crystal was cryoprotected by passing the crystal sequentially into solutions containing 0–16% glycerol with 20% PEG 4000, 50 mm Na/KPO4 pH 6.7. Data sets were collected from a single crystal at SRS Daresbury Station 14.1 (λ =1.488Å), on a ADSC Quantum 4 detector. Crystals of the V317C/T401C variant in the presence of heparin mimetic SR123781 and S195A thrombin were grown from 1:1 drops of protein (6.5 mg/ml in 50mm Tris, 210 mm NaCl, pH 7.4) and precipitant (0.16 m ammonium sulfate, 16% PEG 3350, 10% glycerol). The crystal was cryoprotected in 16% PEG 3350, 0.16 m ammonium sulfate, and 20% glycerol. A data set was collected from a single flash-cooled crystal after annealing by blocking the vapor nitrogen stream three times for 3 s followed by a single 10-s annealing (SRS Station 9.6). Resolution improved from~2.9 to 2.6 Å for the 3-s annealings and to 2.1 Å after the 10-s annealing.
Data Processing and Structure Refinement
Data were processed using Mosflm, Scala, and Truncate (15). The monomeric AT structure was solved by molecular replacement with the program MolRep (16) using the active component of 1E04 (17), and three molecules were placed in the asymmetric unit. After rigid body refinement, strict non-crystallographic symmetry was applied for refinement and rebuilding followed by restrained non-crystallographic symmetry for one round of refinement. The structure of the nonproductive AT-thrombin-heparin complex was solved using MolRep, with S195A thrombin (Protein Data Bank ID code 1JOU, molecule AB (18)) and pentasaccharide-bound AT (Protein Data Bank ID code 1E03) as search models. The heterodimeric structure of the V317C/T401C variant and latent AT was solved by simply conducting rigid body refinement on the protein component of 1E04. All refinement was conducted using the program CNS (19) (version 1.0), and the program XtalView (20) was employed for rebuilding. Data processing and refinement statistics are given in Table 1. The figures were made using Bobscript (21) and Raster3D (22). Buried surface area was calculated using the Protein-Protein Interaction Server.
TABLE 1.
Statistics for crystallographic data processing, refinement, and final models
| Monomer (1T1F) | Active/latent dimer (2BEH) | Complex (2B5T) | |
|---|---|---|---|
| Crystal | |||
| Space group | P21 | P21 | P1 |
| Cell dimensions (Å) | 153.84, 43.71,153.93 | 68.07, 97.27, 87.39 | 59.28, 59.53, 108.41 |
| Angles (α, β,γ) | β = 120.01 | β = 105.03 | 77.5, 78.94, 68.08 |
| Solvent content (%) | 54.4 | 50.7 | 52.8 |
| Data processing statistics | |||
| Wavelength (Å) | 0.87 (SRS 9.6) | 1.488 (SRS 14.1) | 0.87 (SRS 9.6) |
| Resolution (Å) | 44.40-2.75 (2.90-2.75) | 40.00-2.70 (2.85-2.70) | 35.71-2.10 (2.21-2.10) |
| Total reflections | 168,897 (10553) | 151,295 (15202) | 280,653 (15414) |
| Unique reflections | 44,274 (4862) | 30,217 (4359) | 71,994 (9773) |
| Multiplicity | 3.8 (2.2) | 5.0 (3.5) | 3.9 (1.6) |
| 〈I/σ (I)〉 | 5.6 (1.4) | 6.6 (1.6) | 7.9 (1.5) |
| 〈I〉/σ (〈I〉) | 11.9 (1.7) | 18.2 (2.4) | 12.6 (1.6) |
| Completeness (%) | 93.5 (71.8) | 99.7 (99.1) | 92.6 (85.9) |
| Rmerge | 0.112 (0.512) | 0.079 (0.459) | 0.075 (0.440) |
| Model | |||
| Atoms modeled | |||
| Protein | 9756 | 6231 | 8086 |
| Water | 216 | 17 | 389 |
| Carbohydrate | 285 | 123 | 171 |
| Glycerol | 18 | 6 | 72 |
| Iodide | 21 | ||
| Sulfate | 30 | ||
| Heparin | 278 | ||
| Average B-factor (Å2) | 59.70 | 63.60 | 43.30 |
| Refinement statistics | |||
| Resolution range | 38.46-2.75 (2.92-2.75) | 39.10-2.70 (2.87-2.70) | 35.70-2.10 (2.23-2.10) |
| Reflections work/free | 42,369/1,796 (5315/217) | 28,678/1,520 (4,695/245) | 68,987/2,937 (10,600/450) |
| R-factor/R-free (%) | 22.5/23.9 (37.3/37.3) | 21.9/26.5 (34.3/37.1) | 20.9/24.7 (32.3/32.0) |
| Ramachandran plot | |||
| Most favored (%) | 76.3 | 81.5 | 85.2 |
| Additionally allowed (%) | 20.6 | 16.6 | 13.7 |
| Generously allowed (%) | 3.2 | 1.9 | 1.1 |
| Disallowed (%) | 0 | 0 | 0 |
Rates and Stoichiometries of Inhibition
Rates and stoichiometries of protease inhibition for the AT variants were determined as described previously (13). The stoichiometries of inhibition were indistinguishable from one for all AT variants. Rate constants were averaged from at least three determinations.
Heparin Binding Studies
Equilibrium dissociation constants were determined essentially as described previously (13). Briefly, the intrinsic fluorescence of AT (15–50 nm) with increasing concentrations of pen-tasaccharide was measured at 340 nm, exciting at a wavelength of 280 nm, on a PerkinElmer Life Sciences 50B spectrofluorimeter at room temperature. All solutions were made up in 20 mm NaPO4, 100 mm NaCl, 0.1 mm EDTA, 0.1% PEG 8000, pH 7.4 (I = 0.15), and the NaCl concentration was increased to 250 mm to obtain an ionic strength of 0.3.
Thermal Denaturation Studies
The midpoints of thermal denaturation were taken as the X intercept of the second derivative of smoothed melts monitored by circular dichroism. Protein solutions at 0.8 mg/ml in 20 mm NaPO4 buffer, pH 7.4, were subjected to a temperature ramp from 25 to 90 °C on a Peltier controlled Jasco 815 spectropolarimeter, and ellipticity was monitored at a wavelength of 222 nm. At least three runs were averaged.
RESULTS
Disulfide Bond Prevents Latent Transition
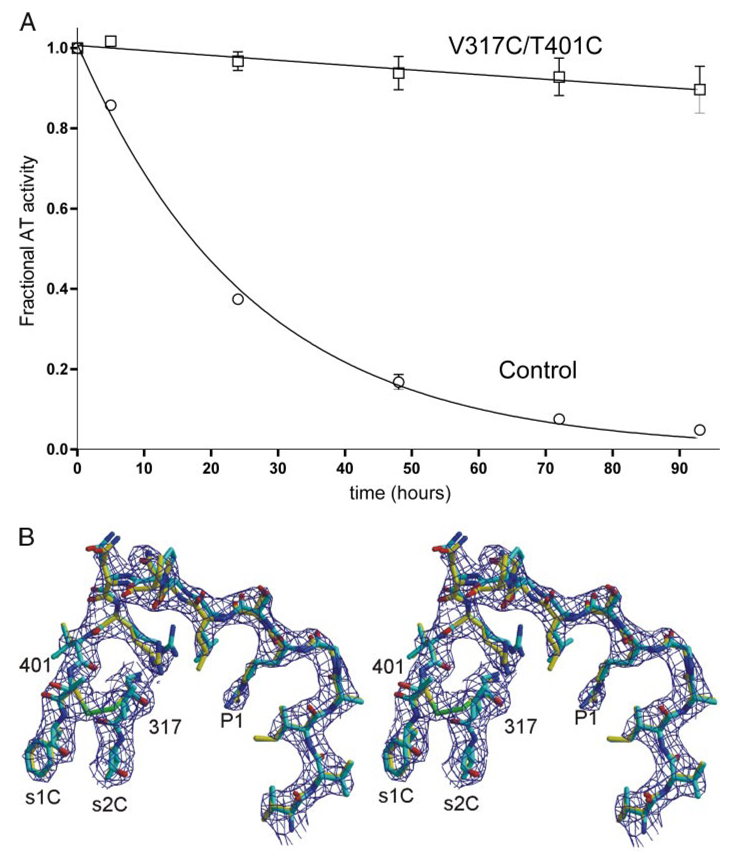
To prevent the conversion of active AT to the inactive latent form, we engineered a disulfide bond between strands 1 and 2 in β-sheet C. Adjacent residues Val-317 and Thr-401 were mutated to Cys, and as predicted, the resultant disulfide bond prevented conversion to the latent form (Fig. 2A). Correct formation of the disulfide bond was also verified by solving the crystal structure of the variant in presence of latent AT. The disulfide bond was observed in the predicted position, and in the context of the heterodimer, the new disulfide bond did not perturb the conformation of strands 1 or 2 of β-sheet C or of the adjacent RCL (Fig. 2B).
FIGURE 2. The engineered disulfide bond prevents the conversion to latent AT.
A, the conversion to latent AT can be followed by monitoring loss of inhibitory activity with time. Under the conditions used, control AT was converted to the latent form with a half-life of 18 h, whereas the V317C/T401C variant was fully resistant to the conformational transition. B, successful formation of the disulfide bond was further verified by solving the crystal structure of the V317C/T401C variant (yellow) in the context of the active/latent dimer. The RCL and parts of strands 1C and 2C are shown with surrounding electron density (contoured at 1 × the root mean squared deviation of the map, 1σ). The structure of native wild-type AT is superimposed for comparison (cyan), and the engineered disulfide bond is shown in green.
The V317C/T401C mutations did, however, have a subtle effect on the activity of the AT variant (Table 2). The pentasaccharide affinity of the variant was decreased by ~2-fold, and the basal rates of thrombin and factor Xa inhibition were both reduced by ~5-fold. The pentasaccharide-accelerated rates were also reduced by 1.6- and 3.8-fold for thrombin and factor Xa, respectively. As the properties of the individual cysteine variants were not determined, it was unclear whether the small reduction in activity was caused by the point mutations or the formation of the novel disulfide bond. The thermal stability of the variant was slightly increased relative to control (1.5 °C). A similar increase in Tm for another AT variant also resulted in a small loss of heparin affinity (5). Furthermore, the region in which the mutations were made (s1C and s2C) is known from crystal structures to be in close contact with proteases in Michaelis complexes with AT (23–25). It is thus unsurprising that these mutations had a small effect on the rates of protease inhibition.
TABLE 2.
Biochemical properties of AT variants
| Antithrombin | Pentasaccharide binding (Kd) | Penta | Rates of inhibition (kapp) | ||||
|---|---|---|---|---|---|---|---|
| I = 0.15 | I = 0.30 | Thrombin | Factor Xa | ||||
| nm | m−1 s−1 | ||||||
| Control | 3.5 ± 0.3 | 26 ± 0.3 | − | 7.3 ± 0.4 × 103 | 4.9 ± 0.5 × 103 | ||
| + | 1.10 ± 0.06 × 104 | 7.0 ± 0.3 × 105 | |||||
| V317C/T401C | 7.1 ± 1.1 | 62 ± 0.3 | − | 1.47 ± 0.04 × 103 | 9.6 ± 0.7 × 102 | ||
| + | 6.9 ± 0.2 × 103 | 1.86 ± 0.09 × 105 | |||||
| E237A | 1.8 ± 0.1 | 15 ± 3 | − | 1.4 ± 0.1 × 104 | 7.6 ± 0.5 × 103 | ||
| + | 1.4 ± 0.2 × 104 | 6.4 ± 0.4 × 105 | |||||
| E237K | 2.0 ± 0.1 | 13.5 ± 0.7 | − | 2.3 ± 0.2 × 104 | 1.4 ± 0.1 × 104 | ||
| + | 1.9 ± 0.2 × 104 | 7.5 ± 0.3 × 105 | |||||
Crystal Structure of MonomericAT
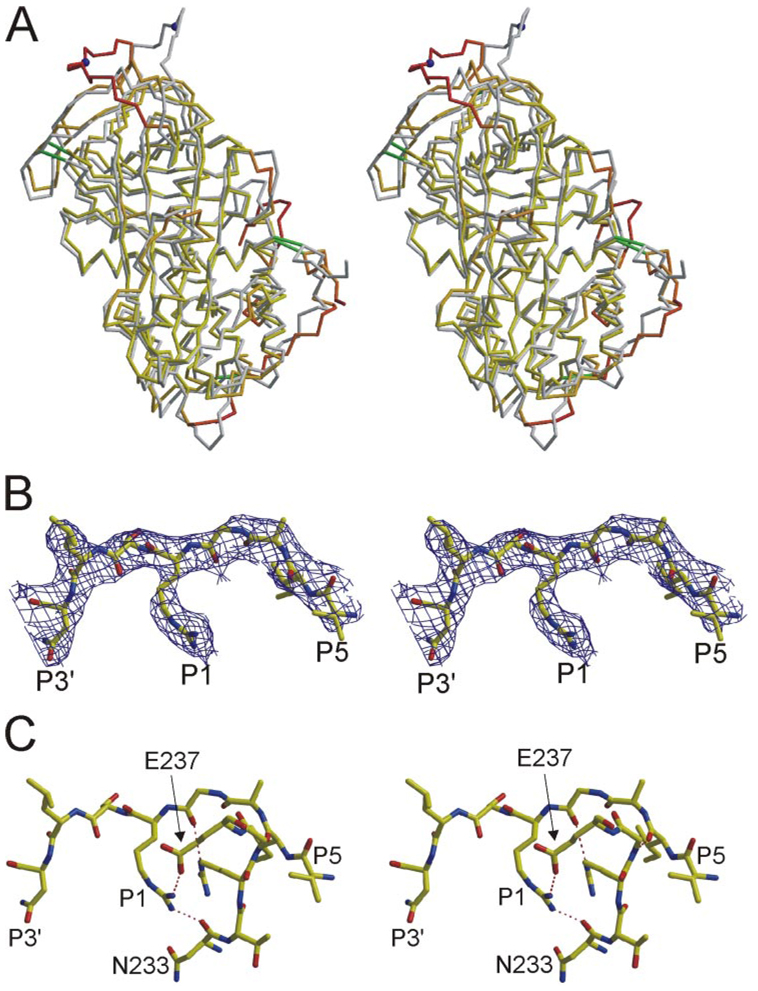
The structure of monomeric AT was solved from a single crystal, and data processing and refinement statistics are given in Table 1. The data were of good quality up to 2.75 Å resolution, and three monomers were found in the asymmetric unit. Due to the presence of pseudo symmetry, the three monomers were identical, and strict noncrystallo-graphic symmetry constraints were used until the last round of refinement. The overall structure is similar to the previous structure of native AT(Protein Data Bank ID code 1E04), with a root mean squared deviation of 1.8 Å for 416 equivalent Cα atoms. However, significant differences were observed in the heparin biding region and the RCL (Fig. 3A).
FIGURE 3. Stereo views of the structure of monomericATreveals a novel RCL conformation and contacts.
A, superimposed Cα traces of native AT from crystals of the heterodimer (gray) and of monomeric AT (oriented as in Fig. 1A) reveal regions of conformational difference. Monomeric AT is colored according to Cα root mean squared deviation (yellow to red, from 1 to 6 Å) with the active component of 1E04 (β-glycoform). Disulfide bonds are shown as green rods, and the P1 residue is indicated by a blue ball. Regions that differ most significantly are the RCL (top) and the heparin binding site (lower right). B, the RCL of monomeric AT (from P5 to P3′) is shown with corresponding electron density (contoured at 1σ). C, extensive intramolecular contacts are observed between the RCL and the body of AT; those involving the P1 Arg-393 are indicated by dashed lines.
Based on the structures of AT solved from crystals of the active/latent dimer, it was thought that the local conformational response to pentasaccharide binding was the elongation of helices A and D and the formation of a new helix P on the N-terminal helix D loop. However, in the structure of the monomeric form of native, AT we found that helix A was already elongated and that helix P was preformed. These apparent conformational differences probably reflect the inherent flexibility of the heparin binding site in native AT and are likely to be influenced by crystal contacts.
The RCL of monomeric AT is found partially incorporated into β-sheet A, as with other structures of native AT, but the remainder of the RCL has shifted position significantly. The P1 Arg-393 residue has shifted 17 Å toward the left in the classical view, and the side chain is oriented toward the body of AT making stabilizing contacts. The electron density of the RCL, shown in Fig. 3B, demonstrates the unequivocal placement of the Arg 393 side chain. Although the P1 Arg 393 makes only a single intramolecular hydrogen bond in the context of the AT heterodimer (Fig. 1D), in monomeric AT the P1 side chain participates in a salt bridge with Glu-237 and a hydrogen bond with the main chain oxygen of Asn-233 (Fig. 3C). The side chain of Arg-393 is in fact buried in a hydrophobic pocket described by Arg-235 and Tyr-253, in a position similar to that observed for Arg-150 of factor Xa in the crystal structure of the AT-factor Xa-pentasaccharide Michaelis complex (23). The interaction interface between the RCL (residues 383–395) and the body of AT is 628 Å2, 27% of which is accounted for by the burying of Arg-393.
Another Crystal Form Reveals Similar RCL Conformation and Contacts
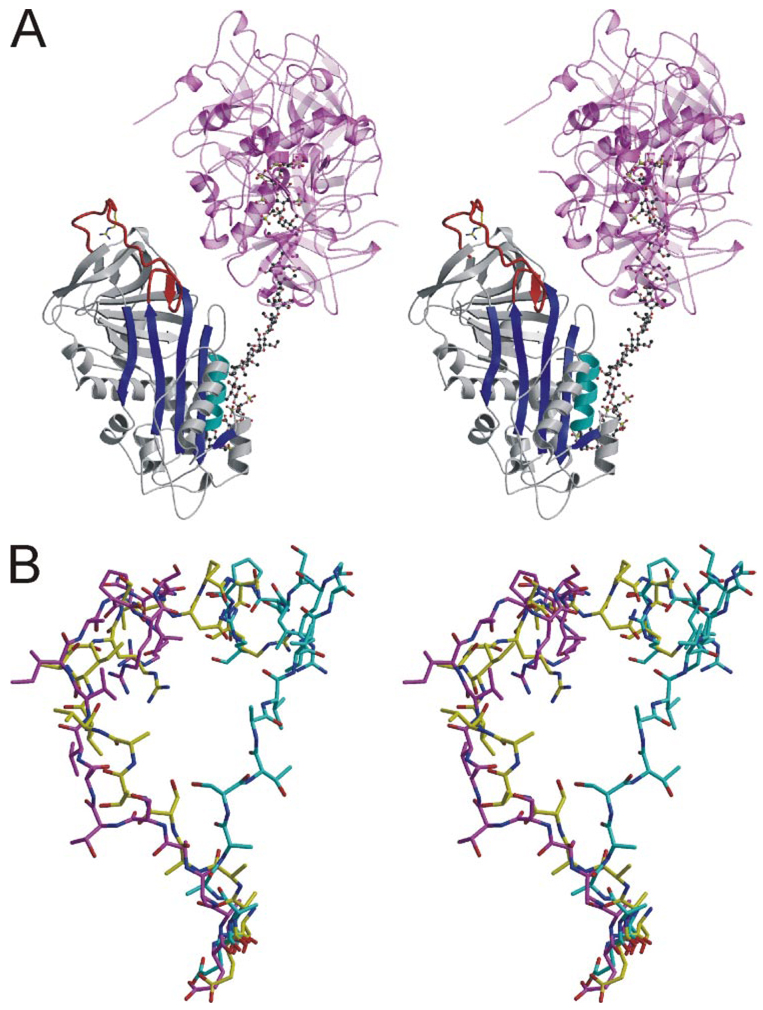
Although biochemical studies have established that the hinge region of AT is expelled from β-sheet A upon binding to its specific heparin pentasaccharide, we previously reported a structure of an AT-pentasaccharide complex in which the hinge region is in a native-like state (26). This “intermediate” structure was solved in the context of the active/latent heterodimer, and thus the RCL was in the same conformation observed for other AT structures. In pursuit of crystals of the ternary complex between AT, S195A thrombin, and heparin, we solved a high-resolution structure (2.1 Å) in which the RCL of AT did not interact with thrombin and the hinge region was inserted into β-sheet A in a native-like fashion (Fig. 4). This serendipitous structure was not useful for the understanding of how thrombin recognizes AT, but it did provide us with a crystal structure of the V317C/T401C AT variant with a native-like RCL in a different crystal form. The conformation of the RCL was similar to that observed in the native monomeric structure (Fig. 4B), with the P1 residue engaged in similar interactions. The salt bridge with Glu-237 and the hydrogen bond with the main chain oxygen of Asn-233 were both observed, strengthening the conclusion that these interactions are sampled in native wild-type AT and were not due to specific crystal contacts.
FIGURE 4. Structure of V317C/T401C AT from a different crystal form reveals similar RCL conformation and contacts.
A, stereo view of a ribbon diagram of the AT (colored as before), heparin (ball-and-stick), and S195A thrombin (semitransparent magenta and pink) complex. The RCL is clearly in a native-like conformation despite the presence of bound heparin. Two thrombin molecules bind to either side of the same site on heparin, and their active sites are occupied by the C terminus of adjacent light chains. B, the RCLs of native AT from the heterodimer (cyan) is compared with those of the V317C/T401C variant in the monomeric form (yellow) and in its nonproductive thrombin complex (magenta). The RCLs are shown from hinge region residue P14 (bottom) to P6′ (top).
Properties of AT Variants Support the Observed Salt Bridge between Arg-393 and Glu-237
Several P1 variants of AT have been characterized, and in each case that Arg-393 was substituted with a neutral amino acid, a small (2–4-fold) increase in heparin affinity resulted (8). This was presumably because the energetic cost of breaking a salt bridge between the P1 Arg and the body of AT was relieved (8). We can predict from the structure of AT in its monomeric form that charge neutralizing mutations on Glu-237 should mimic the effect observed for the P1 variants. In addition to the predicted increase in heparin affinity, we should also observe a small increase in rate of thrombin and factor Xa inhibition, reflecting the greater accessibility of the P1 Arg. We thus created E237A and E237K variants of AT and tested their heparin binding and inhibitory properties (Table 2). As predicted, we observed an ~2-fold increase in pentasaccharide binding affinity for both variants; the second-order rate constants of protease inhibition in the absence of heparin were also increased by ~2-fold. Interestingly, thrombin inhibition was not further accelerated by the pentasaccharide for the two variants, suggesting that the change in P1 accessibility normally caused by the binding of the pentasaccharide is already fully realized for the E237A and E237K variants. The similar magnitude of increase in pentasaccharide affinity through neutralization of either Arg-393 or Glu-237 supports their joint involvement in a salt bridge in native AT.
DISCUSSION
Our effort to determine the crystal structure of native AT in its monomeric state required a strategy to prevent the conformational transition to the latent form. We thus designed and demonstrated the formation of a disulfide bond between residue 317 on strand 2C and residue 401 on strand 1C, which prevented any loss of activity upon prolonged incubation at elevated temperature. Diffraction-quality crystals of this variant were obtained, and a new RCL conformation was observed in the resulting structure. However, the conformations of protein surface loops, such as the RCL of serpins, are generally susceptible to the influence of crystal contacts, and an observed conformation should be considered to represent only one of the possible solution conformers and not necessarily the predominant solution conformer. Therefore, several experimental approaches were undertaken to test the relevance of the RCL conformation and contacts observed in the structure of monomeric AT. One measure of the stability of an observed loop conformation is the B-factors of the main chain and side chain atoms, which give an indication of the flexibility of the residues that make up the surface loop. Interestingly, the B-factors for the RCL of the monomeric variant are higher than average from P12 to P2, but return to normal at the P1, suggesting that the side chain interactions observed for Arg-393 serve to anchor the RCL. This is consistent with the serendipitous structure of the same variant in another crystal form, which showed similar interactions between the side chain of Arg-393 with the same residues on the body of AT, despite a small shift in the position of the RCL. In addition, this structure demonstrates that AT adopts a very similar RCL conformation and forms the same interactions under the influence of different crystal contacts. These observations suggest that the N terminus of the RCL of native AT is somewhat flexible and that the contacts we observed between the side chain of Arg-393 and the body of AT serve to anchor the C-terminal portion of the RCL. However, the appreciable rates of protease inhibition for AT in the absence of activating heparin suggest that the P1 residue is at least partially available for proteolytic attack in native AT. The RCL of native AT thus probably exists in an equilibrium between at least two states, one with a buried and inaccessible P1 side chain and another with a fully solvent-accessible Arg-393 (Fig. 5).
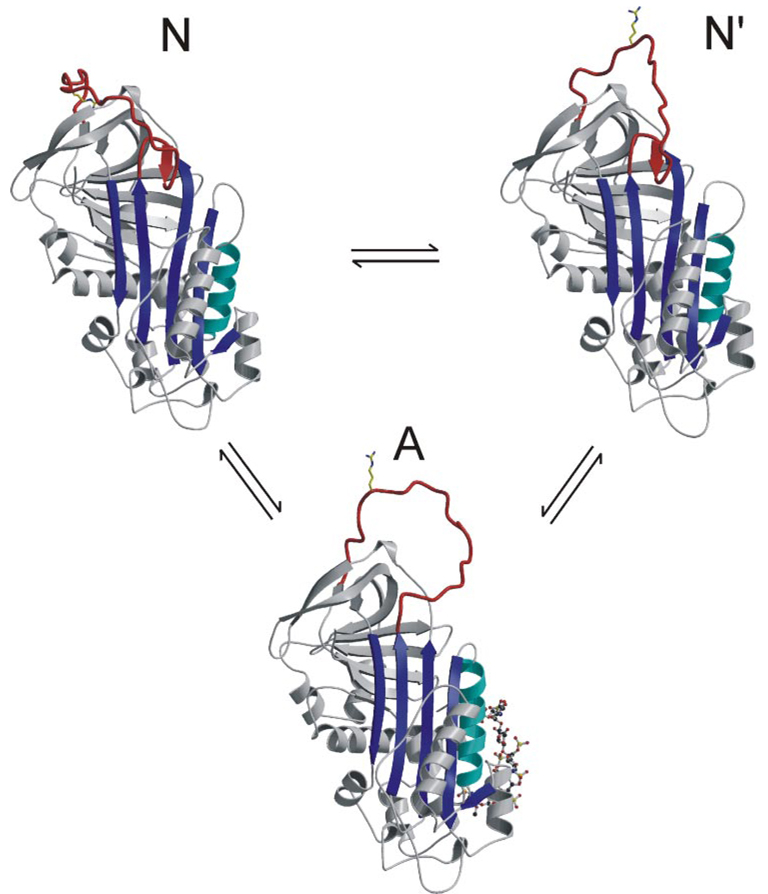
FIGURE 5. Equilibrium model for the conformational states of AT.
Similar to the revised model proposed by Chuang et al. (9), our data support a two-state conformational equilibrium for AT in the absence of heparin. Ribbon diagrams are given to represent the one activated and two native states. State N is the monomeric AT structure presented here with its RCL held close against the body of AT and the P1 side chain sequestered in the acidic pocked used as an exosite for factor Xa binding. This state would be nonreactive toward proteases but is in rapid equilibrium with state N′ (based on the structure of heterodimeric native AT), where there are fewer contacts to constrain the RCL and the P1 residue is free to interact with proteases. Activation of AT by the pentasaccharide results ultimately in the expulsion of the hinge and the liberation of the entire RCL. Monomer A is based on the structure of AT in complex with the pentasaccharide and S195A factor Xa (23).
A model for such a conformational equilibrium was proposed in an elegant paper by Steve Olson and colleagues (9) explaining the heparin binding properties of various P1 variants (similar to Fig. 5). One of the interesting properties of P1 variants is the absolute dependence on charge but not on other properties such as hydrophobicity or potential for hydrogen bonding. Whether His, Leu, Pro, or Trp, all P1 mutations lead to an ~2-fold increase in heparin affinity (8). The R393H variant (AT Glasgow) is of particular interest, because below the pKa of His, where the side chain is positively charged, the affinity returns to normal (8). Similarly, the deimination of P1 Arg leads to a 2-fold increase in affinity, even though the resulting isosteric citrulline side chain contains strong hydrogen bond donor and acceptor groups (10, 27). These data prove that the P1 residue is involved in a charge-dependent interaction in either the native or the activated state. As the activated state, by definition, must have an accessible P1 side chain, the properties of the P1 variants of AT strongly indicate the presence of an intramolecular salt bridge in native AT. The strikingly similar increase in heparin affinity upon neutralization (or reversal) of the negative charge on Glu-237 suggests a direct Arg-393/Glu-237 interaction. The crystal structures presented here reveal a direct salt bridge interaction between Arg-393 and Glu-237 and fully explain the mutagenesis data. In addition, the observed contact explains the previously enigmatic increase in heparin affinity observed for the naturally occurring variant AT-Truro (E237K) (27).
In conclusion, we have presented three structures of an AT variant, which because of an engineered disulfide bond, is resistant to the latent transition. In two unrelated crystals we observed similar interactions between the body of AT and the side chain of the reactive center residue Arg-393. Because the orientation of the P1 residue is of primary importance for recognition of the RCL by target proteases (in the absence of heparin (9)), the observed contacts help explain the poor inhibitory activity of circulating native AT. In addition, the site with which the P1 residue interacts in native AT is the principal exosite for factor IXa and Xa recognition (23), further contributing to the low basal rate of protease inhibition. Our data support the hypothesis that the RCL of native AT exists in dynamic equilibrium between at least two conformations (Fig. 5), one in which the P1 side chain is free from intramolecular contacts and can be recognized by proteases and another in which the RCL is anchored by the specific Arg-393 interactions observed in the crystal structure of monomeric AT described here.
Acknowledgment
We thank Robin Carrell for his comments on this manuscript.
This work was supported by the British Heart Foundation, National Institutes of Health Grant R01 HL68629, and the Medical Research Council (UK).
The atomic coordinates and structure factors (code 1T1F, 2BEH, and 2B5T) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: AT, antithrombin; RCL, reactive center loop; PEG, polyethylene glycol; SRS, Synchrotron Radiation Source.
The proteolytic substrate numbering scheme of Schechter and Berger (28) is used, where residues N-terminal to the scissilebondare numbered from P1 and those C-terminal are numbered from P1′.
REFERENCES
- 1.Olson ST, Chuang YJ. Trends Cardiovasc Med. 2002;12:331–338. doi: 10.1016/s1050-1738(02)00183-4. [DOI] [PubMed] [Google Scholar]
- 2.Huntington JA. In: Chemistry and Biology of Heparin and Heparan Sulfate. Garg HG, Linhardt RJ, Hales CA, editors. Oxford: Elsevier; 2005. pp. 367–398. [Google Scholar]
- 3.Carrell RW, Stein PE, Fermi G, Wardell MR. Structure (Lond.) 1994;2:257–270. doi: 10.1016/s0969-2126(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 4.Schreuder HA, de Boer B, Dijkema R, Mulders J, Theunissen HJ, Grootenhuis PD, Hol WG. Nat. Struct. Biol. 1994;1:48–54. doi: 10.1038/nsb0194-48. [DOI] [PubMed] [Google Scholar]
- 5.Huntington JA, Gettins PG. Biochemistry. 1998;37:3272–3277. doi: 10.1021/bi972182o. [DOI] [PubMed] [Google Scholar]
- 6.Huntington JA, Olson ST, Fan B, Gettins PG. Biochemistry. 1996;35:8495–8503. doi: 10.1021/bi9604643. [DOI] [PubMed] [Google Scholar]
- 7.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen MC, Beresford CH, Carrell RW. FEBS Lett. 1988;231:317–320. doi: 10.1016/0014-5793(88)80841-x. [DOI] [PubMed] [Google Scholar]
- 9.Chuang YJ, Swanson R, Raja SM, Bock SC, Olson ST. Biochemistry. 2001;40:6670–6679. doi: 10.1021/bi002933d. [DOI] [PubMed] [Google Scholar]
- 10.Pike RN, Potempa J, Skinner R, Fitton HL, McGraw WT, Travis J, Owen M, Jin L, Carrell RW. J. Biol. Chem. 1997;272:19652–19655. doi: 10.1074/jbc.272.32.19652. [DOI] [PubMed] [Google Scholar]
- 11.Zhou A, Stein PE, Huntington JA, Carrell RW. J. Biol. Chem. 2003;278:15116–15122. doi: 10.1074/jbc.M211663200. [DOI] [PubMed] [Google Scholar]
- 12.Mushunje A, Zhou A, Carrell RW, Huntington JA. Blood. 2003;102:4028–4034. doi: 10.1182/blood-2003-05-1560. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DJ, Huntington JA. J. Biol. Chem. 2004;279:4913–4921. doi: 10.1074/jbc.M311644200. [DOI] [PubMed] [Google Scholar]
- 14.Garone L, Edmunds T, Hanson E, Bernasconi R, Huntington JA, Meagher JL, Fan B, Gettins PG. Biochemistry. 1996;35:8881–8889. doi: 10.1021/bi960542m. [DOI] [PubMed] [Google Scholar]
- 15.Leslie AWG. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography. No. 26. Warrington, UK: Daresbury Laboratory; 1992. [Google Scholar]
- 16.Vagin A, Teplyakov A. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 17.McCoy AJ, Pei XY, Skinner R, Abrahams JP, Carrell RW. J. Mol. Biol. 2003;326:823–833. doi: 10.1016/s0022-2836(02)01382-7. [DOI] [PubMed] [Google Scholar]
- 18.Huntington JA, Esmon CT. Structure (Camb.) 2003;11:469–479. doi: 10.1016/s0969-2126(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 19.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.McRee DE. J. Mol. Graph. 1992;10:44–46. [Google Scholar]
- 21.Esnouf RM. J. Mol. Graph. Model. 1997;15:132. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 22.Merritt EA, Murphy MEP. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DJ, Li W, Adams TE, Huntington JA. EMBO J. 2006;25:2029–2037. doi: 10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Johnson DJ, Esmon CT, Huntington JA. Nat. Struct. Mol. Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 25.Dementiev A, Petitou M, Herbert JM, Gettins PG. Nat. Struct. Mol. Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DJ, Huntington JA. Biochemistry. 2003;42:8712–8719. doi: 10.1021/bi034524y. [DOI] [PubMed] [Google Scholar]
- 27.Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM. J. Mol. Biol. 2000;301:1287–1305. doi: 10.1006/jmbi.2000.3982. [DOI] [PubMed] [Google Scholar]
- 28.Schechter I, Berger A. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]