Summary
Nucleotide oligomerization domain (NOD)-like receptors (NLRs) are a specialized group of intracellular proteins that play a critical role in the regulation of the host innate immune response. NLRs act as scaffolding proteins that assemble signaling platforms that trigger nuclear factor-κB and mitogen-activated protein kinase signaling pathways and control the activation of inflammatory caspases. Importantly, mutations in several members of the NLR family have been linked to a variety of inflammatory diseases consistent with these molecules playing an important role in host-pathogen interactions and the inflammatory response. In this review, we focus on the role of Nod1 and Nod2 in host defense and in particular discuss recent finding regarding the role of Nlrc4, Nlpr1, and Nlrp3 inflammasomes in caspase-1 activation and subsequent release of proinflammatory cytokines such as interleukin-1β.
Keywords: caspase-1, IL-1, innate immunity, inflammasome, NOD2, NLRs
Introduction
The immune system deals with the daunting task of regulating the encounter of the host with microbes, allowing useful partnerships with commensal bacteria but rejecting potentially harmful pathogens. The innate immune system, the first line of defense, is involved in the initial detection and removal of harmful microbes. Unlike immune reactions induced through the adaptive immune system, innate immune responses are activated within minutes upon encounter with invading organisms. While the mechanisms that allow symbiotic relationships with commensal bacteria remain poorly understood, our knowledge about how the innate immune system senses microbial pathogens has greatly expanded in the last decade. A main conceptual advance has been the discovery of host germline-encoded pathogen recognition receptors (PRRs) that recognize conserved microbial structures often referred to pathogen-associated molecular patterns (PAMPs) (1). These PAMPs often play a critical function in the life of the microbe and include lipopolysaccharide (LPS), a major component of the outer layer of Gram-negative bacteria, peptidoglycan (PGN), the main component of the cell wall of Gram-positive bacteria, flagellin, and microbial nucleic acids. PRRs comprise an array of sensors present in the plasma, plasma membranes, and host cytosol. The existence of multiple PRRs capable of recognizing a single microorganism ensures the induction of immune responses even when a sensor or its signaling pathway is targeted by the pathogen. In addition, the activation of multiple PRRs in response to a pathogen often results in a combinatorial code that specifically tailors the host response to a particular class of microbes (2, 3). There is mounting evidence that certain PRRs are involved not only in sensing microorganisms but also endogenous non-microbial ‘danger’ signals (4). The activation of PRRs by microbial or endogenous stimuli results in the activation of multiple signaling pathways including nuclear factor-κB (NF-κB), mitogen-activated protein kinases (MAPKs), and type I interferon (IFN) response, which lead to the induction of proinflammatory and anti-microbial responses (5).
Most of our knowledge about PRRs derives from studies of Toll-like receptors (TLRs), the first class of cellular PRRs to be identified. Toll receptors were first defined in the fruit fly and localize either at the cell surface or within endosomes (6, 7). In contrast, the nucleotide oligomerization domain (Nod)-like receptors (NLRs) and the retinoid acid-inducible gene-I (RIG-I)-like receptors (RLRs) are intracellular cytosolic sensors. RLRs are helicases that sense primarily viruses (8, 9). This review focuses on NLRs, which are primarily involved in bacterial recognition. An important finding has been the discovery that immune dysregulation linked to genetic variation in NLR genes causes disease or is associated with increased susceptibility to several inflammatory diseases. NLRs were first identified in plants where they play a critical function in disease resistance (R genes) against microbial and parasite pathogens (10, 11). Homologues of the NLRs are present in vertebrates and phylogenetically more primitive organisms such as the sea urchin, whose genome contains at least 203 putative NLRs (12). The evolutionary conservation of the NLRs suggests that they play an important function in host defense.
The NLR family
There are 23 NLR family members in humans and at least 34 NLR genes in mice (Table 1). NLRs are expressed in many cell types including immune cells and epithelial cells, although certain NLR family members are expressed primarily in phagocytes including macrophages and neutrophils. NLRs are multi-domain proteins composed of a variable N-terminal effector region consisting of caspase recruitment domain (CARD), pyrin domain (PYD), acidic domain, or baculovirus inhibitor repeats (BIRs), a centrally located NOD that is critical for activation, and C-terminal leucine-rich repeats (LRRs) that senses PAMPs. The NOD domain (also referred as the NACHT cassette) is closely related to the oligomerization module found in the AAA+ family of adenosine triphosphatases (ATPases) (13). Mutation of conserved residues in the Walker’s A and B boxes of Nod1 and Nod2 proteins abolished signaling, indicating that nucleotide hydrolysis is essential for protein activation (14). Consistently, purified NLRP3 and NLRP12 bind adenosine triphosphate (ATP) and exhibit ATPase activity (15, 16). However, the precise molecular function of ATP hydrolysis in the regulation of NLR proteins is presently unknown.
Table 1.
NLR family members.
| HGNC-approved symbol |
Other names and aliases | Microbial motifs recognized | NLR family | |
|---|---|---|---|---|
| Human | Mouse | |||
| CIITA | Cllta |
NLRA; MHC2TA; C2TA Nlra; MHC2TA; C2TA |
NLRA | |
|
| ||||
| NAIP | NLRB1; BIRC1; CLR5.1 | |||
| Naip1 | Birc1a | |||
| Naip2 | Birc1b | |||
| Naip3 | Birc1c | NLRB | ||
| Naip4 | Birc1d | |||
| Naip5 | Birc1e | Flagellin from Legionella (C- terminal residue) | ||
| Naip6 | Birc1f | |||
| Naip7 | Birc1g | |||
|
| ||||
| NOD1 | NLRC1; CARD4; CLR7.1 | GM-tripeptide γ-D-Glu-DAP(iEDAP) D-lactyl-L-Ala-γ-Glu-meso-DAP-Gly (FK156) heptanolyl-γ-Glu-meso-DAP-Ala (FK565) |
NLRC | |
| Nod1 | Nlrc1; Card4 | |||
| NOD2 | NLRC2; CARD15; CD; | MDP | NLRC | |
|
| ||||
| BLAU; IBD1; PSORAS1; CLR16.3 | MurNAc-L-Ala-g-D-Glu-L-Lys (M-TRILys) | |||
| Nod2 | Nlrc2; Card15 | |||
| NLRC3 | NOD3; CLR16.2 | NLRC | ||
| Nlrc3 | CLR16.2 | |||
| NLRC4 | CARD12; CLAN; | Flagellin from Salmonella, | NLRC | |
| CLR2.1; IPAF | Legionella, Listeria, Pseudomonas | |||
| Nlrc4 | Card12; CLAN; Ipaf | |||
| NLRC5 | NOD27; CLR16.1 | NLRC | ||
| NIrc5 | ||||
|
| ||||
| NLRP1 | NALP1; DEFCAP; NAC; CARD7; CLR17.1 | MDP, Lethal Toxin ? | ||
| Nlrp1a | NALP1a | NLRP | ||
| Nlrp1b | NALP1b | Lethal Toxin | ||
| Nlrp1c | NALP1c | |||
| NLRP2 | NALP2; PYPAF2; NBS1; PAN1; CLR19.9 | NLRP | ||
| Nlrp2 | Pypaf2; Nbs1; Pan1 | |||
| NLRP3 | CIAS1; PYPAF1; Cryopyrin; NALP3; CLR1.1 | bacterial RNA viral RNA uric acid crystals LPS LTA MDP |
NLRP | |
| Nlrp3 | Cias1; Pypaf1; Cryopyrin; Nalp3; Mmig1 | |||
|
| ||||
| NLRP4 | NALP4; PYPAF4; PAN2; RNH2; CLR19.5 | |||
| Nlrp4a | Nalp4a; Nalp-eta; Nalp9D | |||
| Nlrp4b | Nalp4b; Nalp-gamma; Nalp9E | NLRP | ||
| Nlrp4c | Nalp4c; Nalp-alpha; Rnh2 | |||
| Nlrp4d | Nalp4d; Nalp-beta | |||
| Nlrp4e | Nalp4e; Nalp-epsilon | |||
| Nlrp4f | Nalp4f; Nalp-kappa; Nalp9F | |||
| Nlrp4g | Nalp4g | |||
| NLRP5 | NALP5; PYPAF8; MATER; PAN11; CLR19.8 | NLRP | ||
| Nlrp5 | Mater; Op1 | |||
| NLRP6 | NALP6; PYPAF5; PANS; CLR11.4 | NLRP | ||
| Nlrp6 | ||||
| NLRP7 | NALP7; PYPAF3; NOD12; PAN7; CLR19.4 | NLRP | ||
| NLRP8 | NALP8; PAN4; NOD16; CLR19.2 | NLRP | ||
|
| ||||
| NLRP9 | NALP9; NOD6; PAN12; CLR19.1 | |||
| Nlrp9a | Nalp9a; Nalp-theta | NLRP | ||
| Nlrp9b | Nalp9b; Nalp-delta | |||
| Nlrp9c | Nalp9c; Nalp-zeta | |||
| NLRP 10 | NALP10; PAN5; NOD8; PYNOD; CLR11.1 | NLRP | ||
| Nlrp10 | Nalp10; Pynod | |||
| NLRP 11 | NALP11; PYPAF6; NOD17; PAN10; CLR19.6 | NLRP | ||
| NLRP12 | NALP12; PYPAF7; Monarch1; RNO2; PAN6; CLR19.3 | NLRP | ||
| Nlrp12 | Nalp12 | |||
| NLRP13 | NALP13; NOD14; PAN13; CLR19.7 | NLRP | ||
| NLRP14 | NALP14; NOD5; PAN8; CLR11.2 | NLRP | ||
| Nlrp14 | Nalp14; Nalp-iota; GC- LRR | |||
|
| ||||
| NLRX1 | NOD9; CLR11.3 | NLRX | ||
The N-terminal domain of the NLRs mediates signaling through its interaction with downstream factors. CARD and PYD domains belong to the death domain-fold superfamily, which are involved in several cellular processes including apoptosis and inflammation. Both CARD and PYD domains mediate homophilic interactions with other CARD and PYD-containing proteins. BIR-containing proteins were originally identified as regulators of apoptosis in the so-called inhibitor of apoptosis proteins (IAPs). A class of IAPs termed neuronal apoptosis inhibitor proteins (NAIPs) are NLR family members. A nomenclature that subdivides the NLR family into five subfamilies (A–E) based on the class of N-terminal effector domain has been recently proposed and approved by the Human Genome Organization Gene Nomenclature Committee (17). Four subfamilies are defined for the presence of N-terminal CARD, PYD, BIR, and acidic domains (18, 19). An additional subfamily, NLRX, characterized by the presence of a N-terminal domain without significant homology to any known domains and by its localization to the mitochondria, has been recognized (20, 21).
Nod1 and Nod2 signaling
Nod1 and Nod2 were first discovered as mammalian members of the Ced4/Apaf-1 family of apoptosis regulators (22, 23). Nod1 and Nod2 (Fig. 1) are multi-domain proteins consisting of one or two CARD domains respectively, a centrally located NOD domain followed by a number of C-terminal LRRs in contrast to apoptosis protease activating factor-1 (Apaf-1), which contains C-terminal WD40 repeats. Nod1 and Nod2 orthologs are found across higher order vertebrates but are absent in insects or worms. Nod1 is widely expressed in many cell types and organs. Although Nod2 expression is believed to be more restricted, it has been described in macrophages (23), dendritic cells (24), Paneth cells (25), keratinocytes (26), and epithelial cells of the intestine (27), lung (28), and oral cavity (29). Both Nod1 and Nod2 are expressed in the cytosol, but more recently a fraction of Nod1 (30) and Nod2 (31) has been shown to be associated with the plasma membrane. In the case of Nod2, point mutants that prevent membrane association exhibit a decreased ability to activate downstream signaling (31). However, the same mutations are known to affect microbial recognition and secondarily Nod2 activation, so the role of membrane localization in Nod2 signaling remains unclear.
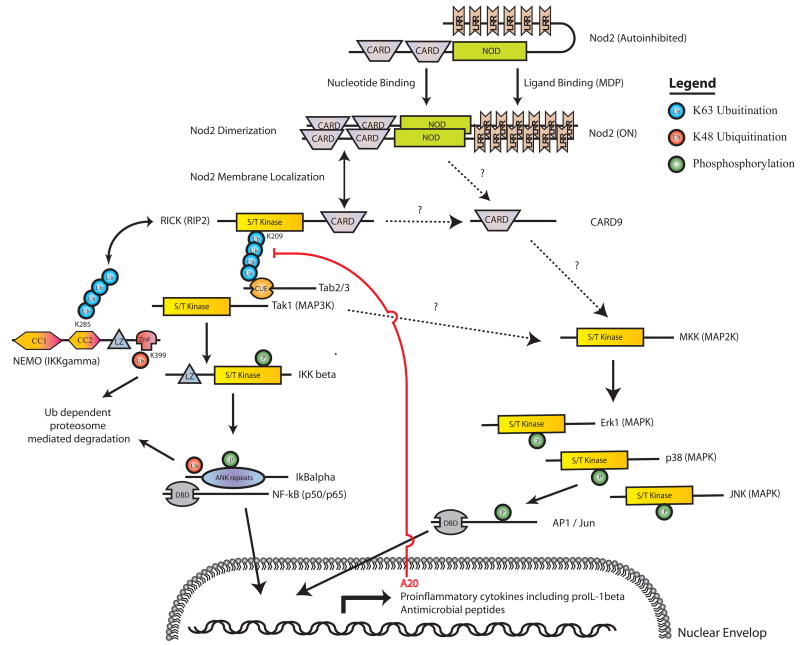
Fig. 1. Schematic representation of the Nod2 signaling pathway.
The NLR Nod2 recognizes bacterial PGN fragments such as MDP and activates NF-κB and MAPK signaling via the indicated proteins leading to the transcriptional upregulation of a variety of proinflammatory cytokines and antimicrobial peptides. Negative feedback regulation occurs upon induction of the deubiquitanase A20. The posttranslational regulation of various components of the pathway is indicated by green circles (phosphorylation) and red or blue circles (ubiquitination). In the case of RICK, K63-mediated polyubiquitination at K209 is essential for recruitment of Tak1. For NEMO, K48-mediated ubiquitination at residue K399 targets it for proteasome-dependent degradation. K63-linked polyubiquitination at amino acid K285 is important for complex formation with RICK (48).
NLRs including Nod1 and Nod2 are thought to be kept in an inactive state by intra-molecular interactions. Experimental evidence supporting this model includes the observation that truncation or mutation of the C-terminal LRR region of a variety of NLRs typically result in a constitutively active form of the receptor (14). In the case of Nod2, a C-terminal fragment consisting of only the LRRs interacts with an N-terminal fragment consisting of the CARD and NOD domains, when co-expressed in cells, an interaction which is disrupted in the presence of the Nod2 bacterial ligand (32). Collectively, the results have suggested a model in which ligand recognition results in a conformation change in the protein relieving these autoinhibitory intramolecular interactions and allowing NOD domain-dependent nucleotide binding and oligomerization. However, evidence for a direct interaction between NLRs including Nod1 and Nod2 and their putative ligands is still lacking, so it is possible that the activation of Nod1 and Nod2 by microbial stimulation is indirect. Understanding of Nod1 and Nod2 activation has been hampered by the lack of structural data of NLR family members which is only limited to the N-terminal effector domains in isolation (33, 34). Clearly, more structural, biochemical, and functional results are needed to confirm these models.
Stimulation of Nod1 or Nod2 results in the activation of NF-κB and MAPKs, which drive the transcription of numerous genes involved in both innate and adaptive immune responses (Fig. 1) (35). Both Nod1 and Nod2 sense bacterial molecules produced during the synthesis, degradation, and remodeling of PGN, a major component of bacterial cell walls. PGN is a polymer composed of glycan chains of alternating N-acylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units cross-linked to each other by short peptides. The cross-linking of two parallel glycan chains is mediated by stem peptides that can be further linked by bridging amino acids (36). Nod2 senses muramyl dipeptide (MDP), which is found in the PGN of nearly all Gram-positive and Gram-negative organisms (36), whereas Nod1 recognizes meso-diaminopimelic acid (meso-DAP)-containing PGN fragments (37, 38). DAP is an usual amino acid residue that is unique to PGN from most Gram-negative bacteria and certain Gram-positive bacteria, including the genus Listeria and Bacillus species (39). Analysis of synthetic compounds revealed the dipeptide γ-D-glutamyl-meso-DAP (iE-DAP) as the core motif that is sufficient to trigger Nod1 activation (37, 38). Nod1 and Nod2 null cells or mice are unable to activate NF-κB and MAPKs as well as to produce cytokines and chemokine in response to ligand stimulation (40–45).
Following microbial sensing, Nod1 or Nod2 directly recruit the serine-threonine kinase RICK [also known as receptor-interacting protein 2 (Rip2)] through CARD-CARD interactions (22, 23). A critical function of Nod1 and Nod2 signaling appears to be the induced proximity of RICK molecules through oligomerization of Nod1 and Nod2 (46). The inability of RICK null cells or mice to activate NF-κB signaling downstream of Nod1/2 ligand stimulation indicates that RICK is required in vivo for signaling (40, 42). Interestingly, the kinase activity of RICK does not appear to be essential for NF-κB activation, although it is required for protein stabilization (47). RICK directly binds to NF-κB essential modulator (NEMO) [also known as inhibitor of NF-κB kinase γ (IKKγ)], the regulatory subunit of IKK, and promotes its ubiquination and activation of the catalytic subunits IKKα and IKKβ (46, 48). Once activated, IKK phosphorylates the inhibitor IκB, leading to its degradation via the proteasome releasing NF-κB and allowing it to translocate to the nucleus (35). In addition, RICK mediates the recruitment of transforming growth factor β-activated kinase (TAK1) in an ubiquitin-dependent manner, which is essential for IKK activation (47, 49). Both Nod1 and Nod2 induce K63-linked polyubiquitination of RICK that is necessary for TAK1 recruitment via its N-terminal ubiquitin-binding CUE domain (47, 49). TAK1, a serine-threonine kinase that forms a complex with the ubiquitin binding proteins TAK1-binding protein 1 (Tab1), Tab2, and/or Tab3, activates IKK by phosphorylation in response to a variety of stimuli including interleukin-1β (IL-1β), tumor necrosis factor (TNF), and several TLR ligands (50–52). In addition to NF-κB signaling, TAK1 is also required for the upregulation of Nod2 in response to MDP stimulation (49). Similar to the NF-κB pathway, TAK1 and RICK are required for Nod1- and Nod2-mediated MAPK activation, although the intermediate steps in this pathway are less well characterized (42, 53, 54). Signaling through NF-κB and MAPKs produces a robust inflammatory response. Stimulation of Nod1 or Nod2 results in the secretion of proinflammatory cytokines and chemokines [IL-6, CXCL8/IL-8, CXCL1/KC, CXCL2/macrophage inflammatory protein-2 (MIP-2), CCL2, CCL5/regulated upon activation, normal T-cell expressed, and presumably secreted (RANTES)] (55–60). Nod1 activation induces neutrophil recruitment in vivo (60), and both Nod1 and Nod2 signaling lead to the production of antimicrobial peptides (26, 28, 61, 62).
Although a number of downstream mediators of Nod1 and Nod2 signaling have been identified, the molecular details of how these signaling pathways are downregulated are less well understood and are just beginning to emerge. For example, an alternatively transcribed CARD domain only isoform of Nod2 appears to function as a dominant negative regulator (63). Additionally, disruption of the Nod2/RICK CARD-CARD interaction appears to be the molecular mechanism by which caspase-12 (64) and MEKK4 (65) downregulate Nod2 signaling. Thirdly, a negative regulatory role for the cell polarity protein, Erbin, has been proposed based on its ability to associate with Nod2 and to inhibit NF-κB activation and subsequent inflammatory cytokine secretion (66, 67). Finally, deubiquination of RICK by the A20 protein has been shown to downregulate Nod1/2-induced NF-κB signaling by inhibiting RICK ubiquitination (47, 68). A20 has a conserved role in dampening TNF and TLR signaling via RIP1 (69–71), indicating that A20 acts broadly to suppress proinflammatory pathways
Nod1 and Nod2 in bacterial infections
Although the Nod1 ligand iE-DAP is primarily found in Gram-negative bacteria, several Gram-positive bacteria including Listeria monocytogenes and Bacillus species also produce the Nod1 agonist (72–74). Most of the Nod1 agonist activity produced by bacteria is found in the culture supernatant of growing bacteria (74). This finding is consistent with the observation that the cell wall of Gram-negative bacteria is remodeled by hydrolases that degrade intact PGN into smaller fragments (75). Gram-negative bacteria have evolved mechanisms for importation and recycling of PGN fragments released during bacterial growth (76). Shigella mutants that are defective in the recycling of PGN fragments exhibit increased Nod1-dependent NF-κB activation (77). Collectively, these results suggest that Nod1 senses bacterial growth through the recognition of PGN fragments released by growing bacteria.
Nod1 and Nod2 are involved in the sensing of numerous pathogenic bacteria. Nod1 detects the Gram-negative Shigella flexneri (78), enteroinvasive Escherichia coli (56), Chlamydia (55, 58, 72), Pseudomonas aeruginosa (57), Campylobacter jejuni (62), and Helicobacter pylori (61, 79). Although there is compelling evidence that Nod1 mediates bacterial sensing, in vivo observations confirming the importance of Nod1 in host defense are less abundant. While Nod1 senses Chlamydia in vitro, studies using vaginally infected mice revealed no in vivo role for Nod1 (55). Similarly, evidence for a role of Nod1 in host defense against enteric pathogens is lacking with the exception of H. pylori, a human pathogen involved in the development of gastritis, duodenal ulcers, and stomach cancer. Mice deficient in Nod1 have greater bacterial loads after intragastric infection with H. pylori (79), which might be explained, at least in part, by impaired expression of the antimicrobial peptide β-defensin 4 (61). Although H. pylori is a noninvasive bacterium, it activates Nod1 by injecting PGN fragments into epithelial cells via a type IV secretion system (79). Consistently, mutant H. pylori lacking the cag pathogenicity island that encodes the secretion apparatus are not detected by Nod1 (79).
The mechanisms by which PGN fragments enter host cells to induce Nod1 and Nod2 activation are poorly understood. There is evidence that Nod1 agonists can enter the cytosol of epithelial cells via pore-forming toxins produced by bacteria. For example, Staphylococcus aureus, Bacillus anthrasis, and Streptococcus pneumonia secrete toxins that mediate cytosolic sensing of Nod1 agonists (80). Neither S. pneumonia nor Haemophilus influenza alone activates Nod1. However, when co-cultured with epithelial cells, the pneumolysin of S. pneumonia promotes the cell uptake of iE-DAP-containing fragments released by H. influenza, leading to Nod1 activation (80). In addition to the pathogen secretion systems and pore-forming toxins described above, there is evidence that Nod1 or Nod2 ligands produced by bacteria enter the cell via endocytosis and epithelial transporters. It has been demonstrated that MDP is internalized by macrophages and localized to acidified vesicles (81), suggesting that uptake of the Nod2 is mediated via endocytosis, although the mechanism involved remains unclear. The transporter PepT1 selectively delivers MDP but not iE-DAP to the cytosol (82), while the reverse is true for the recently described PepT2 transporter (83). Interestingly, PepT1 and PepT2 have different tissue-specific expression patterns, which may lead to differential responses to extracellular Nod1 and Nod2 ligands (84). The expression of PepT1 is largely confined to intestinal epithelial cells, so it is unclear whether this transporter regulates MDP uptake in innate immunity cells such as monocytes and macrophages where Nod2 is primarily expressed. Furthermore, a role for PepT1 and PepT1 in MDP-induced signaling in vivo remains to be investigated.
Similar to Nod1, in vitro studies have identified a variety of pathogens that activate Nod2 including S. pneumonia (85), Mycobacterium tuberculosis (86), S. aureus (87), Salmonella typhimurium (27), and S. flexneri (66). The in vivo relevance of Nod2 during host defense has been clearly demonstrated using Nod2 null mice infected orally with Gram-positive L. monocytogenes, which were more susceptible to infection, had increased bacterial load, and exhibited diminished expression of certain antimicrobial α-defensin peptides relative to wildtype controls (41). Similarly, Nod2-deficient mice exhibited a slight increase in the susceptibility to oral infection with Yersinia pseudotuberculosis (88). Yet, there is no difference in cytokine responses of wildtype and Nod2-deficient macrophages to infection with Y. pseudotuberculosis in vitro (44). Collectively, the results suggest that Nod1 and Nod2 have a redundant role with TLRs in the recognition of bacteria and contribute to the production of proinflammatory and anti-microbial molecules. Consistently, the function of Nod1 and Nod2 appears to be important when TLR signaling is absent or reduced.
Nod1 and Nod2 crosstalk with the innate and adaptive immune system
Recognition of bacteria induces the activation of multiple PRRs including NLRs and TLRs. There is evidence that Nod1 and Nod2 cooperate with TLRs in shaping the host response to bacteria. This is not surprising given that Nod1, Nod2, and TLR signaling converge to common pathways including NF-κB and MAPK activation. A reasonable purpose for having multiple PRRs which induce overlapping signaling pathways is to increase the sensitivity for pathogen detection and to potentiate the cellular response against invading bacteria. Consistently, there is extensive evidence that Nod1 and Nod2 agonists synergize with TLR ligands to produce proinflammatory cytokines and anti-microbial molecules (24, 38, 43, 89). In addition, Nod2 signaling promotes IFN-β production in response to the cytosolic presence of L. monocytogenes or M. tuberculosis, although the mechanism involved is unclear (90). The potentiation of the IFN-β response by Nod2 may serve to boost cytosolic recognition of invasive bacteria. Thus, Nod1 or Nod2 signaling enhances both redundant TLR and non-redundant signaling pathways.
Proinflammatory cytokines are critical for the elimination of bacterial pathogens from the host. However, overproduction of these cytokines via stimulation of innate immune cells with TLR ligands is harmful to the host and can lead to septic shock and multi-organ failure (91, 92). As a form of protection against these deleterious effects, exposure to LPS and other TLR ligands induces a transient state of tolerance to subsequent TLR ligand challenge, thus reducing the negative effects associated with excessive cytokine production. However, TLR tolerization also increases the risk of succumbing to bacterial superinfection in patients (93, 94). Recent studies have revealed that macrophages and mice rendered tolerant to TLR stimulation remain fully responsive to Nod1 and Nod2 agonists (44, 45). In fact, there is a heightened response to MDP in macrophages and mice pre-stimulated with TLR agonists (44, 45). The lack of cross-inhibition between TLR and Nod1/Nod2 signaling is consistent with observations that TLR tolerization targets upstream components of the TLR pathway that are not involved in Nod1/Nod2 signaling (95). In addition, MDP induces refractoriness not only to a subsequent stimulation with MDP but also to iE-DAP stimulation (44). Some studies did not find cross-tolerization between MDP and TLR2 in human monocytes or TLR4 signaling in the production of IL-6 in human monocytes (96). In contrast, other studies found cross-tolerization between MDP and LPS in human cells (96, 97). The discrepancy may be in part due to differences between mice and humans and/or in the experimental systems. Further studies are needed to clarify the discrepancies observed among the different studies.
The priming of immune responses mediated through Nod1 and Nod2 in TLR-tolerant macrophages may serve to prepare the host against intracellular pathogens and to tailor the immune response to harmful invasive bacteria. Consistently, mice deficient in Nod1 and Nod2 are highly susceptible to systemic infection with Listeria when the animals are first exposed to LPS or E. coli (45). A possible implication of these results is that cells that are constantly exposed to microbial stimuli in vivo and are characterized by reduced TLR signaling (e.g. intestinal tissue), can become re-sensitized if intracellular Nod1 and Nod2 signaling is triggered by the presence of invasive bacteria. Indeed, several studies have shown that recognition of pathogenic bacteria in intestinal cells lacking TLRs relies on Nod1 (56, 62, 78). Together these results suggest that the intracellular sensors Nod1 and Nod2 play a critical role in host defense when TLR signaling is reduced such as in intestinal cells or inhibited via tolerization.
MDP and iE-DAP derivatives have been known to act as adjuvants of antigen-specific IgG production for over two decades (98). Analysis of stereoisomeric MDP analogs has revealed a good correlation between Nod2 recognition and adjuvant activity, suggesting that the adjuvant activity of MDP is mediated via Nod2 (87, 99). Indeed, the ability of MDP to induce ovalbumin (OVA)-specific IgG responses is abolished in mice deficient in Nod2 (41). The mechanism by which MDP exerts adjuvant activity is poorly understood, but it has been suggested to be mediated, at least in part, through the expression of costimulatory molecules in monocytes and dendritic cells which mediate differentiation of naive T cells into effector T cells (100–102). In addition, MDP enhances the differentiation of CD4+ T cells into T-helper 17 (Th17) cells (103). Recent studies have provided new insight into the function of Nod1 in the activation of adaptive immunity. For example, Nod1 contributes to adjuvant activity induced by complete Freud’s adjuvant, a crude extract from the cell wall of Mycobacteria (104). Furthermore, it was shown that stimulation with Nod1 agonists was sufficient to drive Th2-dependent antigen-specific adaptive immunity and to promote TLR-mediated Th1, Th2, and Th17 cell responses (104). Intriguingly, Nod1 primes antigen-specific immunity by acting on radiation-resistant non-hematopoietic cells (104), although the mechanism involved remains unknown. Together these studies provide evidence for an instructive role for Nod1 and Nod2 signaling in dictating the adaptive immune response.
Association of Nod1 and Nod2 with disease
There is mounting evidence that deregulation of Nod1 and Nod2 signaling causes or contributes to a variety of human diseases. Genetic studies revealed that several Nod2 variants are associated with susceptibility to Crohn’s disease (CD), a chronic disorder characterized by transmural inflammation of the intestine and particularly of the distal ileum. Although multiple variants of Nod2 have been found be linked to CD, three of them (R702W, G908R, and L1007insC) involving amino acid residues near or within the LRRs of Nod2 are relatively common (105, 106). Individuals homozygous or compound heterozygous for the common Nod2 mutations have about 20-fold increased risk for disease development, whereas heterozygous subjects have only about twofold increase risk (107). Biochemical and functional studies revealed that the human CD-associated Nod2 variants exhibit reduced or loss of ability to activate NF-κB in response to MDP, but they maintain a normal response to LPS stimulation (99, 108). Furthermore, monocytes isolated from CD patients or healthy individuals homozygous or compound heterozygous for the common Nod2 mutations have been reported to exhibit diminished secretion of inflammatory cytokines including TNFα, IL-6, IL-10, and IL-1β after MDP stimulation (105, 109). The selective impairment in MDP recognition exhibited by the CD-associated Nod2 mutations suggests that they result in a loss-of-function phenotype (109–112). The latter is consistent the observation that that homozygosity for the common Nod2 mutations is required for susceptibility to CD (107).
The mechanism by which Nod2 mutations contributes to CD remains poorly understood. Several hypotheses have been proposed to explain the role of Nod2 mutations in the pathogenesis of CD. The first hypothesis argues that Nod2 mutations confer an immunodeficiency state in phagocytic cells and/or the intestinal epithelium that promotes bacterial invasion or impairs the clearance of bacteria. In support of this notion, monocytes harboring mutant Nod2 exhibit impaired microbial sensing and production of anti-microbial molecules which are known to impact on microbial elimination by the host (109, 110, 112). Furthermore, there is increased susceptibility to oral administration of Listeria in Nod2-deficient mice (41). In addition, Nod2 is highly expressed in Paneth cells, which are specialized epithelial cells located at the base of the crypts of the small intestine. Notably, Nod2-deficient mice and CD patients harboring mutant Nod2 have reduced expression of α– defensins, a class of antimicrobial peptides produced by Paneth cells (25, 111, 113). However, it is unclear whether reduced expression of α– defensins is a pathogenic event or merely reflects Paneth cells loss due to damaged epithelium (114). The notion that loss-of-function Nod2 mutations results in increased inflammation appears at first glance counterintuitive, but it is not surprising because bacteria are recognized by multiple proinflammatory pathways. For example, mutant mice deficient in TLR2 or TLR4 exhibit increased inflammatory responses after challenge with pathogenic bacteria (115, 116). Similarly, loss of NEMO, a protein critical for NF-κB activation via multiple proinflammatory pathways, leads to spontaneous colitis presumably triggered by the presence of commensal bacteria in the lamina propria (117). Thus, Nod2 deficiency may lead to increased bacterial invasion and/or impaired clearance of bacteria driving deregulated inflammation at intestinal sites.
The second hypothesis suggests that Nod2 functions as a negative regulator of TLR-mediated signaling, and loss-of-function Nod2 mutations result in dysregulated TLR signaling and increased inflammation. Consistent with this model, IL-12 production induced by TLR agonists was increased in macrophages and dendritic cells deficient in Nod2 (118–120). In this model, lamina propria phagocytic cells are normally exposed to commensal products including TLR ligands, and Nod2 acts as a brake to dampen immune responses, a function which is lost in patients expressing mutant Nod2. The deregulated TLR-mediated responses will result in high IL-12 levels and create an environment conducive to pathological Th1 responses in the intestine. However, several studies have not found increased production of cytokines including IL-12 in Nod2-deficient macrophages or human dendritic cells harboring mutant Nod2 after stimulation with TLR agonists (42, 44, 45, 103, 121–123). Finally, macrophages from knockin mice generated to express the disease-associated L1007insC Nod2 variant exhibit increased NF-κB activation and IL-1β secretion when stimulated with MDP (124), suggesting that this common mutation acts as a gain-of-function mutation in this model. However, unlike this knockin Nod2 mouse model, monocytes from patients or healthy individuals homozygous for the L1007insC mutation display a marked defect in NF-κB activation and IL-1β production in response to MDP (109, 110, 112). Thus, further studies are needed to reconcile these conflicting observations and to understand the link between Nod2 and CD.
Several missense mutations resulting in single amino acid substitutions within the NOD domain of Nod2 cause Blau syndrome (125) and early-onset sarcoidosis (126), two autosomal dominant disorders characterized by early-onset granulomatous inflammation involving the skin, eyes, and joints. In contrast to CD, the Nod2 mutations associated with these diseases exhibit constitutive NF-κB activation and enhanced response to MDP (125, 126), which is consistent with the dominant mode of inheritance of these disorders.
Several Nod1 polymorphisms have been identified that are associated with increased risk of developing atopic eczema and asthma as well as with increased levels of serum IgE in several human populations (127, 128). The mechanism by which Nod1 variants increase the susceptibility to these allergic diseases is unknown, but it might be explained through the interaction of the host with the microbial environment during childhood, which is known to protect against the development of disease (129, 130). Consistently, Nod1 gene polymorphisms have been reported to significantly modify the protective effect against allergic disease afforded by exposure to a farming environment (131). Thus, it is possible that stimulation by bacterial products via Nod1 in the skin and mucosal surfaces might regulate directly or indirectly Th2 polarization and IgE levels as well as susceptibility to allergic disease.
The inflammasome
Caspases are a family of intracellular cysteine proteases that cleave a limited number of substrates after an aspartic acid residue in the P1 position of the scissile bond. Human caspases can be divided into two different groups based on their biological function: the proinflammatory caspases, caspase-1, caspase-4, caspase-5, caspase-11, and caspase-12, and the pro-apoptotic caspases that include caspase-2, caspase-3, caspase-7, caspase-8, caspase-9, and caspase-10 (132). Caspase-1 was the first caspase to be identified and is present in the cytosol of phagocytic cells as an inactive zymogen (133, 134). Upon stimulation by a variety of microbial and endogenous signals, the dormant pro-caspase-1 molecule is self-activated by proteolytic cleavage into the enzymatically active heterodimer composed of a 10 and a 20 kDa chain (135, 136) (Fig. 2). There is conclusive evidence now implicating NLR family members including NLRP1, NLRP3, and NLRP4 as critical factors in the activation of caspase-1 in response to proinflammatory stimuli (137, 138). The activation of caspase-1 is essential for the processing of pro-IL-1β and pro-IL-18 and secretion of their mature biologically active forms (139–141). A critical step in the activation of caspase-1 is the assembly of a large protein complex that includes NLR proteins, the adapter ASC (apoptosis-associated speck-like protein containing a C-terminal CARD) and pro-caspase-1 (142, 143) (Fig. 3). This molecular platform has been termed the inflammasome as an analogy to the apoptosome, the protein platform that directs the activation of the caspase-9 during apoptosis (18, 19). The regulation of the inflammasome formation is poorly understood, although it is thought that it is mediated, at least in part, through homophilic CARD-CARD and PYD-PYD domain interactions between NLRs, ASC, and pro-caspase-1. Inflammasome assembly and initial characterization was originally described in cell extracts using low K+ buffer, but whether NLR-specific inflammasome formation occurs in intact cells under physiological conditions such as bacterial infection remains unknown (144). In this review, we discuss recent literature on the major inflammasomes described, the Nlrp3-inflammasome, the Nlrc4-inflammasome, and the Nlrp1-inflammasome.
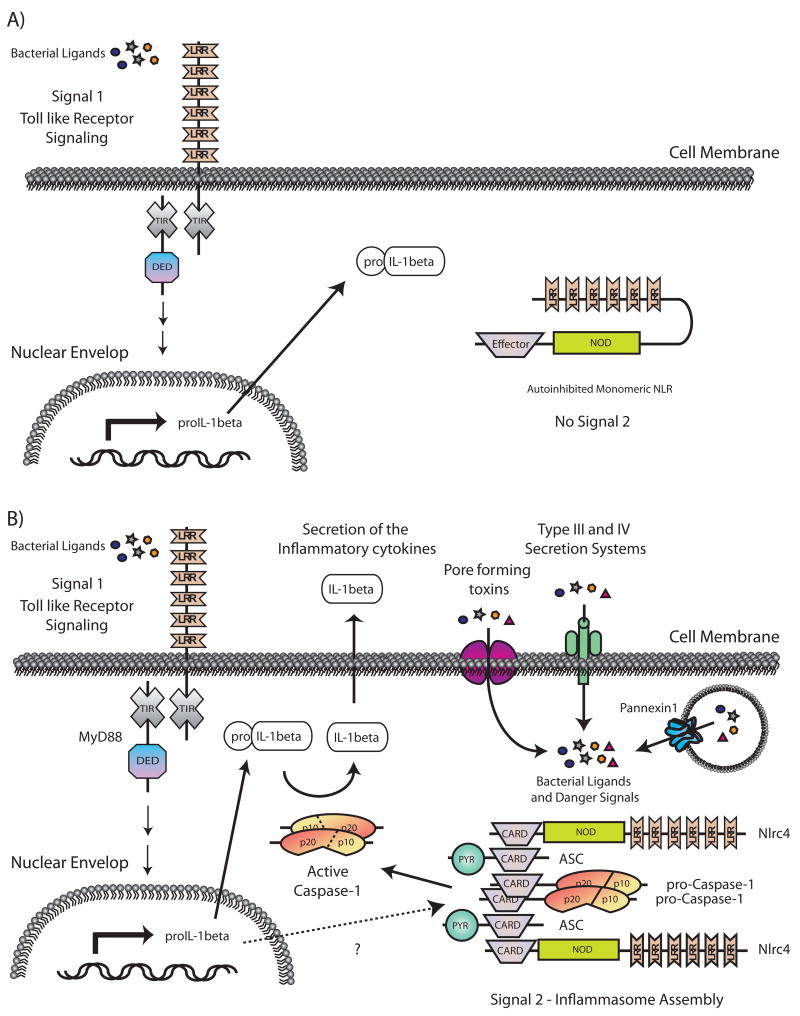
Fig. 2. The two step model for the induction and maturation of the proinflammatory cytokine IL-1β during the innate immune response.
(A) In the absence of NLR signals, TLR signaling (Signal 1) leads to the upregulation of pro-IL-1β transcription. (B) In response to a variety of intracellular signals (Signal 2) such as bacterial ligands and endogenous danger signals, NLRs are released from their auto-inhibited monomeric conformation leading to the assembly of a function inflammasome capable of activating the cysteine protease caspase-1. Active caspase-1 catalyzes the proteolytic processing of proIL-1β (and IL-18 not shown) into active cytokines that are then released from the cell.
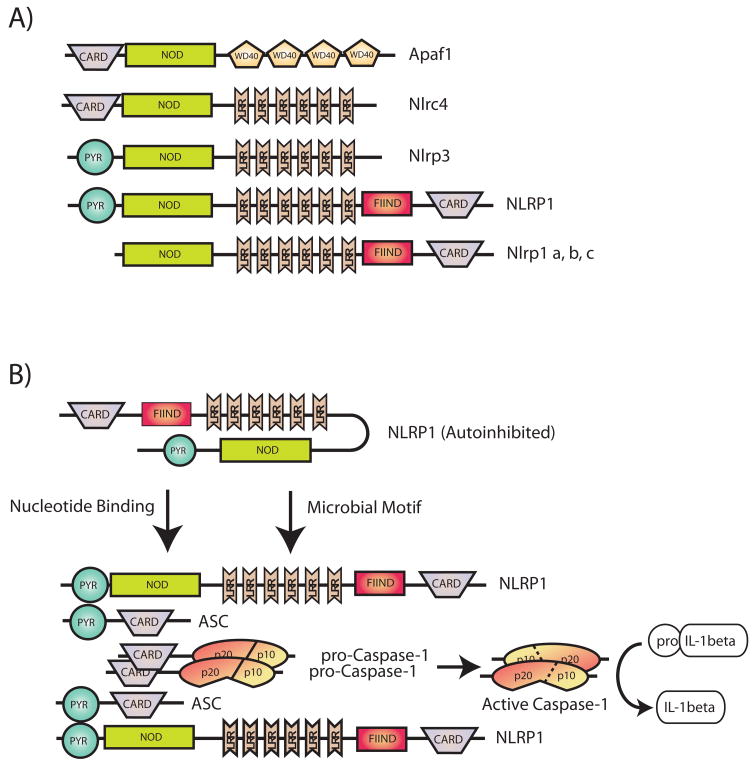
Fig. 3. Schematic representation of the multi-domain architectures of various NLRs and their assembly into a functional inflammasome.
Apaf-1 is a multi-domain scaffolding protein critical for the assembly of the apoptosome (not shown), a multi-protein complex critical for mediating apoptotic cell death. Subsequently, several NLR proteins were identified as Apaf-1-related molecules based on similar domain architectures (A). In response to various intracellular stimuli including microbial moieties and endogenous products released by dying cells, NLRs are released from their auto-inhibited monomeric conformation and act as molecular scaffolds to assemble multi-protein complexes known as inflammasomes. Assembly of the inflammasome leads to caspase-1 activation and proteolytic processing of proinflammatory cytokines such as IL-1β (B). In this review we focus on three of the best characterized inflammasomes: Ipaf1, Nalp3, and Nalp1.
Genetic studies using macrophages from mutant mice deficient in single NLR genes have shown Nlrc4, Nlrp3, and Nlrp1b are required for caspase-1 activation and IL-1β secretion in response to specific microbial stimuli. Phagocytes including monocytes and macrophages express little or no pro-IL-1β, and induction of this cytokine is mediated through transcriptional activation of the IL-1β gene in response to microbial stimulation via TLRs and NOD2, but independently of the inflammasome. Thus, IL-1β secretion requires two steps, namely pro-IL-1β induction and pro-IL-1β processing, which are independently regulated by separate classes of PRRs and intracellular pathways (Fig. 2). Nlrc4, Nlrp3, and Nlrp1b are thought to induce caspase-1 activation in a manner similar to the activation of caspase-9 mediated by Apaf-1. Nlrc4 is composed of an N-terminal CARD domain, a NOD domain and a C-terminal LRR domain while Nalp3 contains an N-terminal Pyrin domain, followed by a NOD domain and LRRs. Human NLRP1 is composed of Pyrin, NOD, LRR, FIIND, and CARD domains (Fig. 3). Biochemical data indicate that Nlrc4 can directly bind caspase-1 through a homotypic CARD-CARD interaction (145). In humans, NLRP3 and NLRP1 associate with caspase-1 through the adapter molecule ASC (144, 146–148). In contrast, Nlrc4 is thought to directly interact with caspase-1 via CARD-CARD interactions, and therefore, it will be predicted that the adapter ASC is not required for the activation of caspase-1 through the Nlrc4 inflammasome. However, genetic studies have revealed that ASC is also required for Nlrc4-dependent caspase-1 activation. For example, activation of caspase-1 induced by microbial stimuli known to involve Nlrc4, such as cytosolic flagellin (149, 150) or infection with Salmonella (151), Shigella (152), Pseudomonas (153, 154), or Legionella (155) is impaired in ASC-deficient macrophages. Notably, both Nlrc4 and ASC are required for caspase-1 activation and the production of IL-1β in response to infection with Salmonella and Pseudomonas. Yet, while Nlrc4 and caspase-1 are required for early induction of cell death, the presence of ASC is dispensable. Thus, ASC does not act exclusively as an adapter for the activation of caspase-1 but also contributes to cell survival. In agreement with this, ASC has been linked to NF-κB activation, although the mechanism involved remains poorly understood (156).
Nlrc4 inflammasome
Initial experiments showed that Nlrc4 mediates caspase-1 activation in macrophages infected with Salmonella (151). Recent studies indicate that a single microbial molecule, flagellin, is required to induce Nlrc4 mediated caspase-1 activation and that Salmonella mutants lacking flagellin or expressing point mutations in flagellin are impaired in their ability to induce caspase-1 activation (149, 150). The observation that cytosolic delivery of purified flagellin, but not other PAMPS, activates caspase-1 in an Nlrc4-dependent manner (149, 157, 158) suggests that the presence of flagellin in the cytosol is sufficient to drive the Nlcc4 inflammasome. In contrast, stimulation with extracellular flagellin is unable to induce the activation of caspase-1 (149, 150). The role of flagellin in promoting Nlrc4-dependent caspase-1 activation was later confirmed with other bacteria including Legionella pneumophila (157) and P. aeruginosa (153, 154, 159). Importantly, cytosolic flagellin or infection with Salmonella (149), Legionella (157), or Pseudomonas (154), induces a comparable activation of caspase-1 in TLR5 and TLR5-deficient macrophages. Collectively, these results indicate the recognition of flagellin is independent of TLR5, the TLR responsible for the detection of extracellular flagellin. In the case of Legionella, but not Salmonella or Pseudomonas, Nlrc4 activation is mediated by Naip5 that contributes to the recognition of the C-terminal portion of flagellin (160). The mechanism by which both Nlrc4 and Naip5 contribute to caspase-1 activation in response to Legionella infection remains unclear, but it has been suggested that Nlrc4 and Naip5 physically interact to regulate caspase-1 activation (160). Nlrc4 also plays a crucial role in the activation of caspase-1 induced by certain aflagellated bacteria, such as Shigella, suggesting that at least another unknown microbial molecule can induce the activation of Nlrc4 (152). The recognition of multiple PAMPs by the same PRR is not surprising because there is evidence that single TLRs are capable of recognizing different PAMPs. For example, TLR2 can recognize bacterial lipoprotein, lipoteichoic acid, and certain forms of LPS (7). Recently, Sutterwala et al. (161) reported that Pseudomonas may also activate caspase-1 through a Nlrc4-dependent, but flagellin-independent, pathway. Consistently, we also found the existence of a minor pathway that leads to caspase-1 activation in the absence of Nlrc4 and flagellin in macrophages infected with Pseudomonas (L.F., unpublished observations). The activation of caspase-1 by pathogenic bacteria requires a functional bacterial secretion systems [type III in Salmonella (149), Shigella (152), and Pseudomonas (154), versus type IV in Legionella (157)]. Bacterial secretion systems are molecular machines used by bacteria to deliver effectors proteins inside the host cell which manipulate cellular behavior and subvert host defense mechanisms. In the case of Salmonella, the type III secretion system promotes the translocation of flagellin into the cytosol of host cells (162). These results suggest that during infection small amounts of flagellin are leaked through the secretion bacterial system into the host cytosol to induce Nlrc4-dependent caspase-1 activation.
One interesting characteristic of the Nlrc4 inflammasome is that it remains active in macrophages tolerant to TLR stimulation. Macrophages exposed to TLR ligands become hyporesponsive to subsequent stimulation with the same or different TLR ligands. This phenomenon, which has been called TLR tolerization, may provide a mechanism to avoid an excessive inflammatory response. When TLR-tolerant macrophages are infected with Salmonella, the activation of caspase-1 is comparable to that obtained in non-tolerized macrophages (149). Furthermore, the production of IL-1β in TLR-tolerant macrophages is about 10 times higher than in naive macrophages stimulated with Salmonella, presumably due to the increased levels of pro-IL-1β expressed in the TLR-stimulated cell. Although the physiological relevance of the latter findings remains unclear, IL-1β production in tolerized cells may be important in tissues such as the intestine which are normally exposed to commensal bacteria and hyporesponsive to TLRs. The necessity to constrain the inflammatory response in the gut is suggested by the observation that mice with genetic defects that prevent TLR-induced tolerization develop an exaggerated inflammatory response when infected with Salmonella (163). Therefore, it will be important to determine the role of the inflammasome after bacterial infection in the intestinal tract and other tissues that are normally exposed to commensal bacteria.
NLRP1 inflammasome
PYR domain-containing NLRP proteins are the largest class of NLRs. NLRP1 was the first member of this subfamily to be identified. In humans, a single NLRP1 gene exists encoding an N-terminal PYR domain, a centrally located NOD domain followed by several LRRs, a FIIND domain, and a C-terminal CARD domain. In mice, three paralogs (Nlrp1a, Nlrp1b, and Nlrp1c) located in tandem in the same chromosomal region have been described, all of which lack the Pyrin and FIIND domains. Human NLRP1 is expressed in a wide variety of tissues but particularly in the thymus and spleen consistent with a functional role in the immune system (164). The importance of NLRP1 is underscored by the finding that a locus containing NLRP1 is responsible for familial vitiligo, a chronic skin disorder characterized by a loss of pigmentation and association with a variety of autoimmune and autoinflammatory conditions such as hypothyroidism and rheumatoid arthritis (165). However, the mechanism by which NLRP1 regulates disease susceptibility remains obscure.
The NLRP1 inflammasome was initially characterized in cell extracts and found to contain NLRP1, caspase-1, caspase-5, and ASC (144). Biochemical reconstitution experiments with purified components revealed that in the presence of MDP and ATP, NLRP1 oligomerizes forming a multi-protein complex that induces caspase-1 activation (166). The authors suggested a model in which caspase-1 activation involves a two-step mechanism, whereby microbial MDP induces a conformational change in NLRP1, which in turn allows it to bind nucleotide and oligomerize, thus creating a platform for caspase activation. Intriguingly, the adapter molecule ASC was not required for caspase-1 activation, but its presence did enhance NLRP1-mediated caspase-1 activation in vitro (166). These studies suggest that MDP activates the NLRP1 inflammasome, although direct binding of MDP to NLRP1 was not demonstrated. The physiological role of NLRP1 as a cytosolic sensor of MDP is controversial, as other NLRs, namely NLRP3 (81, 143) and Nod2 (32), have also been implicated in MDP-induced caspase-1 activation. Clearly, more studies using genetic and biochemical approaches are required to address the role of NLRP1 in MDP-induced caspase-1 activation and its relation to NLRP3 and Nod2.
There is compelling evidence that the mouse Nlrp1b inflammasome is activated in response to B. anthracis. The pathogenicity of B. anthracis is largely mediated by lethal toxin (LT), a dimeric complex consisting of protective antigen (PA), a pore forming toxin, and lethal factor, a protease that is delivered by the PA into the cytosol of infected cells (167). Genetic studies of mouse strains with different susceptibility to LT-induced death identified a single dominant locus designated Ltxs1 (168). Boyden and Dietrich (169) went on to identify mutations in the Nlrp1b gene as the key determinant of LT susceptibility in mice. Functional studies revealed that inhibition of Nlrp1b rendered LT-sensitive macrophages resistant. Furthermore, LT-induced toxicity was restored in macrophages by expressing the susceptible Nlrp1b allele in a LT-resistant background (169). Importantly, LT induces caspase-1 only in macrophages expressing the LT-sensitive Nlrp1b allele and macrophages lacking caspase-1 are protected from LT-induced death, even in the presence of a sensitive Nlrp1b allele (169). These studies establish LT as a critical microbial activator of the Nlrp1b inflammasome. However, further studies are needed to understand the mechanism by which LT activates caspase-1 through Nlrp1b.
NLRP3 inflammasome
Initial studies revealed that the NLRP3 forms an inflammasome with ASC, Cardinal, and caspase-1 in human cells (144). NLRP3 senses multiple PAMPs including LPS, MDP, bacterial and viral RNA, the double-stranded RNA analog polyinosinic-polycytidylic acid (polyI:C) (170–173) as well as the imidazoquinoline antiviral compounds R837 and R848 (170). In addition, NLRP3 has been implicated in the activation of caspase-1 in response to non-microbial signals including uric acid crystals and particulate matter such as asbestos and silica (174–176). Collectively, it has been proposed that activation of the NLRP3 inflammasome requires two separate signals (Fig. 2). The first is provided by a microbial molecule such as LPS or MDP. The second signal is induced by extracellular ATP or pore-forming molecules such as shellfish maitotoxin or bacterial toxins.
Significant insight into the mechanism by which NLRP3 recognizes various microbial molecules came from the observation that the addition of ATP to macrophages pre-stimulated with bacterial PAMPs such as LPS can significantly potentiate caspase-1 activation and IL-1β production (177). Whereas human and mouse macrophages require ATP stimulation to produce large amounts of IL-1β, human monocytes secrete robust levels of IL-1β in the absence of ATP (178, 179). Recently, Rubartelli and coworkers (180) reported that human monocytes stimulated with a variety of microbial ligands release ATP that acts in an autocrine fashion to promote the activation of caspase-1 and the release of IL-1β. The autocrine activity of ATP was also previously described in microglial cells (181) stimulated with microbial ligands and may explain the different sensitivity between monocytes and macrophages. A physiological role for ATP in NLRP3 activation remains controversial, since high concentrations of ATP, which are not normally found in the extracellular milieu, are typically required for enhanced IL-1β production. Extracellular ATP activates the purinergic ATP-gated P2X7 receptor, which actsas a non-desensitizing cation channel to rapidly induce a completecollapse of normal ionic gradients (182). An essential role forK+ efflux in the activation of caspase-1 has been suggested by the observation that pro-IL-1β processing can also be induced by stimuli such as nigericine, a bacterial derived molecule, that also cause a rapid lowering of cytoplasmic K+ (183). However, it is possible that the concentration of K+ itself may not regulate caspase-1 activation since ATP stimulation via the P2X7 receptor is also associated with other ionic fluxes or cellular events that may be what is critically involved in the activation of the NLRP3 inflammasome (184, 185). Moreover, in the absence of PAMP pre-stimulation, ATP does not activate caspase-1, even though it can trigger potent efflux of intracellular K+ (186). Extracellular ATP also results in the opening of a pore mediated by the hemi-channel protein Pannexin-1 (158, 187, 188). Recent studies suggest that P2X7 receptor activation by ATP mediates the entry of microbial products into the cytosol through the Pannexin-1 pore and subsequent activation of NLRP3 and caspase-1. Consistently, addition of ATP induces NLRP3-dependent but TLR and RICK-independent caspase-1 activation in response to several bacterial products and heat-killed bacteria (158). Moreover, the requirement for ATP can be bypassed by cytosolic delivery of microbial molecules with pore-forming bacterial toxins or through the lipophilic DOTAP delivery system (158). Furthermore, there is evidence that microbial ligands contribute to the activation of the NLRP3 inflammasome in a TLR-independent manner. This is best exemplified by the activation of the NLRP3 inflammasome induced by MDP (81) and other microbial ligands (158). MDP induces the activation of NF-κB and MAPKs through Nod2 in a TLR-independent fashion. A consequence of Nod2-dependent gene transcription is the increase in the level of pro-IL-1β. Pro-IL-1β is not, however, cleaved or secreted unless the cells are also stimulated with ATP. ATP induces the translocation of MDP to the cytosol and the activation of caspase-1. Remarkably, MDP-induced caspase-1 activation is Nlrp3 dependent but Nod2 independent (81). The different contribution of Nod2 in the upregulation of proIL-1β and Nlrp3 in the activation of caspase-1 explains why both pathways are required for the production of mature IL-1β (81). The role of Nod2 in caspase-1 activation remains however controversial, in that a recent report concluded that Nod2 was involved in caspase-1 activation in response to MDP and that the absence of Nod2 prevented B. anthracis-induced IL-1β secretion (32).
Several hypotheses have been proposed to explain the role of signal 1, namely microbial molecules, and signal 2, which is provided by extracellular ATP, membrane-damaging molecules, or pore-forming toxins in the activation of the NLRP3 inflammasome. According to one theory, the second signal promotes the translocation of microbial molecules into the host cytosol where they are sensed directly or indirectly by NLRP3 (158, 170, 189). An argument in favor of this hypothesis is that, in the absence of microbial ligands, cell damaging signals such as exogenous ATP do not activate the NLRP3 inflammasome (172, 173). Furthermore, inhibition of the endogenous Pannexin-1 pore which is open downstream of the P2X7 receptor suppresses microbial-induced inflammasome activation (158, 187, 188). A second theory suggests that the inflammasome is kept in an inactive state by high levels of intracellular K+. Two main arguments sustain this hypothesis. First, molecules like nigericine, a K+ ionophore, that reduces the concentration of intracellular K+ promotes caspase-1 activation in the absence of ATP stimulation (183). Second, all stimuli that are known to activate the NLRP3 fail to do so if the cells are incubated with buffers rich in K+, which prevents the decrease in intracellular K+ concentrations induced by ATP stimulation or damage of cellular integrity (186, 190, 191). However, high extracellular K+ concentrations may alter multiple cellular processes, and therefore, the latter experiments must be interpreted with caution. It must also be mentioned that not all bacterial pathogens that invade the cytosol activate NLRP3 since Salmonella, Franciscella, and Listeria, which are capable of delivering bacterial products into the cytosol through type III secretion systems or pore-forming molecules without ATP stimulation activate caspase-1 independently of NLRP3 (158, 189). Thus, activation of the NLRP3 inflammasome appears linked to specific PAMPs, P2X7 receptor activation, and Pannexin-1-dependent pore formation.
There is also evidence that TLRs can modulate the activation of the NLRP3 inflammasome. For example, stimulation of macrophages with TLR agonists not only induces the upregulation of pro-IL-1β but can also potentiate, through a still poorly characterized pathway, the activation of caspase-1 (177). This pathway may be important in patients with familial cold autoinflammatory syndrome, chronic infantile neurologic cutaneous articular syndrome, and Muckle-Wells syndrome, diseases caused by gain-of-function mutations in NLRP3. In normal individuals, stimulation of monocytes with LPS induces the activation of caspase-1 and the production of IL-1β, and this can be increased by stimulation with exogenous ATP. Monocytes from patients who harbor a NALP3 mutation exhibit spontaneous activation of caspase-1 and IL-1β production that can be further increased by LPS stimulation but not ATP (192). Remarkably, these patients show a dramatic improvement in their symptoms when treated with anakinra, a recombinant version of IL-1Rα, an antagonist of IL-1 signaling, thus establishing a cause-effect relationship between IL-1β production induced by NLRP3 mutations and the development of disease (193, 194).
Activation of the NLRP3 inflammasome by uric acid, silica, asbestos, and aluminum hydroxide
Elegant work by Rock and coworkers (195) demonstrated that necrotic cells induce the activation of the adaptive immune system through uric acid. More recently, Tschopp and coworkers (196) showed that the activation of caspase-1 in response to uric acid requires Nlrp3. The capacity of uric acid crystals to activate the Nlrp3 inflammasome is not unique, as other crystals such as calcium pyrophosphate dehydrate (CPPD), silica, and asbestos particles also activate the Nlrp3 inflammasome (196). Further analysis suggests that the activation of caspase-1 induced by uric acid and CPPD requires low cytosolic K+ concentrations (190). The mechanisms by which silica and asbestos trigger activation of the Nlrp3 inflammasome remain unclear. It has been reported that activation of the Nlrp3 inflammasome induced by both silica and asbestos particles is inhibited by interfering with the production of reactive oxygen species (174). There is also evidence that silica crystals can be internalized into phagosomes by macrophages and cause lysosomal destabilization to trigger caspase-1 activation (176). The latter study found that inhibition of either phagosomal acidification or cathepsin B activity but not reactive oxygen species suppresses silica-induced caspase-1 activation (176). It must be mentioned that pre-stimulation with LPS (or another microbial ligand) is needed to observe caspase-1 activation in response to uric acid crystals, asbestos, and silica, which suggests that these molecules may activate the inflammasome, at least in part, by promoting PAMP internalization or some other microbial-induced activity required for caspase-1 activation. Importantly, as uric acid and CPPD are the etiologic factors responsible for the debilitating human conditions gout and pseudogout, it is tantalizing to speculate that their induced pathology may be the result of deregulated IL-1β production. Consistently, IL-1R signaling in non-myeloid cells appears to be required for the proinflammatory effect of uric acid in a mouse model of peritonitis (197). Indeed, preliminary results suggest that patients with gouty attacks improved their clinical conditions when IL-1R signaling is antagonized by IL-1Rα (198).
Aluminiun hydroxide (Alum) is the most widely used adjuvant in human vaccines and has recently been shown to promote the secretion of IL-1β in macrophages and dendritic cells. Several groups have shown that Alum acts like a danger signal inducing the activation of caspase-1 in cells stimulated with microbial ligands such as LPS, and this effect is independent of the P2X7 receptor (199). Interestingly the Nlrp3 inflammasome has been shown to participate in the innate and adaptive immune responses induced by Alum (200–203). For example, using an asthma model, Eisenbarth et al. (201) showed a reduction in the production of immunoglobulin G1 (IgG1) in Nlrp3-deficient mice. Consistently, Li et al. (202) showed a reduction in Alum-mediated IgG1 antibody-specific responses in Nlrp3 null mice. In contrast, we and others (200, 203) did not observe a reduction in antigen-specific IgG antibodies titers following intraperitoneal immunization. These results are difficult to reconcile, but it is possible that the importance of Nlrp3 in the activation of the adaptive immune response depends on the route of administration, the dose of antigen, the Alum formulation, and/or the assay used to measure the adaptive immune response. Collectively, these results indicate that Nlrp3 is not absolutely essential, but it can influence adaptive immune responses. As the production of antibodies induced by Alum is independent of TLRs, IL-1R, and IL-18R signaling (204), a major challenge for future studies will be the elucidation of the pathway(s) through which Alum induces an adaptive immune response and adjuvant activity.
Bacterial Toxins
Several lines of evidence indicate that the activation of the inflammasome induced by extracellular microbial ligands or extracellular bacteria is inefficient unless the cell receives a second stimulus, that may be induced via ATP, membrane-damaging molecules, or certain bacterial pore-forming toxins (205). Several bacterial pore-forming or membrane-damaging toxins have been reported to induce the activation of the Nlrp3 inflammasome including streptolysin O from Streptococcus pyrogens (158), maitotoxin from Gambierdiscus toxicus (173), nigericin from Streptomices hydroscopicus (173), valinomycin from Streptomices sp (206), and aerolysin from Aeromonas hydrophila (206). These microbial molecules can induce membrane damage or mediate pore-formation in a manner reminiscent to that induced by ATP stimulation of the purinergic P2X7 receptor. Thus, they may contribute to the activation of the Nlrp3 inflammasome through multiple mechanisms including via the production of reactive oxygen species (175, 207), ionic imbalances (208, 209), and phagosomal destabilization (176). Alternatively, as proposed by Kanneganti et al. (158), these toxins may promote the passage of microbial molecules into the cytosol. It is interesting to note that in the presence of microbial ligands, the stimulation with maitotoxin and nigericin induces the activation of caspase-1 in a Pannexin-1-dependent manner (187). So, it is possible that danger signals and pore-forming bacterial molecules converge at the level of Pannexin-1 to mediate caspase-1 activation via the Nlrp3 inflammasome.
Another pore-forming toxin that has been implicated in the activation of the inflammasome is listerolysin O (LLO) from L. monocytogenes, an intracellular bacterium. LLO is a pore-forming toxin that allows the bacteria to escape from the vacuole into the cytosol. Initial experiments showed that Listeria-induced caspase-1 activation was independent of TLR2 but requires bacterial LLO and the host adapter ASC (210). Subsequently, Mariathasan et al. (173)showed that the Nlrp3 inflammasome played an essential role in Listeria-induced caspase-1 activation. Later studies, however, could not confirm the essential role of Nlrp3 in Listeria-induced caspase-1 activation (186, 211). One possible explanation is that the activation of caspase-1 induced by Listeria is mediated through two or more NLRs and that some experimental conditions may favor the recognition by Nlrp3 (186, 211). As mice deficient in caspase-1 (212) and ASC (210) showed increased susceptibility to Listeria infection, the contribution of the Nlrp3 inflammasome awaits further in vivo studies that compare mice null in Nlrp3 with mice deficient in ASC or caspase-1.
The inflammasome induces pyroptosis and regulates autophagy
Cell death is conventionally categorized as apoptosis and necrosis, although experimental evidence suggest that these two distinct cell death processes represent two extremes of a continuous spectrum of cellular demise. Apoptosis is an evolutionary conserved form of programmed cell death that plays a crucial role in tissue development and homeostasis. This form of cell death is generally not associated with the generation of the inflammatory response, and there is evidence that the engulfment of apoptotic cells by macrophages may actually elicit an anti-inflammatory response. Proapoptotic caspases play a crucial role in this highly regulated form of cell death that is characterized by the organized dismantling of the cell. In contrast, necrosis, which results from the exposure of cells to a wide variety of toxic insults, is characterized by distortion and rupture of the plasma membrane and organelles. There is mounting evidence that NLRs regulate cell death induced by microbial pathogens. For example, caspase-1 plays an essential role in the induction a specialized form of cell death called pyroptosis that is triggered by infection of macrophages with multiple intracellular bacterial pathogens. Pyroptosis is typically associated with rapid disruption of cell membrane integrity and the maturation of pro-IL-1β (213, 214). Caspase-1 is essential for the early induction of cell death, membrane swelling, and IL-1β production in macrophages infected with Salmonella (149, 215), Pseudomonas (154), Shigella (152), Listeria (216), Legionella (155), or when macrophages are stimulated with anthrax LT (217) or with ATP (218, 219). In the presence of microbial ligands, some toxins such as nigericin (220) and maitotoxin also appear to induce pyroptosis (187). Analysis of macrophages deficient in Nlrc4 revealed that this NLR is required for the induction of pyroptosis triggered by infection of macrophages with Salmonella and Shigella (149, 150, 152), which is consistent with its critical role in caspase-1 activation. Intriguingly, ASC, which is also important for caspase-1 activation, was dispensable for pyroptosis (152). It is important to mention that Salmonella or Shigella-induced cell death is only delayed in the absence of Nlrc4 or caspase-1, implicating that cell death induced by these intracellular pathogens involves alternate cell death mechanisms. Consistently, infection of macrophages with Shigella at high bacteria-macrophage ratios induces necrosis, and this process was shown to be independent of caspase-1 but reliant on Nlrp3 and ASC (221). The biological function of pyroptosis during the bacterial infection in vivo remains poorly understood. It is possible that pyroptosis by means of its rapid kinetics prevents the onset of other cell death pathways, which implies that pyroptosis may actually prevent the anti-inflammatory consequences of apoptotic cell death (222). However, it remains to be established whether pyroptosis plays a role in host defense independent of the production of IL-1β and IL-18.
Autophagy is an intracellular degradation process that allows the cell to recycle their own cytoplasmic constituents by delivering them to the lysosomes for degradation. Microbial ligands are known activators of signaling pathways that increase autophagy (223, 224). Importantly, there is mounting evidence that autophagy contributes to the elimination of microbial pathogens by promoting their degradation in lysosomes (225, 226). However the role of autophagy in host-pathogen interaction is complex, in that pathogens can produce virulence factors that actively inhibit autophagy or can subvert the autophagic machinery to create vacuolar compartment where the bacteria can reside and eventually replicate (225, 226). Recent studies suggest that caspase-1 is a negative regulator of the autophagic response induced by Shigella (152). In macrophages, Shigella is detected by Nlrc4, in a flagellin-independent manner, which in turn induces caspase-1 activation. Caspase-1 not only promotes pyroptosis but also inhibits autophagy. The induction of autophagy was independent of VirG, a virulence factor known to induce autophagy in epithelial cells. Notably, the negative regulation of autophagy mediated by Nlrc4 and caspase-1 was dependent on microbial invasion (152). Given the protective role of caspase-1 and IL-1β in a mouse model of Shigella infection, it appears plausible that the inhibition of autophagy is used by the host to select the most appropriate host-defense mechanism to fight Shigella. Clearly, more studies are needed to understand the link between the inflammasome and autophagy during bacterial infection.
Role of the inflammasome in host defense
Experiments using caspase-1-deficient mice have revealed a role for this caspase in inflammation, innate immunity, and host defense (2). During infection with small number of bacteria, there is evidence that caspase-1 plays a protective role against intracellular bacteria including Salmonella (227, 228), Shigella (229) Listeria (212), Legionella (155, 157), and Francisella (230). As the deficiency of single NLRs do not recapitulate the phenotype observed in caspase-1-deficient mice, these results suggest there is redundancy between NLRs in the recognition of bacteria in vivo. Alternatively, it is possible that caspase-1 mediates activities independent of NLRs in the animal. In contrast to the protective role observed with infection with low number of bacteria, the caspase-1 dependent production of cytokines is detrimental in models of septic shock (231).
Professional phagocytes engulf and destroy bacterial pathogens by phagocytosis. The phagocytic process is essential for the elimination of both intracellular and extracellular bacteria. To survive, intracellular bacteria must avoid phagosome maturation to prevent degradation of the pathogen in lysosomes. Bacteria use different strategies to accomplish this. For example, Salmonella and Legionella halt the maturation of the phagosome, M. tuberculosis prevents the acidification of the phagosome, while Shigella and Listeria escape from the phagosome into the cytosol (232). Recent evidence indicates that microbial ligands can also directly modulate phagocytosis in macrophages and dendritic cells through TLR signaling pathways (233, 234). There is evidence that recognition of cytosolic flagellin via NLRs plays a crucial role in promoting the fusion of late endosomes with lysosomes and in restricting bacterial replication (157). In the absence of flagellin, Nlrc4, or caspase-1, Legionella can establish a vacuolar replicative niche known as the Legionella-containing vacuole. The recognition of flagellin through the Naip5 and Nlrc4 inflammasome is critical for phagosome maturation and restriction of Legionella replication inside macrophages (157). Importantly, mice deficient in Nlrc4 exhibit an increased bacterial burden after pulmonary infection with Legionella (157). The mechanism by which the inflammasome controls phagosome maturation remains unknown.
Concluding remarks
There is now compelling evidence that NLRs are involved in the recognition of conserved microbial components as well non-microbial signals including uric acid crystals and silica. NLR signaling results in the activation of NF-κB, MAPK, and caspase-1 that result in the induction of proinflammatory cytokines, chemokines, and anti-microbial molecules. The intracellular location of NLRs suggests that these PRRs are involved in the recognition of PAMPs derived from bacteria that directly invade the cell or enter the cell through phagocytosis, bacterial secretion systems, pore-forming toxins, or host-derived pores such as Pannexin-1. NLR signaling cooperates with TLR signaling in the production of cytokines in response to bacterial infection. Finally, deregulated NLR signaling caused by either gain-of-function or loss-of-function mutations result in human disease. Future studies need to address the mechanistic aspects of microbial recognition through NLRs, the precise role of NLRs in mucosal and systemic infection, and the mechanism by which genetic variants of NLRs increase the susceptibility to disease.
Table 2.
| Bacteria | Microbial motif/activators | NLR | Signaling pathway | References |
|---|---|---|---|---|
| Bacillus anthracis | anthrax lethal toxin | Nlrp1b | caspase-1 | (169) |
| Bacillus spp. | iEDAP | NOD1 | NF-κB, MAPK | (74) |
| Campylobacter jejuni | iEDAP | NOD1 | NF-κB, MAPK | (62) |
| Chlamydia pneumonia | iEDAP | NOD1 | NF-κB, MAPK | (72) |
| Enteropathogenic Escherichia coli | iEDAP | NOD1 | NF-κB, MAPK | (56) |
| Escherichia coli** | microbial motifs | Nalp3 | caspase-1 | (186) |
| Helicobacter pylori | iEDAP | NOD1 | NF-κB, MAPK | (61, 79) |
| Legionella pneumophila | Flagellin | Nlrc4 | caspase-1 | (157, 160, 235) |
| Listeria monocytogenes | flagellin + microbial motifs | Nlrc4 + Nalp3 | caspase-1 | (173, 186, 210, 211) |
| MDP | NOD2 | NF-κB, MAPK | (41–45, 90) | |
| iEDAP | NOD1 | NF-κB, MAPK | (42–45, 73) | |
| Mycobacterium tuberculosis | microbial motifs | Nlrp3 | caspase-1 | (236) |
| ? | Nlrc4# | caspase-1 | (237) | |
| MDP | NOD2 | NF-κB, MAPK | (86) | |
| Pseudomonas aeruginosa | Flagellin * | Nlrc4 | caspase-1 | (153, 154, 159) |
| iEDAP | NOD1 | NF-κB, MAPK | ||
| Salmonella | Flagellin | Nlrc4 | caspase-1 | (149, 150) |
| Typhimurium | MDP | NOD2 | NF-κB, MAPK | (27, 122) |
| Shigella flexneri | ? | Nlrc4## | caspase-1 | (152) |
| MDP | NOD2 | NF-κB, MAPK | (77) | |
| iEDAP | NOD1 | NF-κB, MAPK | (30, 78) | |
| Staphylococcus aureus | MDP | NOD2 | NF-κB, MAPK | (87) |
| microbial motifs | Nalp3** | caspase-1 | (173) | |
| Streptococcus pneumonia | MDP | NOD2 | NF-κB, MAPK | (85) |
the existence of a flagellin independent has been reported (161).
only observed when macrophages are infected with bacteria deficient in ESX-1
activation of caspase-1 requires permeabilization induced by ATP or bacterial toxins.
A Nalp3 dependent pathway has been reported in macrophages infected with high number of bacteria (221).
Acknowledgments
This work was supported by grants from the National Institutes of Health. We thank all of the colleagues in our laboratory for helpful discussion.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Franchi L, et al. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 3.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 7.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 9.Moore CB, Ting JP. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008;28:735–739. doi: 10.1016/j.immuni.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 11.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 13.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe T, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by monarch- 1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–1850. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan JA, et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci USA. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 19.Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 20.Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 22.Inohara N, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 23.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 24.Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura Y, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- 27.Hisamatsu T, Suzuki M, Reinecker HC, Nadeau WJ, McCormick BA, Podolsky DK. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 28.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Uehara A, et al. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell Microbiol. 2005;7:675–686. doi: 10.1111/j.1462-5822.2004.00500.x. [DOI] [PubMed] [Google Scholar]
- 30.Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10:477–486. doi: 10.1111/j.1462-5822.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- 31.Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-{kappa}B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170:21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu LC, et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci USA. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manon F, Favier A, Nunez G, Simorre JP, Cusack S. Solution structure of NOD1 CARD and mutational analysis of its interaction with the CARD of downstream kinase RICK. J Mol Biol. 2007;365:160–174. doi: 10.1016/j.jmb.2006.09.067. [DOI] [PubMed] [Google Scholar]
- 34.Coussens NP, Mowers JC, McDonald C, Nunez G, Ramaswamy S. Crystal structure of the Nod1 caspase activation and recruitment domain. Biochem Biophys Res Commun. 2007;353:1–5. doi: 10.1016/j.bbrc.2006.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 36.McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280:20177–20180. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 37.Girardin SE, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 38.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 39.Girardin SE, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi K, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 42.Park JH, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 43.Park JH, et al. Nod1/RICK and TLR signaling regulate chemokine and antimicrobial innate immune responses in mesothelial cells. J Immunol. 2007;179:514–521. doi: 10.4049/jimmunol.179.1.514. [DOI] [PubMed] [Google Scholar]
- 44.Kim YG, Park JH, Daignault S, Fukase K, Nunez G. Cross-tolerization between Nod1 and Nod2 signaling results in reduced refractoriness to bacterial infection in Nod2-deficient macrophages. J Immunol. 2008;181:4340–4346. doi: 10.4049/jimmunol.181.6.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YG, Park JH, Shaw MH, Franchi L, Inohara N, Nunez G. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–257. doi: 10.1016/j.immuni.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Inohara N, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa M, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF- kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 49.Kim JY, Omori E, Matsumoto K, Nunez G, Ninomiya-Tsuji J. TAK1 is a central mediator of NOD2 signaling in epidermal cells. J Biol Chem. 2008;283:137–144. doi: 10.1074/jbc.M704746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 2003;22:6277–6288. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shim JH, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 53.Windheim M, Lang C, Peggie M, Plater LA, Cohen P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J. 2007;404:179–190. doi: 10.1042/BJ20061704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.da Silva Correia J, Miranda Y, Leonard N, Hsu J, Ulevitch RJ. Regulation of Nod1-mediated signaling pathways. Cell Death Differ. 2007;14:830–839. doi: 10.1038/sj.cdd.4402070. [DOI] [PubMed] [Google Scholar]
- 55.Welter-Stahl L, et al. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell Microbiol. 2006;8:1047–1057. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 56.Kim JG, Lee SJ, Kagnoff MF. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect Immun. 2004;72:1487–1495. doi: 10.1128/IAI.72.3.1487-1495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Travassos LH, et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280:36714–36718. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- 58.Buchholz KR, Stephens RS. The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect Immun. 2008;76:3150–3155. doi: 10.1128/IAI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werts C, et al. Nod1 and Nod2 induce CCL5/RANTES through the NF-kappaB pathway. Eur J Immunol. 2007;37:2499–2508. doi: 10.1002/eji.200737069. [DOI] [PubMed] [Google Scholar]
- 60.Masumoto J, et al. Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med. 2006;203:203–213. doi: 10.1084/jem.20051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boughan PK, et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: critical regulators of beta-defensins during Helicobacter pylori infection. J Biol Chem. 2006;281:11637–11648. doi: 10.1074/jbc.M510275200. [DOI] [PubMed] [Google Scholar]
- 62.Zilbauer M, et al. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol. 2007;9:2404–2416. doi: 10.1111/j.1462-5822.2007.00969.x. [DOI] [PubMed] [Google Scholar]
- 63.Rosenstiel P, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Natl Acad Sci USA. 2006;103:3280–3285. doi: 10.1073/pnas.0505423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leblanc PM, et al. Caspase-12 Modulates NOD Signaling and Regulates Antimicrobial Peptide Production and Mucosal Immunity. Cell Host Microbe. 2008;3:146–157. doi: 10.1016/j.chom.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 65.Clark NM, Marinis JM, Cobb BA, Abbott DW. MEKK4 sequesters RIP2 to dictate NOD2 signal specificity. Curr Biol. 2008;18:1402–1408. doi: 10.1016/j.cub.2008.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kufer TA, Kremmer E, Banks DJ, Philpott DJ. Role for erbin in bacterial activation of Nod2. Infect Immun. 2006;74:3115–3124. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald C, et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 68.Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 71.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF- kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 72.Opitz B, et al. Nod1-mediated endothelial cell activation by Chlamydophila pneumoniae. Circ Res. 2005;96:319–326. doi: 10.1161/01.RES.0000155721.83594.2c. [DOI] [PubMed] [Google Scholar]
- 73.Boneca IG, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hasegawa M, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–29063. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- 75.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodell EW. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nigro G, et al. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell Microbiol. 2008;10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 78.Girardin SE, et al. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viala J, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 80.Ratner AJ, Aguilar JL, Shchepetov M, Lysenko ES, Weiser JN. Nod1 mediates cytoplasmic sensing of combinations of extracellular bacteria. Cell Microbiol. 2007;9:1343–1351. doi: 10.1111/j.1462-5822.2006.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marina-Garcia N, et al. Pannexin-1-mediated intracellular delivery of muramyl dipeptide induces caspase-1 activation via Cryopyrin/NLRP3 independently of Nod2. J Immunol. 2008;180:4050–4057. doi: 10.4049/jimmunol.180.6.4050. [DOI] [PubMed] [Google Scholar]
- 82.Ismair MG, Vavricka SR, Kullak-Ublick GA, Fried M, Mengin-Lecreulx D, Girardin SE. hPepT1 selectively transports muramyl dipeptide but not Nod1-activating muramyl peptides. Can J Physiol Pharmacol. 2006;84:1313–1319. doi: 10.1139/y06-076. [DOI] [PubMed] [Google Scholar]
- 83.Swaan PW, et al. Bacterial peptide recognition and immune activation facilitated by human peptide transporter PEPT2. Am J Respir Cell Mol Biol. 2008;39:536–542. doi: 10.1165/rcmb.2008-0059OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biegel A, et al. The renal type H+/peptide symporter PEPT2: structure-affinity relationships. Amino Acids. 2006;31:137–156. doi: 10.1007/s00726-006-0331-0. [DOI] [PubMed] [Google Scholar]
- 85.Opitz B, et al. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 86.Ferwerda G, et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:279–285. doi: 10.1371/journal.ppat.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 88.Meinzer U, et al. Nod2 mediates susceptibility to Yersinia pseudotuberculosis in mice. PLoS ONE. 2008;3:e2769. doi: 10.1371/journal.pone.0002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fritz JH, et al. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- 90.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Danner RL, Elin RJ, Hosseini JM, Wesley RA, Reilly JM, Parillo JE. Endotoxemia in human septic shock. Chest. 1991;99:169–175. doi: 10.1378/chest.99.1.169. [DOI] [PubMed] [Google Scholar]
- 92.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 93.Docke WD, et al. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat Med. 1997;3:678–681. doi: 10.1038/nm0697-678. [DOI] [PubMed] [Google Scholar]
- 94.Shahin RD, Engberg I, Hagberg L, Svanborg Eden C. Neutrophil recruitment and bacterial clearance correlated with LPS responsiveness in local gram-negative infection. J Immunol. 1987;138:3475–3480. [PubMed] [Google Scholar]
- 95.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 96.Kullberg BJ, et al. Crohn’s disease patients homozygous for the 3020insC NOD2 mutation have a defective NOD2/TLR4 cross-tolerance to intestinal stimuli. Immunology. 2008;123:600–605. doi: 10.1111/j.1365-2567.2007.02735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci USA. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takada H, Kotani S. Muramyl dipeptide and derivatives. In: Stewart-Tull DES, editor. The Theory of Practical Application of Adjuvants. New York: Wiley & Sons; 1995. pp. 171–202. [Google Scholar]
- 99.Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 100.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Todate A, Suda T, Kuwata H, Chida K, Nakamura H. Muramyl dipeptide-Lys stimulates the function of human dendritic cells. J Leukoc Biol. 2001;70:723–729. [PubMed] [Google Scholar]
- 102.Heinzelmann M, Polk HC, Jr, Chernobelsky A, Stites TP, Gordon LE. Endotoxin and muramyl dipeptide modulate surface receptor expression on human mononuclear cells. Immunopharmacology. 2000;48:117–128. doi: 10.1016/s0162-3109(00)00195-8. [DOI] [PubMed] [Google Scholar]
- 103.van Beelen AJ, et al. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 2007;27:660–669. doi: 10.1016/j.immuni.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 104.Fritz JH, et al. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity. 2007;26:445–459. doi: 10.1016/j.immuni.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 105.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 106.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 107.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn’s disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 108.Ogura Y, Saab L, Chen FF, Benito A, Inohara N, Nunez G. Genetic variation and activity of mouse Nod2, a susceptibility gene for Crohn’s disease. Genomics. 2003;81:369–377. doi: 10.1016/s0888-7543(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 109.van Heel DA, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2- associated Crohn’s disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 110.Li J, et al. Regulation of IL-8 and IL-1beta expression in Crohn’s disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 111.Wehkamp J, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Netea MG, Ferwerda G, de Jong DJ, Girardin SE, Kullberg BJ, van der Meer JW. NOD2 3020insC mutation and the pathogenesis of Crohn’s disease: impaired IL-1beta production points to a loss-of-function phenotype. Neth J Med. 2005;63:305–308. [PubMed] [Google Scholar]
- 113.Lala S, et al. Crohn’s disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 114.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 115.Torres D, et al. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect Immun. 2004;72:2131–2139. doi: 10.1128/IAI.72.4.2131-2139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vazquez-Torres A, et al. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- 117.Nenci A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 118.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 119.Watanabe T, et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–485. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 121.Beynon V, et al. NOD2/CARD15 genotype influences MDP-induced cytokine release and basal IL-12p40 levels in primary isolated peripheral blood monocytes. Inflamm Bowel Dis. 2008;14:1033–1040. doi: 10.1002/ibd.20441. [DOI] [PubMed] [Google Scholar]
- 122.Salucci V, et al. Monocyte-derived dendritic cells from Crohn patients show differential NOD2/CARD15-dependent immune responses to bacteria. Inflamm Bowel Dis. 2008;14:812–818. doi: 10.1002/ibd.20390. [DOI] [PubMed] [Google Scholar]
- 123.Kramer M, Netea MG, de Jong DJ, Kullberg BJ, Adema GJ. Impaired dendritic cell function in Crohn’s disease patients with NOD2 3020insC mutation. J Leukoc Biol. 2006;79:860–866. doi: 10.1189/jlb.0805484. [DOI] [PubMed] [Google Scholar]
- 124.Maeda S, et al. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL- 1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 125.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19– 20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 126.Kanazawa N, et al. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 127.Hysi P, et al. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935–941. doi: 10.1093/hmg/ddi087. [DOI] [PubMed] [Google Scholar]
- 128.Weidinger S, et al. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005;116:177–184. doi: 10.1016/j.jaci.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 129.Ball TM, Castro-Rodriguez JA, Griffith KA, Holberg CJ, Martinez FD, Wright AL. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343:538–543. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- 130.Riedler J, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 131.Eder W, et al. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy. 2006;61:1117–1124. doi: 10.1111/j.1398-9995.2006.01128.x. [DOI] [PubMed] [Google Scholar]
- 132.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 133.Cerretti DP, et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 134.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin- 1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 135.Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- 136.Yamin TT, Ayala JM, Miller DK. Activation of the native 45-kDa precursor form of interleukin-1-converting enzyme. J Biol Chem. 1996;271:13273–13282. doi: 10.1074/jbc.271.22.13273. [DOI] [PubMed] [Google Scholar]
- 137.Franchi L, McDonald C, Kanneganti TD, Amer A, Nunez G. Nucleotide-binding oligomerization domain-like receptors: intracellular pattern recognition molecules for pathogen detection and host defense. J Immunol. 2006;177:3507–3513. doi: 10.4049/jimmunol.177.6.3507. [DOI] [PubMed] [Google Scholar]
- 138.Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 139.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 140.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 141.Ghayur T, et al. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS- induced IFN-gamma production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 142.Lamkanfi M, Kanneganti TD, Franchi L, Nunez G. Caspase-1 inflammasomes in infection and inflammation. J Leukoc Biol. 2007;82:220–225. doi: 10.1189/jlb.1206756. [DOI] [PubMed] [Google Scholar]
- 143.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 144.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 145.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 146.Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Nunez G. Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem Biophys Res Commun. 2003;302:575–580. doi: 10.1016/s0006-291x(03)00221-3. [DOI] [PubMed] [Google Scholar]
- 147.Grenier JM, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 148.Wang L, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 149.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 150.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 151.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 152.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci USA. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Franchi L, Stoolman J, Kanneganti TD, Verma A, Ramphal R, Nunez G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 155.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 156.Masumoto J, et al. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun. 2003;303:69–73. doi: 10.1016/s0006-291x(03)00309-7. [DOI] [PubMed] [Google Scholar]
- 157.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 158.Kanneganti TD, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 159.Galle M, et al. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of Caspase-1 mediated IL-1beta maturation. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2007.00190.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–32345. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 163.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 164.Hlaing T, et al. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J Biol Chem. 2001;276:9230–9238. doi: 10.1074/jbc.M009853200. [DOI] [PubMed] [Google Scholar]
- 165.Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 166.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 167.Leppla SH, Arora N, Varughese M. Anthrax toxin fusion proteins for intracellular delivery of macromolecules. J Appl Microbiol. 1999;87:284. doi: 10.1046/j.1365-2672.1999.00890.x. [DOI] [PubMed] [Google Scholar]
- 168.Roberts JE, Watters JW, Ballard JD, Dietrich WF. Ltx1, a mouse locus that influences the susceptibility of macrophages to cytolysis caused by intoxication with Bacillus anthracis lethal factor, maps to chromosome 11. Mol Microbiol. 1998;29:581–591. doi: 10.1046/j.1365-2958.1998.00953.x. [DOI] [PubMed] [Google Scholar]
- 169.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 170.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 171.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 172.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 173.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 174.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kahlenberg JM, Lundberg KC, Kertesy SB, Qu Y, Dubyak GR. Potentiation of caspase-1 activation by the P2X7 receptor is dependent on TLR signals and requires NF-kappaB-driven protein synthesis. J Immunol. 2005;175:7611–7622. doi: 10.4049/jimmunol.175.11.7611. [DOI] [PubMed] [Google Scholar]
- 178.Kahlenberg JM, Dubyak GR. Differing caspase-1 activation states in monocyte versus macrophage models of IL-1beta processing and release. J Leukoc Biol. 2004;76:676–684. doi: 10.1189/jlb.0404221. [DOI] [PubMed] [Google Scholar]
- 179.Laliberte RE, Perregaux DG, McNiff P, Gabel CA. Human monocyte ATP-induced IL-1 beta posttranslational processing is a dynamic process dependent on in vitro growth conditions. J Leukoc Biol. 1997;62:227–239. doi: 10.1002/jlb.62.2.227. [DOI] [PubMed] [Google Scholar]
- 180.Piccini A, Carta S, Tassi S, Lasiglie D, Fossati G, Rubartelli A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc Natl Acad Sci USA. 2008;105:8067–8072. doi: 10.1073/pnas.0709684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ferrari D, Chiozzi P, Falzoni S, Hanau S, Di Virgilio F. Purinergic modulation of interleukin-1 beta release from microglial cells stimulated with bacterial endotoxin. J Exp Med. 1997;185:579–582. doi: 10.1084/jem.185.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ferrari D, et al. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176:3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 183.Cheneval D, et al. Increased mature interleukin-1beta (IL-1beta) secretion from THP-1 cells induced by nigericin is a result of activation of p45 IL-1beta-converting enzyme processing. J Biol Chem. 1998;273:17846–17851. doi: 10.1074/jbc.273.28.17846. [DOI] [PubMed] [Google Scholar]
- 184.Verhoef PA, Kertesy SB, Lundberg K, Kahlenberg JM, Dubyak GR. Inhibitory effects of chloride on the activation of caspase-1, IL-1beta secretion, and cytolysis by the P2X7 receptor. J Immunol. 2005;175:7623–7634. doi: 10.4049/jimmunol.175.11.7623. [DOI] [PubMed] [Google Scholar]
- 185.Gudipaty L, Munetz J, Verhoef PA, Dubyak GR. Essential role for Ca2+ in regulation of IL-1beta secretion by P2X7 nucleotide receptor in monocytes, macrophages, and HEK-293 cells. Am J Physiol Cell Physiol. 2003;285:C286–299. doi: 10.1152/ajpcell.00070.2003. [DOI] [PubMed] [Google Scholar]
- 186.Franchi L, Kanneganti TD, Dubyak GR, Nunez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J Biol Chem. 2007;282:18810–18818. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- 187.Pelegrin P, Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J Biol Chem. 2007;282:2386–2394. doi: 10.1074/jbc.M610351200. [DOI] [PubMed] [Google Scholar]
- 188.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin- 1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 190.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 191.Wickliffe KE, Leppla SH, Moayeri M. Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell Microbiol. 2008;10:332–343. doi: 10.1111/j.1462-5822.2007.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gattorno M, et al. Pattern of interleukin-1beta secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis Rheum. 2007;56:3138–3148. doi: 10.1002/art.22842. [DOI] [PubMed] [Google Scholar]
- 193.Hawkins PN, Lachmann HJ, McDermott MF. Interleukin-1-receptor antagonist in the Muckle-Wells syndrome. N Engl J Med. 2003;348:2583–2584. doi: 10.1056/NEJM200306193482523. [DOI] [PubMed] [Google Scholar]
- 194.Hawkins PN, Lachmann HJ, Aganna E, McDermott MF. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 2004;50:607–612. doi: 10.1002/art.20033. [DOI] [PubMed] [Google Scholar]
- 195.Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–521. doi: 10.1038/nature01991. [DOI] [PubMed] [Google Scholar]
- 196.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 197.Chen CJ, et al. MyD88-dependent IL-1 receptor signaling is essential for gouty inflammation stimulated by monosodium urate crystals. J Clin Invest. 2006;116:2262–2271. doi: 10.1172/JCI28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.So A, De Smedt T, Revaz S, Tschopp J. A pilot study of IL-1 inhibition by anakinra in acute gout. Arthritis Res Ther. 2007;9:R28. doi: 10.1186/ar2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li H, Nookala S, Re F. Aluminum hydroxide adjuvants activate caspase-1 and induce IL- 1beta and IL-18 release. J Immunol. 2007;178:5271–5276. doi: 10.4049/jimmunol.178.8.5271. [DOI] [PubMed] [Google Scholar]
- 200.Kool M, et al. Cutting Edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 201.Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Franchi L, Nunez G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur J Immunol. 2008;38:2085–2089. doi: 10.1002/eji.200838549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gavin AL, et al. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Freche B, Reig N, van der Goot FG. The role of the inflammasome in cellular responses to toxins and bacterial effectors. Semin Immunopathol. 2007;29:249–260. doi: 10.1007/s00281-007-0085-0. [DOI] [PubMed] [Google Scholar]
- 206.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 207.Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Perregaux D, Gabel CA. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem. 1994;269:15195–15203. [PubMed] [Google Scholar]
- 209.Walev I, Reske K, Palmer M, Valeva A, Bhakdi S. Potassium-inhibited processing of IL- 1 beta in human monocytes. EMBO J. 1995;14:1607–1614. doi: 10.1002/j.1460-2075.1995.tb07149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ozoren N, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- 211.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tsuji NM, et al. Roles of caspase-1 in Listeria infection in mice. Int Immunol. 2004;16:335–343. doi: 10.1093/intimm/dxh041. [DOI] [PubMed] [Google Scholar]
- 213.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Labbe K, Saleh M. Cell death in the host response to infection. Cell Death Differ. 2008;15:1339–1349. doi: 10.1038/cdd.2008.91. [DOI] [PubMed] [Google Scholar]
- 215.Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007;9:2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 216.Hara H, Tsuchiya K, Nomura T, Kawamura I, Shoma S, Mitsuyama M. Dependency of caspase-1 activation induced in macrophages by Listeria monocytogenes on cytolysin, listeriolysin O, after evasion from phagosome into the cytoplasm. J Immunol. 2008;180:7859– 7868. doi: 10.4049/jimmunol.180.12.7859. [DOI] [PubMed] [Google Scholar]
- 217.Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Surprenant A, Rassendren F, Kawashima E, North RA, Buell G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7) Science. 1996;272:735–738. doi: 10.1126/science.272.5262.735. [DOI] [PubMed] [Google Scholar]
- 219.Le Feuvre RA, Brough D, Iwakura Y, Takeda K, Rothwell NJ. Priming of macrophages with lipopolysaccharide potentiates P2X7-mediated cell death via a caspase-1-dependent mechanism, independently of cytokine production. J Biol Chem. 2002;277:3210–3218. doi: 10.1074/jbc.M104388200. [DOI] [PubMed] [Google Scholar]
- 220.Hentze H, Lin XY, Choi MS, Porter AG. Critical role for cathepsin B in mediating caspase-1-dependent interleukin-18 maturation and caspase-1-independent necrosis triggered by the microbial toxin nigericin. Cell Death Differ. 2003;10:956–968. doi: 10.1038/sj.cdd.4401264. [DOI] [PubMed] [Google Scholar]
- 221.Willingham SB, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 225.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 227.Raupach B, Peuschel SK, Monack DM, Zychlinsky A. Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun. 2006;74:4922–4926. doi: 10.1128/IAI.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sansonetti PJ, et al. Caspase-1 activation of IL-1beta and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity. 2000;12:581–590. doi: 10.1016/s1074-7613(00)80209-5. [DOI] [PubMed] [Google Scholar]
- 230.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Scott AM, Saleh M. The inflammatory caspases: guardians against infections and sepsis. Cell Death Differ. 2007;14:23–31. doi: 10.1038/sj.cdd.4402026. [DOI] [PubMed] [Google Scholar]
- 232.Rosenberger CM, Finlay BB. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol. 2003;4:385–396. doi: 10.1038/nrm1104. [DOI] [PubMed] [Google Scholar]
- 233.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 234.Blander JM, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006;7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- 235.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Koo IC, Wang C, Raghavan S, Morisaki JH, Cox JS, Brown EJ. ESX-1-dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol. 2008;10:1866–1878. doi: 10.1111/j.1462-5822.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Master SS, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]