Abstract
Background:
About 20% of familial amyotrophic lateral sclerosis (ALS) is caused by mutations in SOD1 and is typically transmitted as an autosomal dominant trait. However, due to reduced mutation penetrance, the disease may present in a recessive or sporadic manner.
Objective:
To determine the factors responsible for the low penetrance of the SOD1 mutation.
Methods:
Twelve members of a Canadian ALS family of Filipino origin were recruited for the study. SOD1 was sequenced in the proband. SOD1 expression was assessed by real-time-PCR and immunoblotting.
Results:
The proband was a homozygous carrier of a novel 6 bp deletion in exon 2 (ΔG27/P28), the pathologic significance of which was confirmed by immunohistochemistry. Eight living family members are heterozygotes and remain unaffected at ages ranging between 48 and 85 years. Haplotype analysis showed that the deletion is a single founder mutation likely common in the Cagayan province (Philippines). The low penetrance of the mutation is explained by the fact that it enhances the naturally occurring alternative splicing of exon 2 of the SOD1 mRNA, leading to reduced transcription of the mutant allele. Indeed, Western blot analysis demonstrated the low level of SOD1 protein in carriers of the ΔG27/P28 compared to wild-type individuals or a carrier of the A4V SOD1 mutation.
Conclusion:
The enhanced splicing of exon 2 acts as a natural knock-down of the mutant SOD1 allele in the Filipino amyotrophic lateral sclerosis (ALS) family. There is a need for careful investigation of splicing isoforms of SOD1 and other ALS genes as factors influencing the severity of disease.
GLOSSARY
- AD
= Alzheimer disease;
- ALS
= amyotrophic lateral sclerosis;
- CBD
= corticobasal syndrome;
- HCI
= hyaline conglomerate inclusions;
- RT-PCR
= reverse transcriptase–polymerase chain reaction;
- sALS
= sporadic amyotrophic lateral sclerosis;
- SEDI
= SOD1-exposed-dimer-interface antibody.
Amyotrophic lateral sclerosis (ALS) is a progressive and fatal neurodegenerative disease affecting motor neurons of the brain, brainstem, and spinal cord. Approximately 5% of cases are familial ALS with up to 20% of these caused by mutations in the superoxide dismutase-1 gene (SOD1), a ubiquitously expressed free-radical scavenging enzyme.1 ALS caused by SOD1 mutations is not believed to be related to changes in normal enzymatic activity, but is attributed to a gain of toxic function.2 However, the exact mechanism of SOD1 pathogenesis remains uncertain.
Inheritance patterns for patients with familial ALS can be complicated and the risk to currently unaffected mutation carriers can be difficult to ascertain. Hence, it is critical to understand the factors influencing the manifestation of ALS. Over 100 different, mainly missense, SOD1 mutations have been identified in autosomal dominant ALS (http://alsod.iop.kcl.ac.uk/Als/misc/dataDownload.aspx);however, only a third of them have good genetic support for their pathogenic nature (i.e., segregation study). Furthermore, the disease may present in an apparently recessive or sporadic manner due to the considerably reduced penetrance of some SOD1 mutations. The most striking example is the D90A that exhibits both dominant and recessive modes of inheritance of ALS. Several families carrying the D90A mutation require homozygosity for disease, and since loss of SOD1 enzymatic activity has been discounted as a pathologic mechanism, this suggests the presence of modifier genes in the recessive haplotype that may reduce the toxicity of the mutant SOD1 protein.3,4
The factors influencing penetrance remain a mystery and are addressed in this study for the first time. We describe a novel SOD1 mutation in a Canadian family originating from the Philippines, present evidence that the changes in SOD1 alternative splicing underlie the low penetrance of the mutation, and discuss the possibility of a similar mechanism for phenotypic heterogeneity in other ALS families.
METHODS
Subjects.
Twelve members of a Filipino family were recruited at the ALS Clinic, Sunnybrook Health Sciences Centre, Toronto, Canada, and affected individuals were diagnosed with ALS in accordance with revised El Escorial criteria.5 The presence of the SOD1 mutation was assessed in three independent datasets. The previously described UK dataset was recruited through the Queen Elizabeth Hospital, Birmingham, and included 194 patients with sporadic ALS (mean age at onset 58 ± 11; range 18–81 years).6 The Canadian ALS dataset was recently collected at the Sunnybrook Health Sciences Centre and included 60 patients with sporadic (mean age at onset 56 ± 11; range 36–80 years) and 14 patients with familial ALS (mean age at onset 50 ± 15; range 29–73 years). In addition, 179 unrelated neurologically normal Filipino controls (50 ± 5 years of age) were selected from a cohort of women who participated in the Cebu Longitudinal Health and Nutrition Study (www.cpc.unc.edu.projects/cebu). Required consents were obtained from all study participants in accordance with the respective ethical review boards.
Genetic analyses.
A complete mutation analysis of SOD1 including exon/intron boundaries was performed on members of a Filipino family. The frequency of the ΔG27/P28 mutation was evaluated using an AvaII restriction assay. The haplotype analysis was done using genotypes of 15 microsatellite markers (figure 1) spanning 17.2 Mb on chromosome 21 and harboring the SOD1 locus (from D21S1884 at 21.5 Mb to D21S1255 at 38.7 Mb). The haplotypes were constructed using the program MERLIN.7 The exonic splicing enhancer sequences were assessed by using the ESEfinder (http://rulai.cshl.edu/tools/ESE2).8 The cDNAs were generated from total RNA (1.5 μg) of white blood cells or the gray matter region from lumbar spinal cord using the StrataScript first strand synthesis kit (Stratagene, CA). Reverse transcriptase–polymerase chain reaction (RT-PCR) primers were designed for the 5′-UTR and exon 5 of SOD1. Real-time RT-PCR was done by using SYBR Green reagent (TaKaRa Mirus Bio, Madison, WI) on an ABI7500 system (Applied Biosystems, Foster City, CA). Details of the assays are available on the Neurology® Web site at www.neurology.org (see table e-1 and appendix e-1).
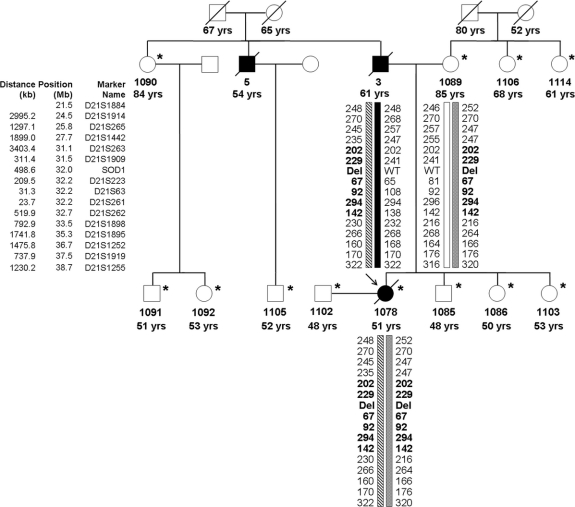
Figure 1 Pedigree of an amyotrophic lateral sclerosis (ALS) family with a SOD1 mutation
The pedigree structure of the Filipino family segregating the ΔG27/P28 mutation (del); the asterisk indicates the 12 individuals for whom DNA samples and genotypes were obtained. Affected individuals are shown as filled symbols and the arrow points to the proband with the homozygous deletion. A slash indicates deceased persons. Below the DNA number is the age at onset for affected individuals, age at examination, or age at death for unaffected individuals. Genotypes are shown only for the key individuals to protect family confidentiality. Markers used for haplotype analysis are shown to the left (founder haplotype is in bold). The inferred haplotype of the proband's father (no. 3) is reconstructed based on the genotypes of his four children available for study.
Protein analyses.
For the immunoblotting analysis, samples of white blood cells and lumbar spinal cord samples from patients were homogenized in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.25% Na deoxycholate, and 1% NP-40) with protease inhibitor cocktail (Roche). The protein concentration of tissue extracts was determined by bicinchoninic acid assay (Sigma) and 2.5 μg protein extracts were separated on 17.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transfer onto PVDF membranes (Millipore). Membranes were blocked with 5% (w/v) nonfat milk powder in Tris-buffered saline and then probed with rabbit polyclonal anti-SOD1 (SOD100, 1:1000, Stressgen) and secondary antibodies (horseradish peroxide-conjugated anti-rabbit IgG, Amersham). Blots were developed with enhanced chemiluminescence reagents (Perkin-Elmer) and digital images were acquired. Blots were re-probed with mouse monoclonal anti-GAPDH antibody (1:5000, Biodesign) to control for equal loading. Quantification of band intensity on the blot was performed by densitometric analysis using ImageJ (NIH). The expression of SOD1 was normalized to that of GAPDH and presented as a relative density. SOD1 activity was measured in erythrocytes from patients with ALS and controls using a superoxide dismutase activity kit (900-157, Stressgen) (see supplementary material).
Immunohistochemistry.
Sections of 6 μm were cut from formalin-fixed paraffin-embedded sections of autopsied lumbar spinal cord tissue, and rehydrated through a graded series of ethanol washes and water, as we have described previously.9 Sections were labeled with antibodies to phosphorylated neurofilaments (SMI31, diluted 1:5,000; Sternberger Monoclonals, Baltimore, MD) or SOD1-exposed-dimer-interface antibody (SEDI antibody, diluted 1:500), a previously described rabbit polyclonal antibody that labels misfolded SOD1.10 For SEDI antibody, sections were pretreated with 10 mM sodium citrate buffer (pH 6.0) before labeling. Labeling was detected using the peroxidase-based Dako EnVision kit with DAB or Permanent Red as chromogenic substrate. All sections were counterstained with hematoxylin.
RESULTS
Clinical findings.
Proband 1078 belongs to a Canadian family of Filipino ancestry with 20 known family members (figure 1), with samples from 12 individuals of this kindred available for study. Overall, three family members have been diagnosed with and died of ALS. The mean age at onset was 55.3 ± 5.1 (range 51–61) years; mean age at death was 59.7 ± 5.8 (range 55–66) years; and mean duration was 4.3 ± 1.5 (range 4–6) years. There is no known history of consanguinity in the family.
Patient 1078 (proband) was diagnosed with ALS at the Sunnybrook ALS Clinic at age 53. The symptoms began at age 51 when the patient experienced progressive leg followed by arm weakness. Bulbar symptoms then followed and the patient died of respiratory failure as a complication of ALS at age 55. Patient 1078 had predominantly lower motor neuron signs with only mildly brisk reflexes in the upper limbs. The autopsy result confirmed the diagnosis of ALS as discussed below.
Patient 3 (the proband's father) was diagnosed with ALS at the Sunnybrook ALS Clinic at age 62 and died 4 years later at age 66 (no autopsy was performed). The ALS symptoms began at age 61 and the disease had a similar course as in patient 1078 with predominantly lower motor neuron findings, which began in the legs and then progressed to the arms and bulbar regions. Patient 5 (the paternal uncle of the proband) was diagnosed with ALS by a neurologist in the Philippines at age 54 and died 4 years later at age 58. No additional clinical details are available.
The 11 remaining family members who were available for the current study are older than 48 years and report good health. A detailed clinical neurologic assessment of these family members was performed by a neurologist, who was blind to genetic status of the participants. All clinical examinations were found to be normal. Hence, none of the living family members demonstrate any signs of ALS, including four individuals older than the mean age at onset (61–85 years old). In addition, the grandparents of the proband died of causes unrelated to ALS at 52, 65, 67, and 80 years of age as result of stroke or pneumonia.
Genetic analyses.
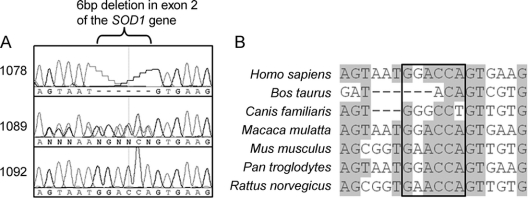
Sequence analysis of the entire open reading frame of SOD1 was performed on the proband 1078. We detected a novel homozygous GGACCA-deletion at genomic nucleotide position 31957983 according to accession number NC_000021.7 (Gly27delGGACCA) (figure 2). This 6-bp deletion in exon 2 removes two amino acids (G27 and P28) in a conserved part of loop II of the SOD1 protein (ΔG27/P28). No other SOD1 variations were identified. The ΔG27/P28 was not found in 179 normal controls of Filipino origin. The mutation is unique to the Filipino family in our ALS dataset, since it was not observed in any of the 254 unrelated patients from two sporadic cohorts of UK (n = 194) and Canadian (n = 60) origin, as well as in 14 independent Canadian patients with familial ALS.
Figure 2 Mutation analysis of a Filipino amyotrophic lateral sclerosis (ALS) family
(A) DNA sequence fluorescent chromatogram of the SOD1 ΔG27/P28 deletion observed in family members. (B) Multiple alignment of the sequence around the ΔG27/P28. The gray highlight indicates areas of conservation (>50% of bases in the alignment matches). The boxed area represents the ΔG27/P28 deletion.
The deletion was independently inherited by the proband's parents originating from the same Filipino province (Cagayan). Segregation analysis revealed the presence of heterozygous ΔG27/P28 mutation in eight unaffected individuals and enabled the reconstruction of the genotype of ALS patient 3 (proband's father) who was an obligate heterozygote, based on the genotypes of his children: 1078 (homozygous carrier) and 1103 (wild-type). Haplotype analysis confirmed this conclusion and excluded nonpaternity in the family (figure 1). Furthermore, it showed that the ΔG27/P28 is a single founder mutation, since all carriers inherited a common 1.6 Mb haplotype harboring SOD1 locus (between markers D21S263 and D21S262).
Sequencing analysis of SOD1 in the two oldest heterozygotes (84 and 85 years old) did not reveal any additional variations; however, the ΔG27/P28 mutation may itself be responsible for the low penetrance. The ΔG27/P28 is predicted to enhance naturally occurring alternative splicing of exon 2 of the SOD1 mRNA, since it eliminates an exonic enhancer sequence for the splicing factor SC35, which promotes exon definition (score 4.7 for GACCAGTG-motif).8 Hence, exon 2 in mutation carriers could be poorly recognized by the splicing machinery, leading to its removal from SOD1 mRNA and therefore decreasing the expression of the mutant allele.
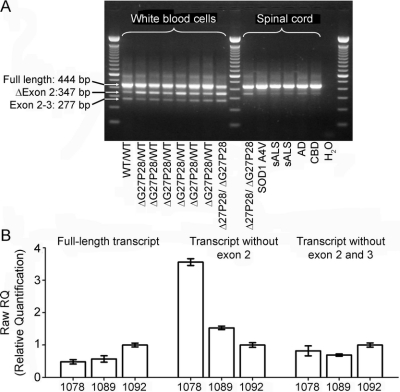
RT-PCR confirmed the prediction above (figure 3). It was performed on white blood cells from eight family members with different mutation status (wild-type, homozygote, and six heterozygotes). Apart from the full-length SOD1 transcript (444 bp), we observed two additional fragments. Sequence analysis showed that the 347 bp band corresponds to SOD1 cDNA without exon 2, while the 277 bp band represents a transcript without exons 2 and 3. Both splicing events create a frameshift after exon 1 resulting in a premature stop codon. The abundance of the transcript lacking exons 2 and 3 was similar in all individuals. In contrast, the expression of the transcript without exon 2 was enhanced in mutation carriers with the highest abundance in the homozygous proband (1:1 ratio between transcripts with and without exon 2), while this isoform was observed at a much lower level in the wild-type individual.
Figure 3 Result of RT-PCR on individuals with different mutation status
(A) The result of conventional reverse transcriptase–polymerase chain reaction (RT-PCR) on blood samples from members of the Filipino family and on spinal cord tissue of six unrelated individuals: the homozygote for ΔG27/P28, heterozygote for A4V SOD1, two patients with sporadic amyotrophic lateral sclerosis (sALS), a patient with Alzheimer disease (AD), and a patient with corticobasal syndrome (CBD). The arrows point to different SOD1 amplification products representing the transcript with all five coding exons (full-length), splice variant lacking exon 2, and splice variant lacking both exon 2 and 3. The RT-PCR result was replicated in three independent experiments. (B) The result of quantitative RT-PCR for Filipino family members, including the homozygote (1078), heterozygote (1089), and wild-type individual (1092). Samples were analyzed in triplicate (error bars represent SD). The ΔG27/P28 homozygote demonstrated the lowest level of full-length SOD1 expression (left graph) and the highest level of expression of the transcript without exon 2 (middle graph). The abundance of the SOD1 fragment lacking both exon 2 and exon 3 was similar in all three individuals (right graph).
In addition, we conducted RT-PCR on lumbar spinal cord tissues of six unrelated individuals including the proband (figure 3A). Again, the highest level of the transcript without exon 2 was observed in the homozygous carrier, whereas it was barely detected in all other samples. The isoform lacking both exon 2 and exon 3 was not identified. The results of the conventional RT-PCR were confirmed by quantitative RT-PCR (figure 3B). Since ΔG27/P28 increases the splicing of exon 2, there is a concomitant reduction in expression of the full-length mutant transcript (twice less in the homozygote than in wild-type).
Protein analyses of SOD1.
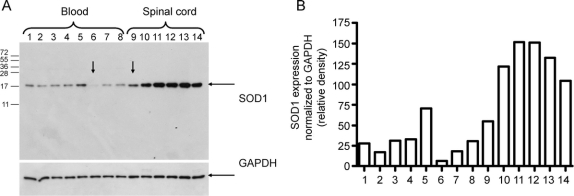
In order to assess the relative expression of SOD1 at the protein level, we performed a Western blot analysis (figure 4A). White blood cells and spinal cord samples from patients with different mutation status demonstrated considerable downregulation of SOD1 in individuals with ΔG27/P28 (especially striking in the homozygous carrier). The splicing isoforms lacking exon 2 would have a predicted molecular weight of 3.7 kDa and 5.5 kDa; however, no such species were detected in either blood or spinal cord samples. Quantification of the band intensities revealed a 30–40% reduction in SOD1 expression in white blood cells from the heterozygous ΔG27/P28 family members relative to wild-type (lanes 1–4 and 7–8 vs lane 5) (figure 4B). This reduction was even more pronounced in the homozygote (lane 6 vs lane 5; ∼8%). The SOD1 level in the spinal cord sample from the homozygous carrier was ∼40% less than in the spinal cord samples of individuals without ΔG27/P28 (lane 9 vs lanes 10–14). In addition, the SOD1 enzyme activity was measured in erythrocytes from the homozygous proband, a heterozygous family member, one patient with sporadic ALS, and two normal controls (figure e-1). The sample from the homozygous carrier demonstrated a reduced activity of SOD1 (by ∼50% of the control level), and the activity of a sample from the heterozygous carrier was also reduced, but to a lesser extent (by ∼30% of the control level).
Figure 4 Immunoblotting analysis of SOD1 expression
(A) Western blot with arrow points to the samples from the proband. Protein lysates of white blood cells from family members heterozygous (lanes 1–4 and 7–8), wild-type (lane 5), and homozygous (lane 6) for the ΔG27/P28 deletion, and lumbar spinal cord extracts from the ΔG27/P28 homozygote (lane 9), familial amyotrophic lateral sclerosis (ALS) A4V case (lane 10), two sporadic ALS cases (lanes 11–12), Alzheimer disease case (lane 13), and corticobasal syndrome case (lane 14) immunoblotted against anti-SOD1 antibody. The membrane was reprobed with anti-GAPDH antibody to show equal loading. (B) Quantification of SOD1 protein expression based on immunoblotting band intensity (relative density normalized to GAPDH).
Immunohistochemistry.
The diagnosis of ALS for the proband was confirmed by neuropathologic analysis (figure 5). The underlying role of SOD1 in causation of ALS was supported by the presence of neurofilamentous hyaline conglomerate inclusions (HCI).11,12 These inclusions had the typical floccular appearance of HCIs and were labeled with antibody to phosphorylated neurofilaments. In addition, sections were labeled with SEDI antibody, targeting misfolded SOD1.10 The HCIs were consistently labeled with the SEDI antibody, displaying a patchy appearance within the inclusions. Collectively, these observations support a diagnosis of ALS caused by mutant SOD1.
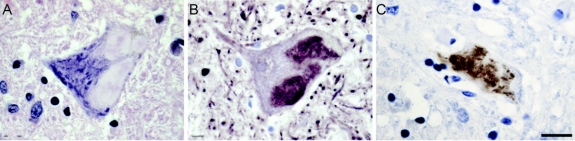
Figure 5 Immunohistochemical labeling in motor neurons of the homozygous mutation carrier
Labeling of hyaline conglomerate inclusions (HCI) in patient 1078 carrying the homozygous ΔG27/P28 SOD1 mutation show specificity for SOD1-related amyotrophic lateral sclerosis. Hematoxylin staining (A) shows floccular appearance of HCI. Labeling of HCIs with phosphorylated neurofilament antibody (B) and an antibody recognizing misfolded SOD1, SEDI antibody (C). Scale bar = 12 μm.
DISCUSSION
This is the first report of a SOD1-linked family of Filipino origin and it would be important to estimate the mutation prevalence in Filipino patients with ALS, since the deletion was independently inherited by the proband's parents originating from the Cagayan province. In agreement with a single founder, the extended 1.6 Mb haplotype around SOD1 is common to all carriers of ΔG27/P28. The absence of any clinical signs of ALS in eight heterozygotes and the occurrence of a homozygous carrier could indicate that ΔG27/P28 is associated with a recessive mode of inheritance; however, patient 3 was likely a heterozygote and yet developed ALS. Nevertheless, the disease in patient 3 was less severe than in the homozygous proband with ALS onset, delayed by a decade and extended duration (figure 1). There are several possible explanations for why patient 3 developed ALS, while other heterozygotes have not (e.g., male gender as a disease risk factor13 [figure 1], trans-acting suppressors of alternative splicing, or a second mutation in another ALS gene). The absence of a sample for patient 3 precludes such an investigation; however, the ongoing collection of Filipino cases could recover similar families.
The low penetrance of ΔG27/P28 could be explained by the fact that the deletion enhances alternative splicing of exon 2 and consequently reduces the expression of the mutant allele, which is supported by the results of RT-PCR and Western blot (figures 3 and 4). In agreement with these data, SOD1 activity in the erythrocytes of mutation carriers was considerably reduced compared to the control samples (figure e-1).
The possibility for the pathologic significance of the SOD1 transcript lacking exon 2 (or both exon 2 and 3) is diminished by the fact that it was observed in subjects with and without neurologic diseases (including ALS).14,15 Furthermore, these transcripts containing a premature termination codon are predicted to encode a protein truncated after exon 1; however, our result (figure 4) and two independent studies using different antibodies14,15 failed to detect such proteins, suggesting either its rapid degradation or the absence of translation. It was reported that nonsense-mediated mRNA decay underlies the clearance of SOD1 mRNA with a premature termination codon in exons 1–4, but not in exon 5.16 This is why among the reported SOD1 ALS mutations, stop codon mutations exclusively occur in exon 5. It is possible that nonsense-mediated decay could be involved in the degradation of the alternatively spliced SOD1 mRNA, which could be more efficient in spinal cord tissue than in blood cells, since the level of SOD1 transcript without exon 2 was lower in the spinal cord (figure 3).
Thus, the enhanced splicing of exon 2 acts as a natural knock-down of the mutant allele, since half of the mutant transcript would not be translated. It is tempting to speculate that a similar mechanism could play a role in the phenotypic modification of other ALS kindreds, including the Scandinavian families with D90A mutation.3,4 Since pedigrees with D90A have retained a recessive inheritance regardless of considerable outbreeding, it was suggested that SOD1-linked variations account for the modifying effect.4 Indeed, the pathologic mutation could be part of a particular SOD1 haplotype that includes polymorphisms affecting splicing elements, the promoter region, or microRNA target sites and as a result causing decreased expression of the mutant allele. In general, SOD1 could be vulnerable to alternative splicing because the invariant donor splice site of the first intron has an atypical GC-motif instead of the GT-motif required for efficient pre-mRNA splicing.17
Over 74% of human genes generate alternative transcripts with a bias toward genes expressed in the nervous system.18 Multiple reports have indicated that splice variants of proteins associated with key neuropathologies are linked with disease; however, the current study for the first time suggests a protective effect of alternative splicing in neurodegenerative disease. As such, strategies aimed at inducing splicing may have therapeutic potential by reducing mutant SOD1 expression. Hence, there is a need for careful investigation of alternative splicing of SOD1 and other ALS genes as factors influencing severity or phenotypic heterogeneity of either sporadic or familial ALS.
Supplementary Material
Address correspondence and reprint requests to Dr. Ekaterina Rogaeva, Centre for Neurodegenerative Diseases, Department of Medicine, University of Toronto, 6 Queen's Park Crescent West, Toronto, Ontario, Canada, M5S 3H2 ekaterina.rogaeva@utoronto.ca
Supplemental data at www.neurology.org
*These authors contributed equally.
Supported by grants from the Krembil Scientific Development Seed Fund (E.R., J.R., L.Z.), Japan-Canada, the Temerty Family Foundation (L.Z.), and Canadian Institutes of Health Research (CIHR) Joint Health Research Program (E.R.); J.R. holds a Canada Research Chair and is funded by grants from the American ALS Association, US Muscular Dystrophy Association, the ALS Society of Canada, and the CIHR.
Disclosure: The authors report no disclosures.
Received July 31, 2008. Accepted in final form December 16, 2008.
REFERENCES
- 1.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993;362:59–62. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci 2001;2:806–819. [DOI] [PubMed] [Google Scholar]
- 3.Andersen PM, Nilsson P, Ala-Hurula V, et al. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat Genet 1995;10:61–66. [DOI] [PubMed] [Google Scholar]
- 4.Parton MJ, Broom W, Andersen PM, et al. D90A-SOD1 mediated amyotrophic lateral sclerosis: a single founder for all cases with evidence for a Cis-acting disease modifier in the recessive haplotype. Hum Mutat 2002;20:473. [DOI] [PubMed] [Google Scholar]
- 5.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 6.Xiao S, Sato C, Kawarai T, et al. Genetic studies of GRN and IFT74 in amyotrophic lateral sclerosis. Neurobiol Aging 2008;8:1279–82. [DOI] [PubMed] [Google Scholar]
- 7.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 8.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a Web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003;31:3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanelli T, Xiao S, Horne P, Bilbao J, Zinman L, Robertson J. Evidence that TDP-43 is not the major ubiquitinated target within the pathological inclusions of amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 2007;66:1147–1153. [DOI] [PubMed] [Google Scholar]
- 10.Rakhit R, Robertson J, Vande Velde C, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med 2007;13:754–759. [DOI] [PubMed] [Google Scholar]
- 11.Hays AP, Naini A, He CZ, Mitsumoto H, Rowland LP. Sporadic amyotrophic lateral sclerosis and breast cancer: hyaline conglomerate inclusions lead to identification of SOD1 mutation. J Neurol Sci 2006;242:67–69. [DOI] [PubMed] [Google Scholar]
- 12.Ince PG, Tomkins J, Slade JY, Thatcher NM, Shaw PJ. Amyotrophic lateral sclerosis associated with genetic abnormalities in the gene encoding Cu/Zn superoxide dismutase: molecular pathology of five new cases, and comparison with previous reports and 73 sporadic cases of ALS. J Neuropathol Exp Neurol 1998;57:895–904. [DOI] [PubMed] [Google Scholar]
- 13.Nelson LM. Epidemiology of ALS. Clin Neurosci 1995;3:327–331. [PubMed] [Google Scholar]
- 14.Kawata A, Kato S, Shimizu T, et al. Aberrant splicing of human Cu/Zn superoxide dismutase (SOD1) RNA transcripts. Neuroreport 2000;11:2649–2653. [DOI] [PubMed] [Google Scholar]
- 15.Hirano M, Hung WY, Cole N, Azim AC, Deng HX, Siddique T. Multiple transcripts of the human Cu,Zn superoxide dismutase gene. Biochem Biophys Res Commun 2000;276:52–56. [DOI] [PubMed] [Google Scholar]
- 16.Han-Xiang D, Hujun J, Ronggen F, et al. Molecular dissection of ALS-associated toxicity of SOD1 in transgenic mice using an exon-fusion approach. Hum Mol Genet 2008;17:2310–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res 1982;10:459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol 2004;22:535–546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.