Abstract
3-acetamido-3,6-dideoxy-α-D-glucose or Quip3NAc is an unusual deoxyamino sugar found in the O-antigens of some Gram-negative bacteria and in the S-layers of Gram-positive bacteria. It is synthesized in these organisms as a dTDP-linked sugar via the action of five enzymes. The focus of this investigation is on QdtB from Thermoanaerobacterium thermosaccharolyticum E207–71, a PLP-dependent aminotransferase that catalyzes the penultimate step in the production of dTDP-Quip3NAc. For this analysis, the enzyme was crystallized in the presence of its product, dTDP-Quip3N, and the structure was solved and refined to 2.15-Å resolution. QdtB is a dimer, and its overall fold places it into the well-characterized aspartate aminotransferase superfamily. Electron density corresponding to the bound product reveals the presence of a Schiff base between C-4′ of the PLP cofactor and the amino nitrogen of the sugar. Those amino acid side chains involved in binding the dTDP-sugar into the active site include Tyr 183, His 309, and Tyr 310 from subunit 1 and Lys 219 from subunit 2. Notably there is a decided lack of interactions between the pyranosyl C-4′ hydroxyl of the dTDP-sugar and the protein. In keeping with this observation, we show that QdtB can also turn over dTDP-3-acetamido-3,6-dideoxy-α-D-galactose. This investigation represents the first structural analysis of a sugar-modifying aminotransferase with a bound product in its active site that functions at the C-3′ rather than the C-4′ position of the hexose.
Deoxyamino sugars represent an interesting and unusual class of carbohydrates synthesized by a variety of bacteria, fungi, and plants (1). These types of carbohydrates can be found, for example, attached to macrolide antibiotics such as erythromycin. A past structural analysis of the 50S ribosomal subunit from Deinococcus radiodurans complexed with erythromycin highlights the importance of the deoxyamino sugar in ribosome binding (2). Indeed, antibiotics such as carbomycin A, spiramycin, tylosin, and azithromycin, all of which contain derivatives of deoxyamino sugars, have been shown to function as anti-microbial agents by binding to ribosomes and blocking protein synthesis (3). Deoxyamino sugars, however, are not confined solely to antibiotics. They are, in fact, widespread in Nature where they are often found in the O-antigens of Gram-negative bacteria or in the glycan portions of bacterial cell surface layers (S-layers) (4, 5). In all cases, whether these deoxyamino sugars are attached to macrolide antibiotics or are constituents of the bacterial O-antigens or S-layers, they are synthesized via biochemical pathways that require the initiating sugar to be attached to a nucleotide, typically dTDP (1, 6). Additionally, regardless of the positions of the amino groups on the carbohydrate scaffolds, they are attached to the sugars via the actions of pyridoxal 5′-phosphate-(PLP)-dependent aminotransferases (6).
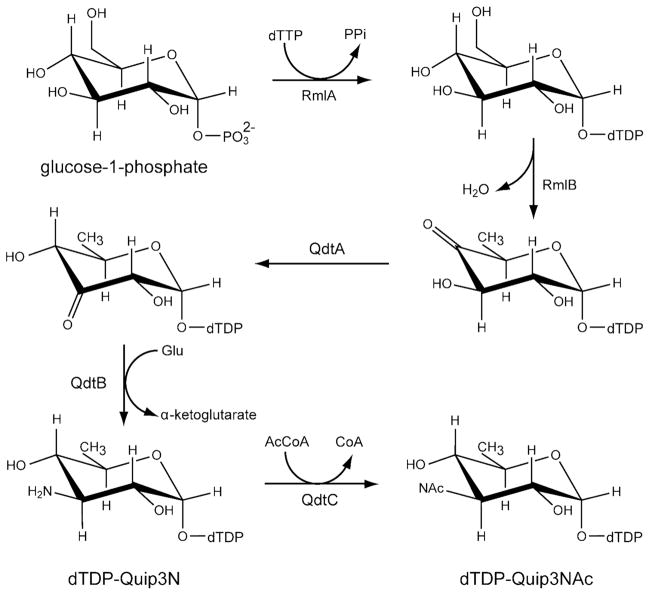
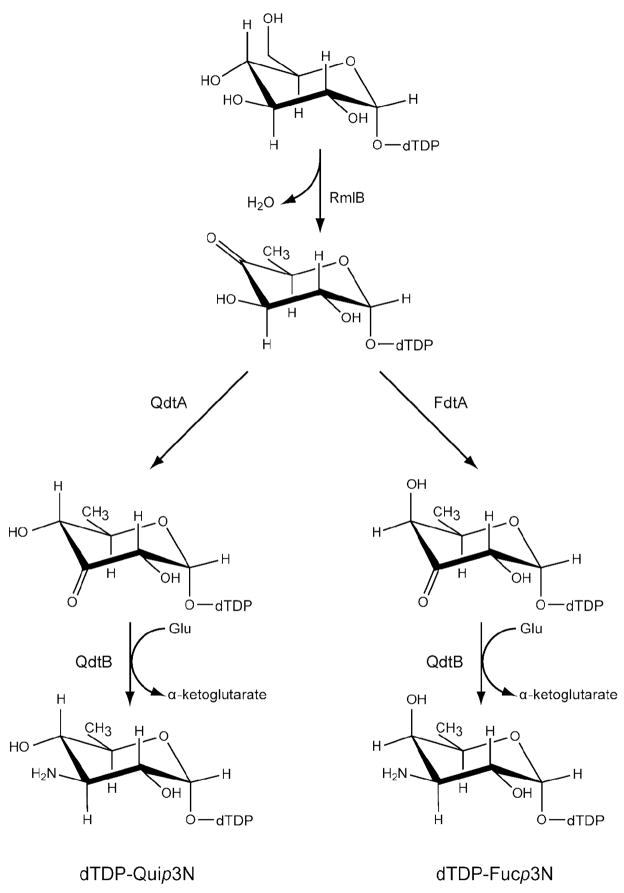
The PLP-dependent aminotransferases have been the focus of significant structural and functional studies in the past due to the remarkable reactions they catalyze, ranging from racemizations, decarboxylations, transaminations, and eliminations, among others (7). Only recently, however, have the structures of sugar-modifying aminotransferases been determined (8–13). The focus of this investigation is QdtB, a sugar-modifying aminotransferase from Thermoanaerobacterium thermosaccharolyticum E207–71 (14). QdtB plays a key role in the biosynthesis of 3-acetamido-3,6-dideoxy-α-D-glucose (Quip3NAc), a carbohydrate that has been observed in the O-antigens of some Gram-negative bacteria including Escherichia coli O114 (15) and in the S-layers of Gram-positive bacteria (16). As indicated in Scheme 1, five enzymes are required for the formation of dTDP-Quip3NAc (16). In the first step of the pathway, glucose-1-phosphate is attached to the nucleotide via the action of RmlA, a glucose 1-phosphate thymidylyltransferase. Subsequently, the 6′-hydroxyl group is removed and the C-4′ hydroxyl group is oxidized to a keto moiety by RmlB, a 4,6-dehydratase. The third step in the pathway is catalyzed by QdtA, a 3,4-ketoisomerase that produces dTDP-3-keto-6-deoxy-D-glucose. This nucleotide-linked sugar is then aminated at the C-3′ position of the hexose by the action of QdtB. In the final step of the pathway, dTDP-QuipN, the product of the QdtB reaction, is acetylated by QdtC, a CoA-dependent enzyme.
Scheme 1.
Here we present the three-dimensional structure of QdtB complexed with its dTDP-sugar product. This investigation reveals the detailed manner in which the nucleotide-linked sugar is positioned into the active site. In particular, the lack of a specific interaction between the protein and the C-4′ hydroxyl group of the hexose prompted us to examine the substrate specificity of QdtB. Activity assays demonstrate that QdtB can also use dTDP-3-keto-6-deoxy-D-galactose as a substrate. In addition, we show that another aminotransferase, namely DesV of the dTDP-desosamine biosynthetic pathway (17), can use the QdtB substrate in place of its natural substrate, dTDP-3-keto-4,6-dideoxy-D-glucose. Taken together, these investigations shed new light on the active site subtleties of the sugar aminotransferases that lead to aminations of the hexose rings at different positions and with different stereochemistries.
MATERIALS AND METHODS
Cloning, Expression, and Purification
Genomic DNA from T. thermosaccharolyticum E207–71 was isolated by standard procedures. The qdtB gene was PCR-amplified using primers that introduced 5′ NdeI and 3′ XhoI sites. The purified PCR product was A-tailed and ligated into the pGEM-T (Promega) vector for screening and sequencing. A QdtB-pGEM-T vector construct of the correct sequence was then appropriately digested and ligated into a pET31 (Novagen) plasmid for gene expression leading to protein being produced with a C-terminal 6xHis-tag.
The QdtB-pET31 plasmid was used to transform Rosetta(DE3) E. coli cells (Novagen). The cultures were grown in Luria-Bertani media at 37 °C with shaking until an optical density of ~0.8 at 600 nm was reached. The cultures were then cooled to 16 °C, isopropyl β-D-thiogalactopyranoside was added to a final concentration of 1.0 mM, and protein production was allowed to proceed for 18 hrs. QdtB was purified by standard procedures with Ni-NTA resin. The original lysis buffer contained 3 mM PLP. A typical yield for recombinant QdtB was ~25 mg per liter of cells.
Purified QdtB samples were dialyzed against 10 mM Tris-HCl and 200 mM NaCl at pH 8. Following dialysis, the samples were concentrated to approximately 15 mg/ml and frozen in liquid nitrogen.
Enzymatic Synthesis of dTDP-Quip3N
dTDP-Quip3N was synthesized starting from dTDP-glucose. A typical 10 mL reaction contained the following: 20 mM HEPPS (pH 8.5), 5 mM MgCl2, 60 mg dTDP-glucose, 200 mg glutamate, 4 mg RmlB, 3 mg QdtA, and 15 mg QdtB (Scheme 1). The reaction was allowed to proceed at 37° C for 7 hours. All enzymes were removed via filtration with a 10-kDa cutoff Centriprep concentrator, and the enzyme-free reaction products were diluted by 1:4 with water. Purification was achieved with an ÄKTA™ Purifier HPLC (GE Healthcare) equipped with a Resource-Q 6 mL anion exchange column (GE Healthcare) and using a 120 mL gradient from 0–250 mM ammonium bicarbonate at pH 8.5. The desired product peak was identified by ESI mass spectrometry (Mass Spectrometry/Proteomics Facility at the University of Wisconsin-Madison). Fractions containing the amino sugar product were pooled and lyophilized until all traces of the buffer were removed. Typical yields of this compound were ~50% based on the starting amount of dTDP-glucose.
Originally it was planned to conduct steady-state kinetic measurements with QdtB. However, the substrate for QdtB, which is the product of the QdtA reaction, is unstable, and attempts to isolate it were unsuccessful.
Crystallization of QdtB
Crystallization conditions were initially surveyed with either the protein alone or that incubated with 20 mM dTDP-Quip3N. The initial surveys were conducted by the hanging drop method of vapor diffusion and employed a sparse matrix screen developed in the laboratory. Diffraction quality crystals were ultimately grown via hanging drop by mixing in a 1:1 ratio the protein incubated with 20 mM dTDP-Quip3N and 18–20% monomethylether poly(ethylene glycol) 5000 at pH 7.5. Prior to x-ray data collection, the crystals were transferred to a stabilization solution containing 25% monomethylether poly(ethylene glycol) 5000, 400 mM NaCl, 20 mM dTDP-Quip3N, and 15% ethylene glycol. X-ray data were measured at 100K using the SBC-3 CCD-detector at the Structural Biology Center Beamline 19-BM (Advanced Photon Source, Argonne National Laboratory, Argonne, Illinois). These data were processed and scaled with HKL3000 (18). Relevant x-ray data collection statistics are presented in Table 1.
Table 1.
X-ray Data Collection Statistics
| resolution limits | 30.0-2.15 (2.25-2.15)b |
| number of independent reflections | 60951 (7491) |
| completeness (%) | 93.7 (93.0) |
| redundancy | 8.4 (5.9) |
| avg I/avg σ(I) | 22.6 (3.1) |
| Rsym (%)a | 5.2 (25.1) |
Rsym = (Σ|I − Ī|/ΣI) × 100.
Statistics for the highest resolution bin.
The structure of QdtB was solved via molecular replacement with the software package Phaser (19, 20) and using the structure of DesV (11) as the search model. Two subunits were positioned into the asymmetric unit, and these were partially refined by least-squares analysis with TNT (21). The electron densities corresponding to these two subunits were subsequently averaged with DM (22). On the basis of this averaged electron density map, a complete subunit of QdtB was rebuilt with the appropriate sequence using the graphics program Coot (23). The averaged model was placed back into the unit cell, and alternate cycles of refinement with TNT and model building with Coot reduced the R-factor to 21.5% for all measured data from 30.0 to 2.15-Å resolution. Relevant refinement statisticsare presented in Table 2.
Table 2.
Least-Squares Refinement Statistics
| Resolution Limits (Å) | 30.0-2.15 |
| aR-factor (overall) %/no. reflections | 21.5(60951) |
| R-factor (working) %/no. reflections | 21.3(54989) |
| R-factor (free) %/no. reflections | 26.9(5962) |
| No. Protein Atoms | 5876b |
| No. Hetero-atoms | 280c |
| Average B values (Å2) | |
| Protein Atoms | 50.6 |
| PLP Derivatives | 45.7 |
| Solvents | 45.2 |
| Weighted RMS Deviations from Ideality | |
| Bond Lengths (Å) | 0.012 |
| Bond Angles (deg) | 2.32 |
| Trigonal Planes (Å) | 0.007 |
| General Planes (Å) | 0.013 |
| d Torsional Angles (deg) | 19.5 |
R-factor = (Σ|Fo − Fc|/Σ|Fo|) × 100 where Fo is the observed structure-factor amplitude and Fc is the calculated structure-factor amplitude.
These include Glu 16, Tyr 17, and Gln 258 in subunit 1 and Asp 36 in subunit 2 adopting multiple conformations.
These include 2 PLP/dTDP-sugars and 180 waters.
The torsional angles were not restrained during the refinement.
Aminotransferase Activity Assays
For assaying the production of dTDP-Quip3N (Scheme 1), a reaction was set up with the following: 20 mM HEPPS (pH 8.5), 5 mM MgCl2, 4 mg dTDP-glucose, 25 mg glutamate, 0.4 mg RmlB, 0.2 mg QdtA, and 2.0 mg QdtB. The reaction was allowed to proceed at 37° C for 5 hours. All enzymes were removed via filtration with a 10-kDa cutoff Microcon concentrator, and the enzyme-free reaction products were diluted by 1:5 with water. Purification was achieved by HPLC chromatography using a 1 mL Resource-Q column and a 20 mL gradient from 0–250 mM ammonium bicarbonate at pH 8.5. The desired amino-sugar product peak was identified by mass spectrometry (ESI mass spectrometry parent ion m/z: 546.3 amu).
To test whether QdtB could use dTDP-3-keto-6-deoxy-D-galactose as a substrate, assays were conducted as described above, but substituting 0.2 mg of FdtA from Aneurinibacillus thermoaerophilus DSM10155 for QdtA.
DesV from Streptomyces venezuelae is a sugar aminotransferase that is involved in dTDP-desosamine production (17). The substrate for DesV is dTDP-3-keto-4,6-deoxy-D-glucose. To test whether DesV could accept a sugar with a C-4′ hydroxyl, assays were conducted as described above, but this time substituting 2.0 mg of DesV for QdtB.
RESULTS AND DISCUSSION
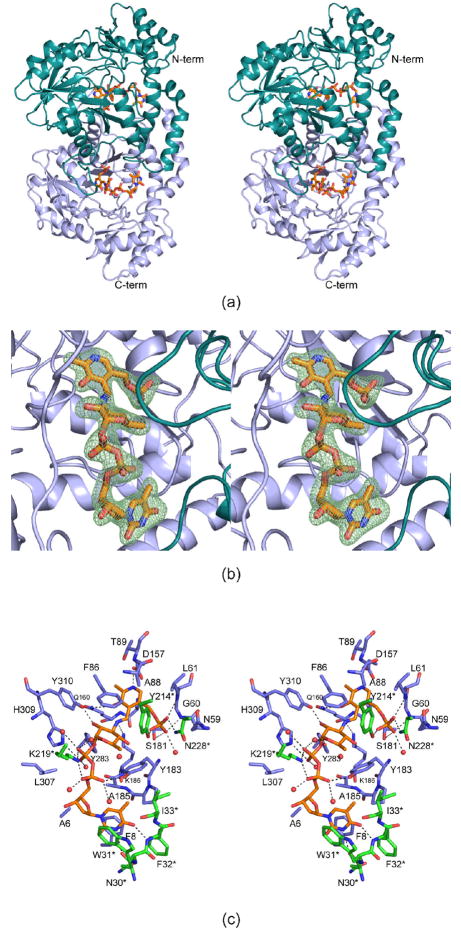
QdtB crystallized in the space group P61 with unit cell dimensions of a = b = 109.1 Å, c = 177.9 Å, and one dimer per asymmetric unit. The structure was solved and refined to a nominal resolution of 2.15 Å with 86.5%, 12.6% and 0.9% of the amino acids lying within the core, allowed, and generously allowed regions of the Ramachandran plot. Shown in Figure 1a is a ribbon representation of QdtB, and as can be seen the subunit:subunit interface is extensive with a total buried surface area of ~4800 Å 2. Each subunit of the dimer contains ten β-strands, seven of which form a mostly parallel β-sheet and the other three fold into an anti-parallel β-sheet. These β-sheets are surrounded by a total of eleven α-helices. Overall the electron density was well ordered for both subunits of the dimer, and in each Tyr 310 adopted a cis-peptide conformation. The side chain of Tyr 310 hydrogen bonds to the hexose ring of the sugar ligand as described below. Given that the α-carbons for the individual subunits of the dimer correspond with a root-mean-square deviation of 0.29 Å 2, the following discussion refers only to subunit 1 in the x-ray coordinate file unless otherwise indicated.
Figure 1.
Molecular architecture of QdtB. A ribbon representation of the QdtB dimer is displayed in (a). Subunits 1 and 2 of the dimer are highlighted in light blue and cyan, respectively. The ligands are drawn in stick representations. Electron density corresponding to the Schiff base between the PLP cofactor and the dTDP-sugar is presented in (b). The map was calculated with coefficients of the form (Fo−Fc), where Fo was the native structure factor amplitude, and Fc was the calculated structure factor amplitude. Atoms corresponding to the PLP and the dTDP-sugar were omitted from the calculation. A close-up view of the active site is shown in (c). Only those residues located within ~3.8 Å of the PLP and the dTDP-sugar are depicted. Amino acid residues corresponding to subunits 1 and 2 are colored in blue and green, respectively whereas the PLP and the dTDP-sugar are shown in gold bonds. Water molecules are represented as red spheres. Possible hydrogen bonding interactions are indicated by the dashed lines. All figures were prepared with the software package PyMOL (25).
The observed electron density corresponding to the dTDP-sugar ligand, displayed in Figure 1b, shows that a Schiff base between C-4′ of PLP and the amino nitrogen of the sugar has been trapped within the active site cleft. The pyranosyl group assumes the 4C1 conformation whereas the ribosyl moiety adopts the C2′-endo pucker. A close-up view of those residues located within ~3.8 Å of the PLP and dTDP-sugar is presented in Figure 1c. The thymine ring forms hydrogen bonds with the carbonyl oxygen of Asn 30 and the backbone amide nitrogen of Phe 32 from subunit 2, and it also participates in parallel stacking interactions with the side chains of Phe 8 and Trp 31 from subunits 1 and 2, respectively. The α-phosphoryl oxygens of the dTDP-sugar ligand lie within 3.2 Å of the side chains of Tyr 183 from subunit 1 and Lys 219 from subunit 2. In addition, the side chain of His 309 from subunit 1 participates in a hydrogen bond with a β-phosphoryl oxygen. Both Asp 157 and Gln 160 from subunit 1 anchor the pyridoxal ring to the protein whereas the side chain of Ser 181, and the backbone amide nitrogens of Gly 60 and Leu 61 lie within 3.2 Å of the 5′-phosphate group. The hydrogen-bonding pattern surrounding the 5′-phosphate group is completed via interactions with Tyr 214 and Asn 228 from subunit 2. There is only one specific protein interaction between the pyranosyl ring of the ligand and the protein, namely a hydrogen bond between the C-2′ hydroxyl and Oη of Tyr 310 from subunit 1. Five water molecules surround the dTDP-sugar ligand with one serving as a bridge between a β-phosphoryl oxygen and the ring oxygen of the hexose. Other than Lys 186, which normally forms a Schiff base with the PLP cofactor (the internal aldimine), there are no catalytic bases within the general area of the hexose ring.
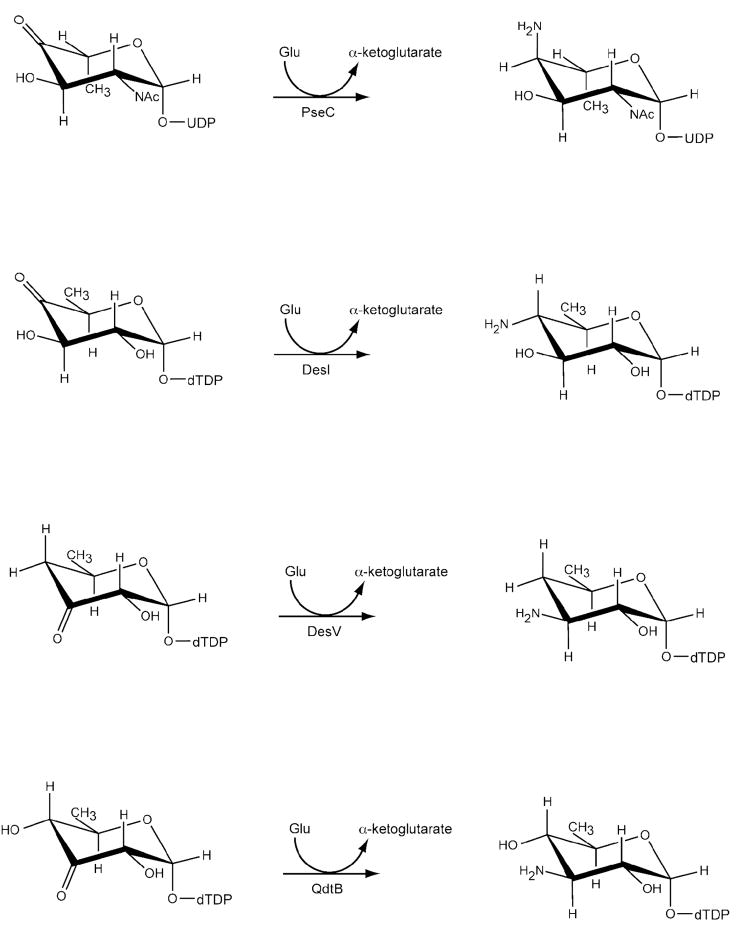
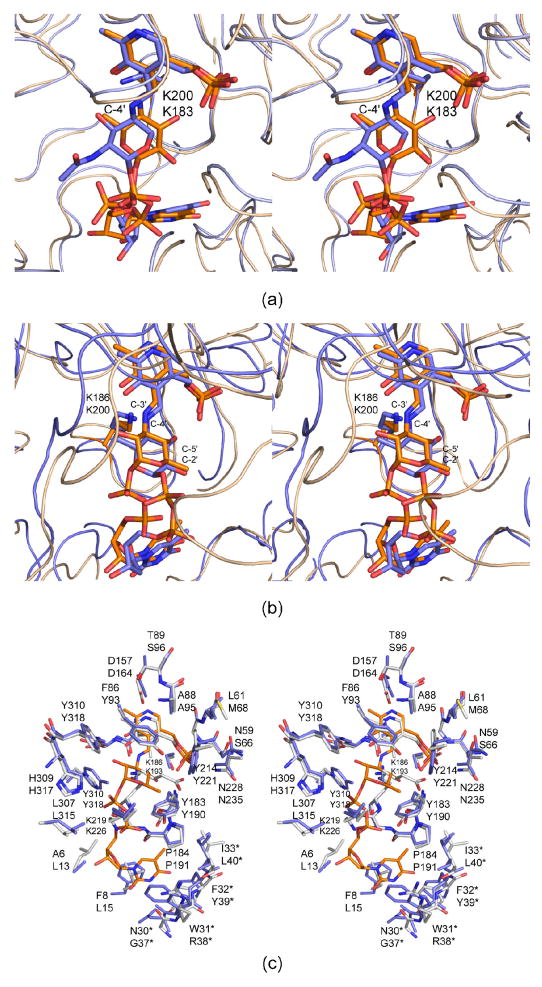
Previous x-ray crystallographic studies on sugar-modifying aminotransferases have focused primarily on those that attach amino groups to the C-4′ rather than the C-3′ position as is the case for QdtB. Historically, the structural analysis of PseC, an aminotransferase from Helicobacter pylori, provided the first detailed glimpse of the active site geometry for a sugar aminotransferase (9). PseC is involved in the biosynthesis of pseudaminic acid, and it catalyzes the reaction outlined in Scheme 2, namely amino transfer to the axial position of the sugar C-4′. For this x-ray analysis, the protein was complexed with UDP-4-amino-4,6-dideoxy-β-L-AltNAc. Following this elegant x-ray analysis, the molecular architecture of DesI from S. venezuelae was reported from this laboratory with its product, dTDP-4-amino-4,6-dideoxyglucose, bound in the active site (12). DesI is involved in the production of dTDP-desosamine, and its specific reaction is presented in Scheme 2. Note that this enzyme transfers the amino group to the equatorial position at the sugar C-4′. Strikingly, a superposition of the active sites for PseC and DesI revealed a nearly 180° difference in the hexose orientations of their respective ligands, thereby explaining the equatorial versus axial amino transfer exhibited by DesI and PseC, respectively (Figure 2a).
Scheme 2.
Figure 2.
Comparison of QdtB with other sugar-modifying aminotransferases. A superposition of the DesI structure (gold) onto the PseC structure (slate) is presented in (a). Lys 200 and Lys 183 belong to DesI and PseC, respectively. These are the residues that normally hold the cofactor in place as the internal aldimine. Note the nearly 180° rotation of the pyranosyl groups in the active sites. A superposition of the QdtB model (gold) onto the DesI structure (slate) is given in (b). The top and bottom labels correspond to QdtB and DesI, respectively. Shown in (c) is a superposition of the active sites for QdtB (blue and yellow) and DesV (white). The coordinates for DesV are those for the trapped ketimine intermediate. The top and bottom labels refer to QdtB and DesV, respectively.
Like DesI, QdtB catalyzes equatorial transfer, albeit at the sugar C-3′ rather than the C-4′ position. A close-up view of the differences in sugar binding observed between QdtB and DesI is presented in Figure 2b. As might be expected, given that these two enzymes are 31% identical and 51% similar, the protein regions surrounding the hexose moieties in these aminotransferases are exceedingly similar. However, as can be seen in Figure 2b, the phosphoryl groups of the nucleotide-linked sugars differ markedly in their dihedral angles, which allows for the different positioning of the hexose groups into their respective active sites. The loops defined by Glu 217 to Tyr 224 in QdtB (subunit 2) and Asp 230 to Ala 237 in DesI (subunit 2) contain very different amino acid residues and adopt markedly different conformations. In particular, Lys 219 in QdtB interacts with the phosphoryl groups of the dTDP-sugar (Figure 1c), and this residue is a proline in DesI. The loop in QdtB reaches down into the active site region whereas the corresponding loop in DesI splays outwards. As a consequence, in QdtB, the phosphoryl groups are pushed more towards the interior of the protein.
The biosynthesis of dTDP-desosamine requires two sugar-modifying aminotransferases, the first being DesI and the second DesV. Like QdtB, DesV transfers the amino group to the sugar C-3′ (Scheme 2) (24), and both of these enzymes catalyze equatorial transfer. Thus far, however, only the structures of DesV in the internal aldimine form or with a trapped ketimine intermediate have been reported (11). On the basis of amino acid sequence alignments (DesV and QdtB are 45% identical and 65% similar), we predict that the dTDP-sugar binding mode observed in QdtB serves as an excellent model for substrate binding to DesV as well. A superposition of their two active sites is presented in Figure 2c. Note that the loop enveloping the thymine ring of the dTDP-sugar in QdtB (Asn 30 to Ile 33) has a nearly identical conformation to that found in DesV (Gly 37 to Leu 40). Also, the key side chains involved in binding the dTDP-sugar to the protein in QdtB, namely His 309, Tyr 310, and Tyr 183 in subunit 1 and Lys 219 in subunit 2 are conserved in DesV as His 317, Tyr 318, and Tyr 190 in subunit 1 and Lys 226 in subunit 2. Like Tyr 310 in QdtB, Tyr 318 in DesV adopts a cis-conformation, and the loop in QdtB that is formed by Glu 217 to Tyr 224 has nearly the same conformation as that observed in DesV (Arg 224 to Thr 231).
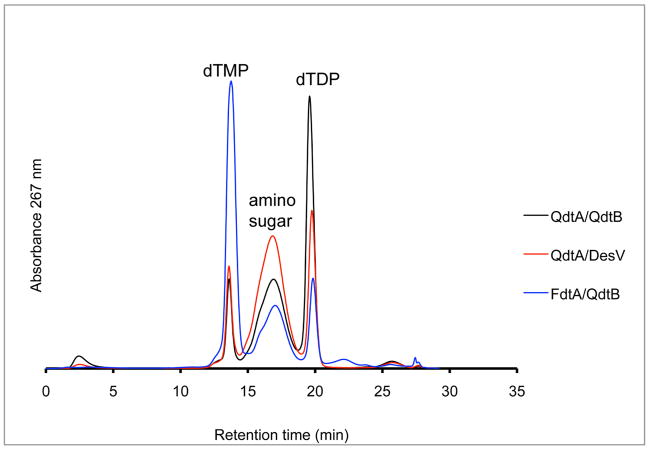
It is notable that whereas the side chain of Tyr 310 in QdtB hydrogen bonds to the C-2′ hydroxyl of the dTDP-sugar, there are no interactions between the sugar and the C-4′ hydroxyl. In light of this decided lack of interactions between the protein and sugar C-4′ hydroxyl group and given the similarity between the QdtB and DesV active sites, we were curious as to whether DesV could accept dTDP-3-keto-6-deoxy-D-glucose as a substrate. The normal substrate for DesV is missing a hydroxyl group at the C-4′ position (compare the QdtB and DesV substrates in Scheme 2). To test for this, a reaction mixture was set up as described in the Materials and Methods section, but substituting DesV for QdtB. As shown in Figure 3, the substitution of DesV for QdtB in the reaction mixture still led to the formation of an amino sugar product peak with the same retention time. We were also curious as to whether QdtB could accept dTDP-3-keto-6-deoxy-D-galactose as a substrate, which differs in configuration from the natural QdtB substrate around the C-4′ hydroxyl. For this experiment, a reaction mixture was set up whereby the enzyme FdtA from A. thermoaerophilus DSM10155 was substituted for QdtA. As indicated in Scheme 3, both QdtA and FdtA are 3,4 isomerases, and function on the same substrate. But the product of FdtA is dTDP-3-keto-6-deoxy-D-galactose rather than dTDP-3-keto-6-deoxy-D-glucose (Scheme 3). The reaction mixture was set up and allowed to proceed. The HPLC traces for the FdtA/QdtB reaction (Figure 3) again clearly showed the development of a peak having the same retention time as that for the QdtA/QdtB reaction product. These experiments indicate that both QdtB and DesV can function on dTDP-linked sugars with differing configurations from their natural substrates about the C-4′ hydroxyl positions.
Figure 3.
Aminotransferase activity assays. The HPLC elution profile, monitored at 267 nm, for the reaction mixture containing dTDP-glucose and the enzymes RlmB, QdtA, and QdtB is indicated by the black line. The peaks corresponding to dTDP (ESI mass spectrometry parent ion m/z: 401.2 amu) and dTMP (ESI mass spectrometry parent ion m/z: 321.2 amu) result from breakdown of the sugar intermediates (Scheme 3). The HPLC elution profile for the reaction in which DesV from the dTDP-desosamine biosynthetic pathway was substituted for QdtB is displayed in red. Note that an amino sugar is still formed thus indicating that DesV is able to accept as a substrate the QdtA reaction product. The normal substrate for DesV does not have a C-4′ hydroxyl group. Finally, the HPLC elution profile for the reaction in which FdtA was substituted for QdtA is indicated by the blue line. Note that FdtA results in a sugar intermediate having the opposite configuration about the C-4′ hydroxyl as compared to the QdtA product (Scheme 3).
Scheme 3.
In summary, whereas the structures of sugar-modifying aminotransferases that function at the C-4′ position have been solved with bound ligands, this report represents the first glimpse of one with a bound dTDP-sugar that functions at the C-3′ position. Importantly, the QdtB structure and its comparison to other sugar aminotransferases highlights the fact that the orientation of the pyranosyl group in the active site is not solely a function of those amino acid residues immediately surrounding the Schiff base. Rather, the orientation of the pyranosyl group in the active site results from interactions that extend to those residues involved in binding the pyrophosphoryl and nucleotide groups of the nucleotide-linked sugars.
Acknowledgments
Results in this report were derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by the University of Chicago, Argonne, LLC, for the U. S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Dr. Norma Duke for assistance at the beamline. The insightful conversations of Dr. W. W. Cleland and Mr. Paul D. Cook are gratefully acknowledged.
Footnotes
X-ray coordinates have been deposited in the Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, N. J. (accession no. 3FRK).
Abbreviations: CoA, Coenzyme A; dTDP, 2-deoxythymidine diphosphate; ESI, electrospray ionization; HEPPS, 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid; HPLC, high performance liquid chromatography; NiNTA, nickel-nitrilotriacetic acid; PCR, polymerase chain reaction; PLP, pyridoxal-5′-phosphate; Tris, 2-amino-2-hydroxymethyl-propane-1,3-diol.
This research was supported in part by an NIH grant (DK47814 to H. M. H.)
References
- 1.Nedal A, Zotchev SB. Biosynthesis of deoxyaminosugars in antibiotic-producing bacteria. Appl Microbiol Biotechnol. 2004;64:7–15. doi: 10.1007/s00253-003-1535-9. [DOI] [PubMed] [Google Scholar]
- 2.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 3.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messner P, Schaffer C. Prokaryotic glycoproteins. Fortschr Chem Org Naturst. 2003;85:51–124. doi: 10.1007/978-3-7091-6051-0_2. [DOI] [PubMed] [Google Scholar]
- 6.Timmons SC, Thorson JS. Increasing carbohydrate diversity via amine oxidation: aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr Opin Chem Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 8.Noland BW, Newman JM, Hendle J, Badger J, Christopher JA, Tresser J, Buchanan MD, Wright TA, Rutter ME, Sanderson WE, Muller-Dieckmann HJ, Gajiwala KS, Buchanan SG. Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: a 4-amino-4-deoxy-L-arabinose lipopolysaccharide-modifying enzyme. Structure (Camb) 2002;10:1569–1580. doi: 10.1016/s0969-2126(02)00879-1. [DOI] [PubMed] [Google Scholar]
- 9.Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 2006;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 10.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 11.Burgie ES, Thoden JB, Holden HM. Molecular architecture of DesV from Streptomyces venezuelae: a PLP-dependent transaminase involved in the biosynthesis of the unusual sugar desosamine. Protein Sci. 2007;16:887–896. doi: 10.1110/ps.062711007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgie ES, Holden HM. Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine. Biochemistry. 2007;46:8999–9006. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook PD, Holden HM. GDP-perosamine synthase: structural analysis and production of a novel trideoxysugar. Biochemistry. 2008;47:2833–2840. doi: 10.1021/bi702430d. [DOI] [PubMed] [Google Scholar]
- 14.Novotny R, Pfoestl A, Messner P, Schaffer C. Genetic organization of chromosomal S-layer glycan biosynthesis loci of Bacillaceae. Glycoconj J. 2004;20:435–447. doi: 10.1023/B:GLYC.0000038290.74944.65. [DOI] [PubMed] [Google Scholar]
- 15.Feng L, Wang W, Tao J, Guo H, Krause G, Beutin L, Wang L. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J Clin Microbiol. 2004;42:3799–3804. doi: 10.1128/JCM.42.8.3799-3804.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfostl A, Zayni S, Hofinger A, Kosma P, Schaffer C, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose. Biochem J. 2008;410:187–194. doi: 10.1042/BJ20071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Y, Zhao L, Liu HW, Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci U S A. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;D60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 20.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 21.Tronrud DE, Ten Eyck LF, Matthews BW. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr Sect A. 1987;43:489–501. [Google Scholar]
- 22.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Szu PH, He X, Zhao L, Liu HW. Biosynthesis of TDP-D-desosamine: identification of a strategy for C4 deoxygenation. Angew Chem Int Ed Engl. 2005;44:6742–6746. doi: 10.1002/anie.200501998. [DOI] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]