Summary
Nearly 6% of eukaryotic protein sequences contain ankyrin repeat (AR) domains, which consist of several repeats and often function in binding. AR proteins show highly cooperative folding despite a lack of long-range contacts. Both theory and experiment converge to explain that formation of the interface between elements is more favorable than formation of any individual repeat unit. IκBα and Notch both fold upon binding, perhaps gaining folding energy in the binding event. The simple architecture combined with identification of consensus residues that are important for stability, has enabled systematic perturbation of the energy landscape by single point mutations that affect stability, or by addition of consensus repeats. The folding energy landscapes appear highly plastic, with small perturbations re-routing folding pathways.
Introduction
Nearly 20% of the proteins estimated to be coded in the human genome contain multiple repeating units of 30 – 40 amino acids. One commonly occurring type of repeat, the Ankyrin Repeat (AR) is found in all three phyla, and is present in some 6% of eukaryotic protein sequences [1,2]. Ankyrin repeats are nearly always found in a tandem array, suggesting that the repeating elements only function in the context of similar repeats. Sequence and structural characterization of these proteins have revealed a conserved amino acid pattern that forms a repeating structural array, with non-conserved amino acids preferentially located on the surface [3,4]. Architecturally, the conserved scaffold presents a wide variety of protein binding surfaces. This natural property has been successfully mimicked by constructing synthetic repeat-protein libraries in which specific, high affinity binders can be found [5]. Usually, these designed AR proteins are thermodynamically more stable than their natural counterparts. Recently, in an affinity maturation experiment on a designed AR protein, binding improved when mutations were introduced that reduced the "foldedness" of the domain [6], suggesting that there is an intimate coupling between the folding of the repeating array and the functional binding/recognition process. In addition, two natural AR proteins, IκBα and the Notch intracellular domain, have been shown to undergo folding transitions upon binding to targets, the NF-κB and CSL transcription factors, respectively [7–9].
The structural simplicity of repeat proteins has provided fertile soil for theoretical and experimental exploration of their folding landscapes (for a recent review, see Kloss et al., 2007[10]). In contrast to globular proteins, all the interactions important for folding repeat proteins are close in the amino acid sequence space [11]. The repeat structure simplifies the topological characterization of their energy landscapes. We will here review experimental and theoretical attempts at a quantitative description of these landscapes.
Main Text
Structure and Folding of Globular vs. Repeat Proteins
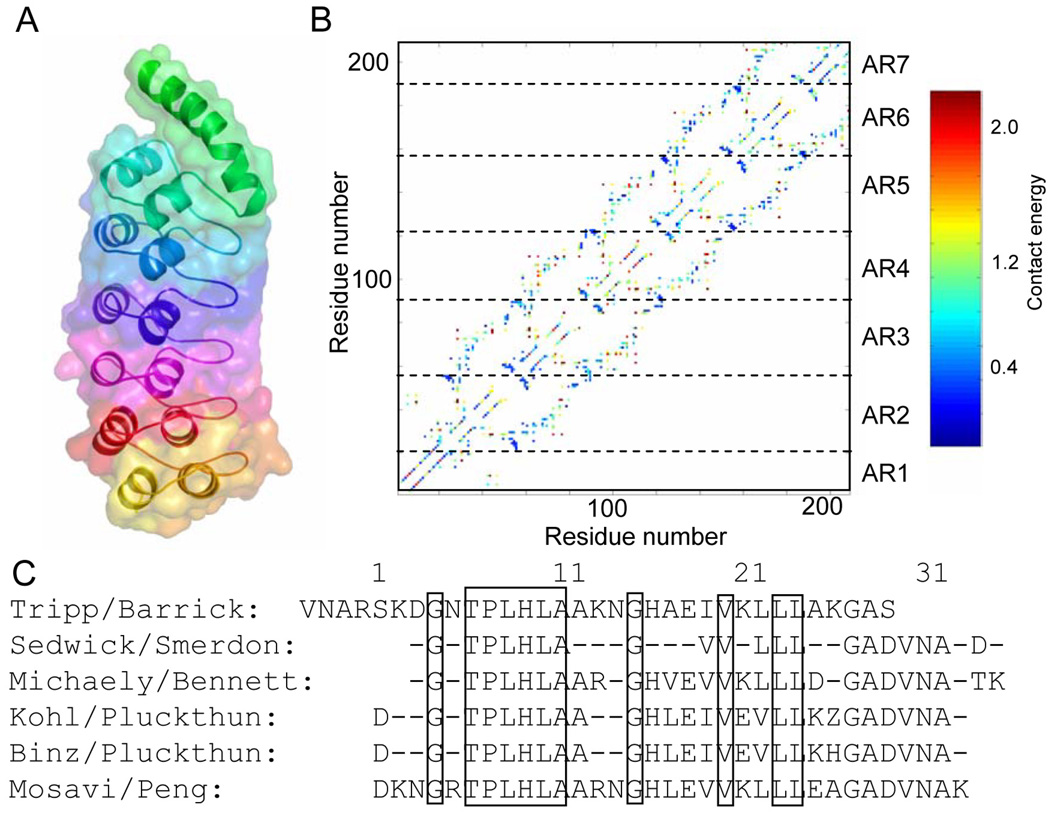
A hallmark of globular proteins is that, when folded, distant segments of the polypeptide chain are in close proximity. Such interconnected, long-range topologies lead to two related problems that limit our understanding of how the energy is distributed in a globular protein: “dissection” and "comparison". In globular proteins, numerous contacts among distant chain segments are likely to promote cooperativity in folding and prevent structural fragments from folding out of context [12]. This prevents "dissection" of the energetics in different structural elements of globular proteins, which is required to experimentally map the energy landscape. Repeat proteins bypass this "dissection" problem because they lack long-range contacts, and it is relatively easy to modify, add, or remove repeating units [13,14]. Along with roughly linear architecture, ease of "dissection" in repeat proteins permits separation of local and nearest-neighbor energetic contributions, especially when probed experimentally with length variation by deletion or insertion of repeats. A "comparison" problem arises when secondary and tertiary structural elements are arranged in an irregular and highly variable manner. Both the structural elements and their local environments are very heterogeneous, making it difficult to attribute folding stability to various "parts" of the protein through direct "comparison". Repeat proteins avoid the "comparison" problem because their architecture is simple. In ankyrin repeat proteins, each repeating unit is made of two short (10–11 residue) α-helices connected by alternating short and extended β–turns (Figure 1A). The repeat structures are very similar across the domain with backbone RMSD values from repeat to repeat typically below 1Å. In addition, the interaction of adjacent repeats is conserved across the domain. These similarities can be visualized clearly as regular patterns in contact maps (Figure 1B). Thus, comparisons between repeats can be readily made that are not confounded by the context in which the entire AR or individual secondary structural element resides.
Figure 1.
A) Structure of the Notch AR domain, 1OT8.pdb. A ribbon trace of the backbone is shown colored from red (AR1) to blue (AR7), the space-filling model is shown shaded. B) Contact map of the Notch AR domain colored according to A. C) Consensus sequences of stably folded ARs.
The robust AR consensus sequence
The abundance of AR sequences means that consensus sequences are robustly determined (Figure 1C). This consensus sequence information has been used to design AR proteins with identical repeats, simplifying the folding problem and smoothing the energy landscape. The resulting consensus-designed AR domains adopt structures that closely resemble their naturally occurring counterparts, with RMSDs less than 1 Å [15–17]. Designed AR domains have very high thermodynamic stability compared to their naturally-occurring counterparts [16,18]. Remarkably, the robust consensus of AR domains combined with sequence variation of non-conserved residues has allowed the construction of libraries of AR domains that bind many different targets [5,19]. These designed binding proteins can be selected for extremely tight binding to a wide variety of proteins making them an alternative to antibodies [6,20]. The ready success of these efforts helps establish the primary role that the conserved residues play in folding stability. Several studies have demonstrated the importance of consensus sequences in stabilizing naturally-occurring AR proteins as well. Mutation of conserved residues nearly always results in reduction of stability whereas mutation of non-conserved surface residues can have various effects [21–25]. Further, the marginally stable AR domain of IκBα is stabilized by mutating residues to conform to the consensus [26] and addition of consensus repeats also greatly stabilizes the Notch AR domain [27].
Cooperativity of folding in AR domains
Small, globular domains often display equilibrium “two-state” folding reactions in which only the folded and unfolded thermodynamic states are significantly populated. The high cooperativity of the folding of globular proteins resembles a phase transition and may arise, in part, from interactions between residues distant in sequence space [28]. Although repeat proteins lack such distant interactions, many experiments show that AR protein folding is also highly cooperative [15,22,23,29]. p16INK4A, a tumor suppressor protein containing four ARs, displayed a steep, cooperative unfolding transition when monitored by circular dichroism (CD) spectroscopy [30–32], tryptophan fluorescence, and gel filtration chromatography [23]. Coincidence of distinct probes supports an all-or-none transition between the native (N) state and denatured (D) ensemble consistent with only two populated states at equilibrium. More recently, another four-AR protein, Myotrophin, has also been shown to satisfy the spectroscopic test for cooperative equilibrium two-state folding [15,33].
The x-ray structure of the Notch receptor AR domain contains six well-structured AR repeats (2–7) but shows substantial disorder of the first repeat [21]. This domain also appears to unfold via a cooperative mechanism with highly coincident urea and thermal unfolding transitions when monitored by CD and by tryptophan fluorescence (local probe of repeat five). Moreover, the van’t Hoff enthalpy estimated from fitting a two-state model to the thermal transition is the same as the calorimetric enthalpy, supporting the view that intermediate states are not significantly populated at equilibrium [29,34]. Finally, the m-value (the sensitivity of unfolding free energy to urea) and ΔCp (the change in heat capacity on thermal unfolding), which are both correlated with the size of the cooperative unit, match predicted values for the unfolding in a single concerted transition [29].
In contrast to Notch and p16INK4A, two other naturally occurring AR domains appear to undergo multistate unfolding. The five-repeat p19INK4D AR domain shows a third species that forms in the transition region, as monitored by heteronuclear NMR [35,36]. A more dramatic multistate equilibrium unfolding transition has been seen for a large, twelve-AR fragment of ankyrinR (named D34). Urea unfolding transitions of D34 show clear multistate unfolding, populating a well-resolved intermediate at moderate urea concentrations [37]. Point-substitutions in D34 suggest that the C-terminal repeats are structured in this intermediate, but the N-terminal repeats are not.
In IκBα, one or more ARs are partly unstructured when IκBα is free in solution but become structured when it binds to its target protein, NF-κB. Hydrogen exchange studies showed that ARs 1, 5, and 6 are highly dynamic in the unbound state, but adopt consolidated structure on binding to NF-κB [7,38]. A similar disorder-order transition is seen for the first repeat of the Notch AR domain when binding the CSL transcription factor [8,9,21]. In both cases, this folding upon binding may provide part of the binding energy for the interaction [39].
Folding cooperativity in AR domains strongly depends on interactions of the repeats with their nearest neighbors [13,40]. Both experiments and simulations indicate that this can be understood if the domains fold up by a mechanism in which formation of the interface between elements is more favorable than formation of any individual repeat unit [13,41]. Indeed, structures of AR domains show high surface complementarity between repeats (Figure 1A), burying an average of 1490 Å2 at the interrepeat interfaces, compared with 1510 Å2 buried upon folding of individual repeats [10]).
Folding simulations of natural AR domains of different lengths suggest that as the number of repeats increases, the cooperativity tends to break down, presumably because the increasing entropy advantage of introducing a broken interface anywhere between repeats is weighed against a fixed energy cost [41]. The expected breakdown of strict cooperativity was recently observed in experiments on the 12 AR domain, D34 [37], and in Notch ankyrin constructs bearing internal duplications [42].
AR folding landscapes from theory and experiment—equilibrium folding
The energy landscape theory of protein folding argues that three-dimensionally connected globular proteins must fold along a landscape that is funneled to the native state [28,43]. On the other hand, the one-dimensionality of repeat proteins weakens this necessity [44]. The comparison between experiments and folding simulations based on perfectly funneled model landscapes has revealed how fine details of the energetic contributions can strongly influence folding [41,45]. Preferred folding routes determined experimentally and theoretically can be directly compared.
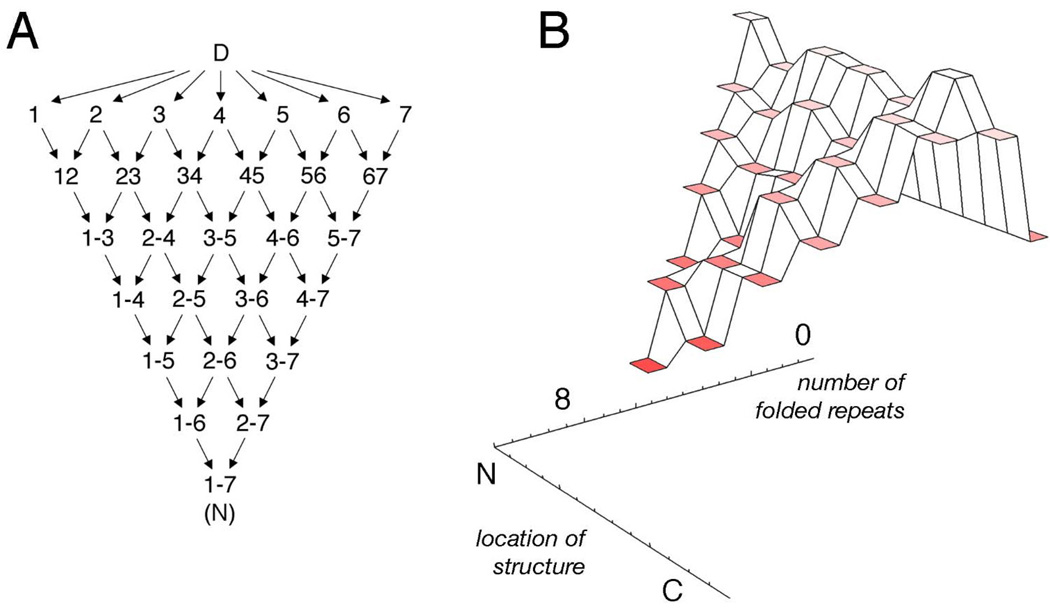
Two approaches to experimentally probe the equilibrium folding landscape of AR domains have been applied; truncation and mutation. Due to their simplified topologies, dissection by truncation is highly informative when applied to repeat proteins. This approach was used to identify the two C-terminal repeats as the minimally folded unit in p16INK4A (four ARs) [14]. Consistent with native state amide exchange data, dissection of the six-repeat IκBα showed that repeats 1 to 4 accounted for all of the cooperative folding transition [26,38]. Dissection of the larger Notch AR identified a four-AR repeat segment (repeats 2–5) as the minimal folding unit [46]. In this case, nine overlapping truncated constructs were used to determine the stability distribution at the single-repeat level and these data were used to create a heterogeneous model with a free energy coefficient associated with each repeat:
| (1) |
where the xi terms are simple binary variables reflecting the presence or absence of each repeat. The nine deletion constructs were all well-fitted by the linear equation (1), with an unbiased correlation coefficient of 0.95 [46]. This analysis provided a direct map of the free energy landscape for the Notch ankyrin domain (Figure 2) [46]. Levels on this landscape correspond to species with a single contiguous block of folded repeats (Figure 2B). Energy levels are depicted as a function of the number of folded repeats and as a function of where structure is localized (see schematic, Figure 2A). Thus, moving in this space from the denatured ensemble to the native state corresponds to coalescing structure in neighboring repeats, and it can be done in a number of different ways, especially early in folding. Free energy decreases in the direction of the native state as repeats are added to existing (i.e. folded) repeats. In this regard, the landscape is funneled. An important conclusion from the model is that the folding of each repeat is intrinsically unstable, but the formation of the inter-repeat interface is highly stabilizing. Thus, conformations that have non-contiguous blocks of repeats are strongly disfavored. Although ‘internal’ energies vary from repeat to repeat, there is an overall evenness on a length scale of two to three repeat blocks. This uniformity, together with the favorable energy of interface formation are what may underlie the appearance of two-state equilibrium folding transition.
Figure 2.
A) A schematic of the manner in which the free energy landscape of Notch was experimentally determined by obtaining an overall thermodynamic stability for each of the repeats in the domain. B) The free energy landscape of the Notch AR domain as experimentally determined.
Perturbation of the free energy folding landscape by mutation has also been highly informative. Essentially all results suggest that local destabilization can re-route folding. In the Notch AR domain, destabilizing substitutions [40], or substitution of highly stable consensus repeats in place of the C-terminal repeats, cause sufficient unevenness to break down the cooperativity of folding of the domain and bias the folding towards the much more stable consensus repeats [27]. Substitutions in the four ARs of Myotrophin result in similar landscape biasing [22]. Most remarkably, folding studies of D34, a 12-AR domain, show that it is composed of roughly two six-repeat subdomains, which fold in an equilibrium three-state manner (U->I->N). Mutations in the N-terminal repeats reduced the stability, but the cooperativity of the native to intermediate (N->I) transition was not greatly affected. Thus, in the N-terminal subdomain mutants, the N->I transition could be distinguished from the intermediate to unfolded (I->U) transition. In contrast, mutations in the C-terminal repeats dramatically increased the cooperativity of the N->I transition and correspondingly decreased the cooperativity of the I->U transition. When the mutation was closest to the C-terminus, nearly all the repeats unfolded in the N -> I transition [37]. All of these examples point to the observation that AR domains fold in a highly cooperative manner because of inter-repeat stabilization and balanced energetics amongst folding subdomains. Thus, the folding free energy landscapes of AR domains, are highly "plastic" such that substitutions can subtly alter the free energies of partially folded species dramatically altering the cooperativity of the folding reaction [47]. In extreme cases such as D34, single mutations can actually form different intermediate species [37].
AR folding landscapes from theory and experiment—kinetics of folding
A comprehensive description of the folding transition state structures and folding pathways requires a full kinetic characterization of the folding pathway, a goal that is best achieved by integration of theory and experiment in an iterative process. The identities of the early-folding repeats depend on the fine details that underlie the ‘unevenness’ of the landscape, and both simple folding models and experiments show a preference for a discrete nucleation event followed by a further propagation of structure. This preference argues against a large number of parallel routes involving structure formation in different regions, since small energetic differences will strongly bias the routes. Starting at the most basic level of theory, folding simulations based on perfectly funneled model landscapes revealed how finer details of the energetic contributions contribute to folding, and recapitulate experimental results [41,45]. Remarkably, these most simple Go-type models in which every contact is given the same energetic weight, predicted that short AR domains would fold in an apparent two-state manner while also revealing kinetic intermediates. A good example is the 4-AR domain of the tumor suppressor p16INK4A, for which both equilibrium and kinetic unfolding has been analyzed [23]. This domain displays highly cooperative two-state equilibrium folding, and phi-value analysis revealed that the two C-terminal repeats fold first [48]. The perfectly funneled model landscape simulations recapitulated this bias and predicted a high energy kinetic intermediate comprising only the two C-terminal repeats [41]. In the case of IκBα, these "perfectly funneled" models were sufficient to predict a cooperative folding transition involving roughly the first four ARs while the fifth and sixth repeats were predicted to fold in a separate event [41]. These results, based on the native contacts observed for IκBα in the crystal structure of the NF-κB-bound form, accurately predicted the experimental folding on binding results [7]. On the other hand, such a simplified model is not accurate enough to reproduce fine details of the Notch AR domain. The models predicted that Notch would fold along two parallel routes nucleating at either terminal repeat pairs, a result that was contradicted by experiment (see below).
One remarkable feature of AR domains is how slowly they fold. Compared to expectations based on contact order [49], they fold at least three orders of magnitude slower [46]. One possibility for the rate limiting step, prolyl isomerization, has been effectively dismissed as the cause of slow folding in AR domains despite the large number of prolines at consensus positions [50]. Another possible explanation for this phenomenon is revealed by the experimental determination of the free energy landscape, which endows a high thermodynamic instability for folding individual repeats. If the rate limiting step for folding involves formation of two adjacent repeats without docking the inter-repeat interface, such a barrier would be entropically unfavorable and would be traversed very slowly. Indeed, a designed AR domain, which has both stable individual repeats and strong interfaces, folds much faster [51]. Folding simulations give a deeper understanding of the kinetic bottleneck and recapitulate the slow folding rates of AR domains. In these simulations, the barrier that limits folding speed is associated with an imbalance between the energetic gain of contact formation and entropy cost of folding that generates an effective free energy barrier along the funnel [52]. In globular proteins, the entropic cost is related to the "loop entropy" of forming contacts between residues remote in sequence space so it correlates with contact order, but in AR domains the entropic cost seems to originate elsewhere.
Although AR domains fold with a surprisingly high degree of cooperativity, kinetic studies show a more complex picture where kinetic two-state folding is more the exception than the rule. Additional kinetic events in refolding and unfolding result in nonlinear chevrons plots, where the denaturant dependence of the rate constant is said to “roll-over” [53]. Such is the case for the four-AR proteins Myotrophin [22] and p16INK4A and the five-AR protein p19INK4D. Moreover, the multiphasic kinetics observed for some AR proteins indicates that additional species have to be invoked in the mechanisms. For example, the larger Notch AR domain shows a single non-proline refolding phase, but two unfolding events associated with a roll-over in the unfolding arm of the chevron plots. At high urea concentrations, the two unfolding steps (N->I and I->D) have similar rates, but at low urea concentrations, the first folding step (D->I) is much faster than the subsequent (I->N) step, and thus constitutes the rate-limiting process. In addition to reproducing the kinetic data, the fitted equilibrium constants and denaturant dependences from this three-state model reproduce the species observed at equilibrium (involving just the lowest energy N and D states), supporting both the three-state kinetic and two-state equilibrium treatment [46]. Similar results were recently obtained for p19INK4D [36].
Experimental data as well as theory converge in showing that small energetic perturbations can strongly affect the folding kinetics of repeat proteins. Experimentally, a so-called phi–value analysis ideally involves probing the entire protein by substitutions on single amino acids (one at a time) and then measuring the relative effect of the mutation on the folding and unfolding rates as compared to the effect on the overall equilibrium stability [54]. For repeat proteins, high sensitivity of the folding kinetics to mutations in certain repeats, but not others, indicates that those repeats contribute to the free energy of the transition state ensemble [55], effectively suggesting that structure consolidation is ‘polarized’ towards certain parts of the protein domain. The Itzhaki group was the first to demonstrate the effectiveness of this approach in their work on p16INK4A where phi-value analysis revealed that the C-terminal two ARs fold before the N-terminal two as was discussed earlier [48]. They also recently carried out an extensive site-directed perturbation analysis on the folding of the four-AR Myotrophin domain [22]. This protein was initially shown to undergo a simple two-state transition at equilibrium [33], but the kinetics revealed a richer mechanism in which the population of a high energy intermediate was proposed. In turn, when this mechanism was probed by site-directed mutagenesis the data couldn’t be fitted even with this model, but a parallel folding route was invoked to explain the results [22]. Thus, the mutations were interpreted as changing the relative free energies of the transition state ensembles of parallel routes, nucleating at different AR pairs. Folding simulations with perfectly funneled models of this protein also suggest that multiple folding routes are energetically accessible, although a quantitative description of the effect of point mutations remains elusive (Ferreiro et al, unpublished).
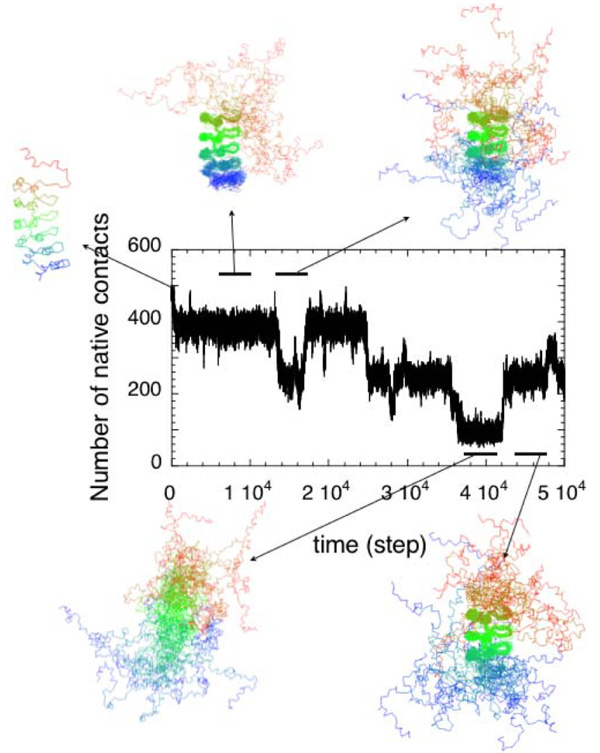
Folding simulations of the Notch AR domain using the simplest Go-type energy function for which all contacts are equi-energetic predicts folding nucleation at either end of the array, presumably for entropic reasons [41]. In contrast, experiments show a preferred single route through the central repeats [56]. This apparently is a case where the most simple level of theoretical representation is inadequate to represent the true balance between entropy loss and energy gain. In order to further investigate this discrepancy, a more detailed energy function was used, which includes different energy weights for the different types of contacts according to the Miyazawa-Jernigan description [57]. For most globular proteins, folding landscapes simulated with such "flavored" energy functions do not differ substantially from landscapes obtained from the simple homogeneous Go-models (Cho and Wolynes in preparation). For the Notch AR domain, however, the "flavors" made all the difference. The resulting simulations show a predominant folding intermediate in which the middle repeats fold first consistent with the experimentally observed mechanism (Figure 3, Ferreiro et al., unpublished observations). This result highlights again the plasticity of AR domain folding and reveals just how subtle the energetic terms are that determine preferred routes of folding in AR domains.
Figure 3.
Folding simulation of Notch using the Go-model energy function in which contacts are weighted according to the Miyzawa-Jernigan energies. The trajectory at an intermediate temperature is shown with snapshots of structures along the trajectory.
Conclusions
We envision that in the next years, AR proteins will continue to be ideal models for folding and binding. The possibility of manipulating their energy landscape suggests that the fine balance between folding and coupling among the repeats may be of functional significance. In particular, it is striking how both IκBα and Notch appear to fold upon binding to their protein targets. In the next few years, the same strategies that have been used to probe the protein folding of AR domains will hopefully be used to systematically probe the energetics of protein binding to actually measure the contribution of protein folding to the binding energy in these systems. Moreover, local perturbation of the energetics (by mutation, covalent modification, or binding of other macromolecules) will likely affect the folding of contiguous repeats, providing the means to differentially regulate binding events mediated by AR domains.
Acknowledgements
D.U.F. is a Jane Coffin Childs Fellow. Support also came from PO1 GM071862 to E. A. K.
REFERENCES
- 1.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjorklund AK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comput Biol. 2006;2:e114. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci U S A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrer P, Stumpp MT, Binz HK, Pluckthun A. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 2003;539:2–6. doi: 10.1016/s0014-5793(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 5.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 6.Zahnd C, Wyler E, Schwenk JM, Steiner D, Lawrence MC, McKern NM, Pecorari F, Ward CW, Joos TO, Pluckthun A. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB.DNA interaction fold upon binding to NF-kappaB. Proc Natl Acad Sci U S A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 10.Kloss E, Courtemanche N, Barrick D. Repeat protein folding: New insights into origins of cooperativity, stability, and topology. Arch Biochem Biophys. 2007 doi: 10.1016/j.abb.2007.08.034. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Main ER, Jackson SE, Regan L. The folding and design of repeat proteins: reaching a consensus. Curr Opin Struct Biol. 2003;13:482–489. doi: 10.1016/s0959-440x(03)00105-2. [DOI] [PubMed] [Google Scholar]
- 12.Chow CC, Chow C, Raghunathan V, Huppert TJ, Kimball EB, Cavagnero S. Chain length dependence of apomyoglobin folding: structural evolution from misfolded sheets to native helices. Biochemistry. 2003;42:7090–7099. doi: 10.1021/bi0273056. [DOI] [PubMed] [Google Scholar]
- 13.Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc Natl Acad Sci U S A. 2004;101:14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Peng Z. A minimum folding unit in the ankyrin repeat protein p16(INK4) J Mol Biol. 2000;299:1121–1132. doi: 10.1006/jmbi.2000.3803. [DOI] [PubMed] [Google Scholar]
- 15.Mosavi LK, Williams S, Peng Z. Equilibrium Folding and Stability of Myotrophin: A Model Ankyrin Repeat Protein. J Mol Biol. 2002;320:165–170. doi: 10.1016/S0022-2836(02)00441-2. [DOI] [PubMed] [Google Scholar]
- 16.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 17.Stumpp MT, Forrer P, Binz HK, Pluckthun A. Designing repeat proteins: modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J Mol Biol. 2003;332:471–487. doi: 10.1016/s0022-2836(03)00897-0. [DOI] [PubMed] [Google Scholar]
- 18.Tripp KW, Barrick D. Folding by consensus. Structure. 2003;11:486–487. doi: 10.1016/s0969-2126(03)00078-9. [DOI] [PubMed] [Google Scholar]
- 19.Forrer P, Binz HK, Stumpp MT, Pluckthun A. Consensus design of repeat proteins. Chembiochem. 2004;5:183–189. doi: 10.1002/cbic.200300762. [DOI] [PubMed] [Google Scholar]
- 20.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Pluckthun A, et al. Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe AR, Itzhaki LS. Rational redesign of the folding pathway of a modular protein. Proc Natl Acad Sci U S A. 2007;104:2679–2684. doi: 10.1073/pnas.0604653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang KS, Guralnick BJ, Wang WK, Fersht AR, Itzhaki LS. Stability and folding of the tumour suppressor protein p16. J Mol Biol. 1999;285:1869–1886. doi: 10.1006/jmbi.1998.2420. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Kohl A, Binz HK, Pluckthun A, Grutter MG, van Gunsteren WF. Molecular dynamics study of the stabilities of consensus designed ankyrin repeat proteins. Proteins. 2006;65:285–295. doi: 10.1002/prot.20991. [DOI] [PubMed] [Google Scholar]
- 25.Binz HK, Kohl A, Pluckthun A, Grutter MG. Crystal structure of a consensus-designed ankyrin repeat protein: implications for stability. Proteins. 2006;65:280–284. doi: 10.1002/prot.20930. [DOI] [PubMed] [Google Scholar]
- 26.Ferreiro DU, Cervantes CF, Truhlar SM, Cho SS, Wolynes PG, Komives EA. Stabilizing IkappaBalpha by "consensus" design. J Mol Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J Mol Biol. 2007;365:1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Zweifel ME, Barrick D. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding. Biochemistry. 2001;40:14357–14367. doi: 10.1021/bi011436+. [DOI] [PubMed] [Google Scholar]
- 30.Tevelev A, Byeon IJ, Selby T, Ericson K, Kim HJ, Kraynov V, Tsai MD. Tumor suppressor p16INK4A: structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry. 1996;35:9475–9487. doi: 10.1021/bi960211+. [DOI] [PubMed] [Google Scholar]
- 31.Boice JA, Fairman R. Structural characterization of the tumor suppressor p16, an ankyrin- like repeat protein. Protein Sci. 1996;5:1776–1784. doi: 10.1002/pro.5560050903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B, Peng Z. Defective folding of mutant p16(INK4) proteins encoded by tumor-derived alleles. J Biol Chem. 1996;271:28734–28737. [PubMed] [Google Scholar]
- 33.Lowe AR, Itzhaki LS. Biophysical characterisation of the small ankyrin repeat protein myotrophin. J Mol Biol. 2007;365:1245–1255. doi: 10.1016/j.jmb.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 34.Bradley CM, Barrick D. Limits of cooperativity in a structurally modular protein: response of the Notch ankyrin domain to analogous alanine substitutions in each repeat. J Mol Biol. 2002;324:373–386. doi: 10.1016/s0022-2836(02)00945-2. [DOI] [PubMed] [Google Scholar]
- 35.Zeeb M, Rosner H, Zeslawski W, Canet D, Holak TA, Balbach J. Protein Folding and Stability of Human CDK Inhibitor p19INK4d. J Mol Biol. 2002;315:447–457. doi: 10.1006/jmbi.2001.5242. [DOI] [PubMed] [Google Scholar]
- 36.Löw C, Weininger U, Zeeb M, Zhang W, Laue ED, Schmid FX, Balbach J. Folding Mechanism of an Ankyrin Repeat Protein: Scaffold and Active Site Formation of Human CDK Inhibitor p19INK4d. J Mol Biol. 2007;373:219–231. doi: 10.1016/j.jmb.2007.07.063. [DOI] [PubMed] [Google Scholar]
- 37.Werbeck ND, Itzhaki LS. Probing a moving target with a plastic unfolding intermediate of an ankyrin repeat protein. Proc Natl Acad Sci U S A. 2007;104:7863–7868. doi: 10.1073/pnas.0610315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. Thermodynamics reveal that helix four in the NLS of NF-kappaB p65 anchors IkappaBalpha, forming a very stable complex. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Street TO, Bradley CM, Barrick D. Predicting coupling limits from an experimentally determined energy landscape. Proc Natl Acad Sci U S A. 2007;104:4907–4912. doi: 10.1073/pnas.0608756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreiro DU, Cho SS, Komives EA, Wolynes PG. The energy landscape of modular repeat proteins: topology determines folding mechanism in the ankyrin family. J Mol Biol. 2005;354:679–692. doi: 10.1016/j.jmb.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 42.Tripp KW, Barrick D. The tolerance of a modular protein to duplication and deletion of internal repeats. J Mol Biol. 2004;344:169–178. doi: 10.1016/j.jmb.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Bryngelson JD, Wolynes PG. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luthey-Schulten Z, Ramirez BE, Wolynes PG. Helix-coil, liquid crystal, and spin glass transitions of a collapsed heteropolymer. J Phys Chem. 1995;99:2177–2185. (1995) Journal of Physical Chemistry 99, 2177–2185. [Google Scholar]
- 45.Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Q Rev Biophys. 2005;38:405–410. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- 46.Mello CC, Bradley CM, Tripp KW, Barrick D. Experimental characterization of the folding kinetics of the notch ankyrin domain. J Mol Biol. 2005;352:266–281. doi: 10.1016/j.jmb.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 47.Ferreiro DU, Komives EA. The plastic landscape of repeat proteins. Proc Natl Acad Sci U S A. 2007;104:7735–7736. doi: 10.1073/pnas.0702682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang KS, Fersht AR, Itzhaki LS. Sequential unfolding of ankyrin repeats in tumor suppressor p16. Structure (Camb) 2003;11:67–73. doi: 10.1016/s0969-2126(02)00929-2. [DOI] [PubMed] [Google Scholar]
- 49.Ivankov DN, Garbuzynskiy SO, Alm E, Plaxco KW, Baker D, Finkelstein AV. Contact order revisited: influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley CM, Barrick D. Effect of multiple prolyl isomerization reactions on the stability and folding kinetics of the notch ankyrin domain: experiment and theory. J Mol Biol. 2005;352:253–265. doi: 10.1016/j.jmb.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 51.Devi VS, Binz HK, Stumpp MT, Pluckthun A, Bosshard HR, Jelesarov I. Folding of a designed simple ankyrin repeat protein. Protein Sci. 2004;13:2864–2870. doi: 10.1110/ps.04935704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: what determines the structural details of the transition state ensemble and "en-route" intermediates for protein folding? an investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 53.Baldwin RL. On-pathway versus off-pathway folding intermediates. Fold Des. 1996;1:R1–R8. doi: 10.1016/S1359-0278(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 54.Matouschek A, J.T. K, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 55.Fersht AR, Itzhaki LS, elMasry NF, Matthews JM, Otzen DE. Single versus parallel pathways of protein folding and fractional formation of structure in the transistion state. Proc Natl Acad Sci U S A. 1994;91:10426–10429. doi: 10.1073/pnas.91.22.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bradley CM, Barrick D. The notch ankyrin domain folds via a discrete, centralized pathway. Structure. 2006;14:1303–1312. doi: 10.1016/j.str.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 57.Miyazawa S, Jernigan RL. A new substitution matrix for protein sequence searches based on contact frequencies in protein structures. Protein Eng. 1993;6:267–278. doi: 10.1093/protein/6.3.267. [DOI] [PubMed] [Google Scholar]