Abstract
The early branching eukaryote Entamoeba histolytica is a human parasite that is the etiologic agent of amebic dysentery and liver abscess. The sequencing of the E. histolytica genome combined with the development of an E. histolytica microarray has resulted in the identification of several distinct gene expression profiles associated with virulence. The function of many modulated transcripts is unknown and their role in pathogenicity unclear. They however represent a pool of potential virulence factors that could be targets for the development of novel therapeutics. Efficient tools and methods to characterize these novel virulence-associated genes and proteins would be beneficial. Here we report the use of the Gateway® cloning system to generate the E. histolytica expression vector pAHDEST. To test the usefulness of this system, the vector was used to construct a plasmid containing a recombinant version of the locus EHI_144490, which encoded a protein of unknown function. The recombinant gene was expressed and the recombinant protein, which was Strep-Myc-tagged, showed a cytoplasmic localization in transfected trophozoites. This expression vector with the Gateway® system should facilitate investigation into the functions of novel proteins in E. histolytica.
Keywords: Entamoeba histolytica, Gateway® Vector, EHI_144490
Entamoeba histolytica is an enteric aerotolerant parasite that can colonize the large intestine, and invade through the intestinal epithelium to cause amebic colitis and liver abscess [1]. The sequencing of the E. histolytica genome has provided new opportunities to identify virulence factors. Distinct gene expression profiles that may be associated with pathogenicity have been identified by microarray analysis via the comparison of transcripts of cultured HM1: IMSS E. histolytica trophozoites to amebae isolated from mouse ceca [2]. These analyses have produced fairly large data sets [2–7]. An efficient and high throughput method is needed for cloning and analyzing the function of the proteins encoded by these transcripts.
The Gateway® (Invitrogen) system has been used successfully to stably express open reading frames (ORFs) from P. falciparum by recombinational cloning, which like E. histolytica, contains an AT-rich genome [8–10]. The Gateway® system also has the potential to rapidly transfer cloned inserts from one vector to another [11]. To construct the E. histolytica ‘Destination’ plasmid we used the well-characterized plasmid pGir308 as the backbone of the new construct [12]. This vector uses the E. histolytica ferredoxin promoter, which drives strong expression of cloned genes in vivo and in vitro [2, 13]. The vector also carries a hygromycin resistance gene, which permits the use of this vector in co-infection studies with other E. histolytica shuttle vectors that carry G418-selectable markers [14].
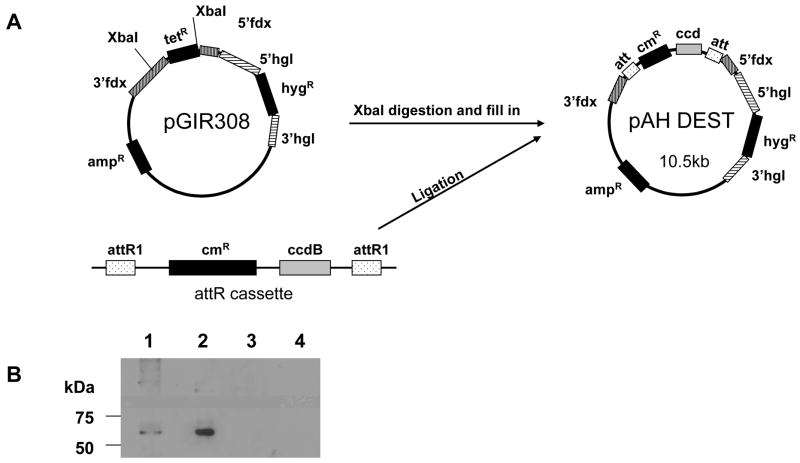
The vector pGir308 was digested with XbaI to remove the structural gene encoding the TetR protein, and the Klenow fragment of DNA polymerase used to fill-in the sticky ends. The Gateway® attR cassette was then subcloned between the blunt ends as per the manufacturer’s instructions (Invitrogen). The Gateway® attR cassette with att sites enabled recombinational cloning [15–17], and also contained a chloramphenicol resistance marker and a ccdB death gene [18]. The resultant vector, pAH-DEST could only be maintained in bacterial strains resistant to the effects of ccdB (Figure 1-A).
Fig. 1.
(A) Construction of the E. histolytica Gateway® expression vector pAH-DEST. The tetR cassette from pGIR308 was replaced with the Gateway® (Invitrogen cat. # 11828-029) cassette containing attR recombination sites flanking a ccdB gene and a chloramphenicol-resistance (cmR) gene. The new expression vector contained an ampicillinR gene for bacterial selection. (B) Affinity purification of recombinant EHI_144490. The cell lysate from trophozoites expressing Strep-Myc tagged recombinant protein was affinity-purified using a Strep-Tactin column (IBA, Gmbh) as per the manufacturer’s instructions and immunoblotted. The blot was probed with anti-Myc antibody. As expected a predominant 68kDa band was seen in whole cell lysate (lane 1) as well as in the purified fraction (lane 2). Minor higher molecular mass bands that might represent protein aggregation or post-translational modifications were also identified. No proteins recognized by the anti-Myc antibody were seen in lysate (lane 3) or purified fraction (lane 4) controls prepared from trophozoites transfected with pAH DEST vector alone.
To evaluate the high-throughput Gateway® cloning strategy with pAH-DEST as a cloning and E. histolytica expression vector, we selected the open reading frame EHI_144490. This locus was designated EHI_144490 in the new annotation of the E. histolytica genome (http://pathema.tigr.org), but was deposited in Genbank by the earlier E. histolytica genome sequencing project release as XM_650868 [19]. The EHI_144490 transcript was reduced by a factor of 6.25 in trophozoites isolated from mouse ceca 29 days after infection when compared to cultured amebae. (p<0.0001)[2]. Bioinformatics analysis indicated that locus EHI_144490 was part of an E. histolytica gene family (Ehis-1) of conserved hypothetical proteins. Ehis-1 consists of fifteen genes, ten of which were represented by unique gene sets on the Affymetrix array E_his-1a520285. Five family members were expressed at above background levels, and all were significantly down-regulated at day 29 post challenge in the mouse model of amebiasis (XM_650868, XM_646771, XM_645239, XM_649852, XM_647536)[2]. EHI_144490 (XM_650868) was one of the most highly expressed genes in this family. Members of Ehis-1 share a region similar to the sigma factor 54 ATPase domain. This domain has been shown to interact with RNA polymerase in bacteria (EMBL-EBI InterPro # IPR002078) and is present in a number of bacterial proteins involved in two-component signal transduction [20]. The Ehis-1 family proteins also contain a domain with similarity to the SEPTIN domain (InterPro Family # IPR000038). The cytoplasmic septin proteins are involved in cell division and binding of GTP in higher eukaryotes and are localized at the cleavage furrow in dividing cells [21, 22]. Thus the function(s) and locations of this family of proteins are unclear.
The coding sequence of EHI_144490 was PCR amplified from HM1:IMSS genomic DNA. The primers used were designed to introduce two N-terminal tags in tandem: Strep (WSHPQFEK) and c-myc (EQKLISEEDL). These tags permit identification and one step purification of the recombinant protein. The PCR product was then cloned into the Gateway® entry vector-pCR8/GW/TOPO (Invitrogen). The att recombination sites in the vector allowed for rapid recombination into a variety of destination vectors. The LR Clonase system (Invitrogen) was then used to transfer the recombinant EHI_144490 cassette from the entry clone into pAH-DEST destination plasmid using the manufacturer’s instructions. While aberrant recombination events have been reported to occur only at extremely low frequencies our experience suggests that resequencing the final expression cassette is an essential step in quality control [17]. The pAHDEST-EHI_144490 expression plasmid generated as described above was used to transfect HM1: IMSS trophozoites using the standard protocol [23, 24]. Approximately 4 × 106 trophozoites carrying pAH DEST-EHI_144490 were then used for protein purification. A Strep-Tactin spin column (IBA) was used to affinity purify the recombinant protein from total cell lysate. As expected the eluted protein showed a 68 kDa band on a western blot when probed with anti c-myc antibody (Fig. 1-B).
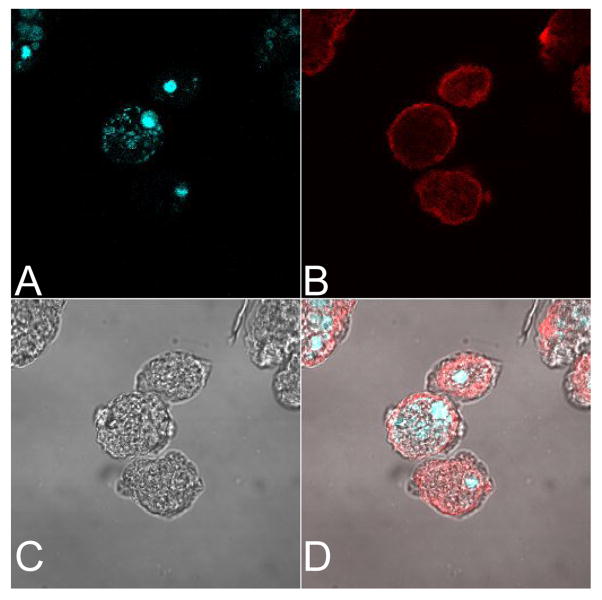
To determine the subcellular location of the product of EHI_144490 the transfected amebae were grown at 37°C in TYI-S-33 medium containing 50μg/ml hygromycin. These trophozoites (106) were then bound to glass cover slips in a 24-well plate for 30 min at 37°C in TYI-S-33 medium. Adherent amebae were fixed in 3% paraformaldehyde for 30 min at room temperature (RT) followed by permeablization using 0.2% Triton X-100 in PBS for 1 min. Nonspecific binding was blocked with 20% goat serum and 5% bovine serum albumin (Sigma) in PBST for 2h at RT. The cover slips were then incubated with anti-cMyc antibody (Santa Cruz) for 2h at RT followed by three washes with PBST. Cya-3 conjugated donkey anti-mouse secondary antibody (Santa Cruz) was added for 30 min at RT followed by DAPI staining. The cover slips were washed three times and mounted using Vectashield (Sigma) mounting medium. Confocal images were visualized using a Zeiss LSM 510 laser scanning microscope (Carl Zeiss). This experiment was repeated with two independent constructs and transfected strains. The recombinant protein was located in the cytoplasm (Fig. 2).
Fig. 2.
Cytoplasmic location of recombinant EHI_144490. HM1: IMSS trophozoites were transfected with the Gateway® construct expressing recombinant EHI_144490. Nuclei were stained with DAPI (blue, panel A). The recombinant protein was detected with monoclonal anti-Myc antibody and Cya 3-conjugated donkey anti-mouse antibody (panel B). The bright field image is shown in panel C, and panel D shows the merged images. The recombinant protein was cytoplasmic (red, panel B and D). Amebae probed with secondary antibody alone did not show staining (data not shown). The images were obtained using a Zeiss LSM 410 laser scanning confocal microscope.
In conclusion, a Gateway®-based vector applicable for high-throughput cloning and expression of recombinant proteins in E. histolytica trophozoites was constructed. The vector was used to epitope tag and show that open reading frame EHI_144490 encoded a protein with a cytoplasmic location. This new expression system should facilitate the study of unique proteins in E. histolytica.
Acknowledgments
This work was supported by NIH grant AI-37941. The Pathema-Entamoeba Database is an NIAID Bioinformatics Resource Center (BRC).
Abbreviations
- ORF
open reading frame
- myc
myelocytomatosis viral oncogene homolog
- strep
streptavidin
- ampR
ampicillin resistance
- cmR
chloramphenicol resistance
- tetR
tetracycline resistance
- fdx
ferredoxin
- hgl
heavy subunit of the galactose and N-acetylgalactosamine lectin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haque R, Mondal D, Duggal P, Kabir M, Roy S, Farr BM, Sack RB, Petri WA., Jr Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect Immun. 2006;74:904–909. doi: 10.1128/IAI.74.2.904-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA., Jr Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Ackers JP, Mirelman D. Progress in research on Entamoeba histolytica pathogenesis. Curr Opin Microbiol. 2006;9:367–373. doi: 10.1016/j.mib.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Davis PH, Schulze J, Stanley SL., Jr Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol Biochem Parasitol. 2007;151:118–128. doi: 10.1016/j.molbiopara.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 5.MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrenkaufer GM, Eichinger DJ, Singh U. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics. 2007;8:216. doi: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 8.Aguiar JC, LaBaer J, Blair PL, Shamailova VY, Koundinya M, Russell JA, Huang F, Mar W, Anthony RM, Witney A, Caruana SR, Brizuela L, Sacci JB, Jr, Hoffman SL, Carucci DJ. High-throughput generation of P. falciparum functional molecules by recombinational cloning. Genome Res. 2004;14:2076–2082. doi: 10.1101/gr.2416604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Skinner-Adams TS, Hawthorne PL, Trenholme KR, Gardiner DL. GATEWAY vectors for Plasmodium falciparum transfection. Trends Parasitol. 2003;19:17–18. doi: 10.1016/s1471-4922(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 11.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 12.Ramakrishnan G, Vines RR, Mann BJ, Petri WA., Jr A tetracycline-inducible gene expression system in Entamoeba histolytica. Mol Biochem Parasitol. 1997;84:93–100. doi: 10.1016/s0166-6851(96)02784-3. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist CA, Mann BJ, Petri WA., Jr Control of ferredoxin and Gal/GalNAc lectin gene expression in Entamoeba histolytica by a cis-acting DNA sequence. Infect Immun. 1998;66:2383–2386. doi: 10.1128/iai.66.5.2383-2386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vines RR, Purdy JE, Ragland BD, Samuelson J, Mann BJ, Petri WA., Jr Stable episomal transfection of Entamoeba histolytica. Mol Biochem Parasitol. 1995;71:265–267. doi: 10.1016/0166-6851(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 15.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 16.Nunes-Duby SE, Matsumoto L, Landy A. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in lambda excisive recombination. Cell. 1989;59:197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- 17.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard P, Gabant P, Bahassi EM, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 19.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillen N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 20.Morett E, Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita M. The septins. Genome Biol. 2003;4:236. doi: 10.1186/gb-2003-4-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haarer BK, Pringle JR. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olvera A, Olvera F, Vines RR, Recillas-Targa F, Lizardi PM, Dhar S, Bhattacharya S, Petri W, Jr, Alagon A. Stable transfection of Entamoeba histolytica trophozoites by lipofection. Arch Med Res. 1997;28(Spec No):49–51. [PubMed] [Google Scholar]
- 24.Asgharpour A, Gilchrist C, Baba D, Hamano S, Houpt E. Resistance to intestinal Entamoeba histolytica infection is conferred by innate immunity and Gr-1+ cells. Infect Immun. 2005;73:4522–4529. doi: 10.1128/IAI.73.8.4522-4529.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]