Summary
Fertility and embryonic viability are measures of efficient germ cell growth and development. During oogenesis and spermatogenesis, new proteins are required for both mitotic expansion and differentiation. Qualitative and quantitative changes in protein synthesis occur by translational control of mRNAs, mediated in part by eIF4E, which binds the mRNAs 5′ cap. IFE-1 is one of five eIF4E isoforms identified in C. elegans. IFE-1 is expressed primarily in the germ line and associates with P granules, large mRNPs that store mRNAs. We isolated a strain that lacks IFE-1 [ife-1(bn127)] and demonstrated that the translation of several maternal mRNAs (pos-1, pal-1, mex-1 and oma-1) was inefficient relative to that in wild-type worms. At 25°C, ife-1(bn127) spermatocytes failed in cytokinesis, prematurely expressed the pro-apoptotic protein CED-4/Apaf-1, and accumulated as multinucleate cells unable to mature to spermatids. A modest defect in oocyte development was also observed. Oocytes progressed normally through mitosis and meiosis, but subsequent production of competent oocytes became limiting, even in the presence of wild-type sperm. Combined gametogenesis defects decreased worm fertility by 80% at 20°C; ife-1 worms were completely sterile at 25°C. Thus, IFE-1 plays independent roles in late oogenesis and spermatogenesis through selective translation of germline-specific mRNAs.
Keywords: Germ line, Protein synthesis, Maternal mRNAs, Oogenesis, Spermatogenesis, CED-4/Apaf-1, Meiosis, Polysomes
Introduction
Gamete development involves crucial periods of both gene regulation and cell differentiation. During this time stem cells undergo multiple rounds of mitotic divisions followed by differentiation into either sperm or oocytes via meiotic divisions. Transcription is active early in gametogenesis allowing the cell to make mRNAs for concurrent and future stages. Later as the cell enters meiosis transcription is silenced. mRNAs that have been stored during the proliferative phase become the sole means of producing new proteins and are regulated by translational control. Proper development of gametes is essential for fertilization and therefore efficient propagation of the organism (Tadros and Lipshitz, 2005).
Progression through gametogenesis is mediated (in part) by the translation of maternally or paternally derived mRNAs, specifically their coordinated recruitment to ribosomes. The mRNA binding step and subsequent formation of the 48S pre-initiation complex are rate-limiting in translation and thus often the target of regulation. Eukaryotic translation initiation factor 4E (eIF4E) first recognizes the mRNA 7-methyguanosine cap structure, then associates with eIF4G, the scaffolding protein of the complex, bringing mRNA into contact with other initiation factors and the 40S ribosomal subunit (Gingras et al., 1999). In many systems, including mammals and yeast, the binding of eIF4E and eIF4G is inhibited by 4E-binding proteins (4EBPs) that bind the same site bound by eIF4G (Proud, 2006). 4EBPs found in germ cells and embryos have been shown to inhibit eIF4G binding in a similar fashion (Barnard et al., 2005; Dahanukar et al., 1999; Wilhelm and Smibert, 2005). Several 4EBPs also bind indirectly to mRNA 3′ end sequences. This mode of translational control has been most extensively demonstrated during oogenesis in Xenopus laevis, in which a cap-dependent translation inhibitor called Maskin prevents translation of cyclin, cdk2 and c-mos mRNAs by preventing eIF4E interaction with eIF4G. Maskin also associates with the cytoplasmic polyadenylation element binding protein (CPEB) to modulate poly(A) tail length. The eIF4E-Maskin-CPEB complex represents a dormant mRNP that is poised to recruit the resident mRNAs upon a physiological (usually cell cycle) signal. During Xenopus oocyte maturation the hormone progesterone triggers both cytoplasmic poly(A) tail elongation and the release of Maskin to allow eIF4E-bound mRNAs to associate with ribosomes and thereby promote synthesis of cell cycle regulators for meiotic progression (Barnard et al., 2005; Richter, 2007). Translational control by 4EBPs such as Maskin are evident in other species including Drosophila and mammals (Mendez and Richter, 2001).
Multiple isoforms of eIF4E exist in every species so far examined. Eight eIF4E isoforms are expressed in Drosophila, three in mammals and sea urchins, two in Xenopus, Arabidopsis and zebrafish, and five in C. elegans (Keiper et al., 2000; Minshall et al., 2007; Morales et al., 2006; Robalino et al., 2004; Rodriguez et al., 1998). Of these isoforms, germ-cell-specific isoforms have been found in mouse, Drosophila and C. elegans (Amiri et al., 2001; Evsikov et al., 2006). Tissue-specific expression and function have been demonstrated for several of the five eIF4E isoforms (named IFE-1 to IFE-5) in the nematode Caenorhabditis elegans. IFE-2 and IFE-4 are expressed in somatic tissues and function in aging and nerve/muscle development, respectively (Dinkova et al., 2005; Syntichaki et al., 2007). IFE-4 is required for the translation of a small number of mRNAs required for neuromuscular function. The remaining IFEs (-1, -3 and -5) are expressed primarily in the germ line. Of them, IFE-1 is the only isoform that binds PGL-1, an RNA binding protein that is the hallmark of P granules. P granules are large ribonucleoprotein complexes (mRNPs) found in both spermatocytes and oocytes that store maternal/paternal mRNAs (e.g. pos-1, mex-1, nos-2 and gld-1) as well as numerous mRNA binding proteins, helicases and translation factors (Amiri et al., 2001; Jud et al., 2008; Schisa et al., 2001). P granules segregate to the germ lineage to transport mRNAs and proteins during oogenesis and early embryogenesis (Seydoux and Strome, 1999). Therefore translational control becomes the predominant mode of gene regulation during late gametogenesis. As an integral part of P granules, IFE-1 associates with stored maternal mRNAs and may mediate their eventual recruitment during development.
Previous work has shown that ife-1 mRNA expression begins in the L3 larval hermaphrodite gonad, and coincides with the onset of meiosis (Amiri et al., 2001). In the C. elegans hermaphrodite, spermatogenesis during the L4 stage is followed by oogenesis during adulthood. Depletion of IFE-1 by RNAi dramatically decreased fertility by arresting the development of primary and secondary spermatocytes. ife-1(RNAi) oocytes, however, could be fertilized by wild-type sperm, suggesting IFE-1 has a less critical role in oogenesis.
The present study further delineates the role of IFE-1 in the germ line by closely examining its requirement for the development of both types of gametes. Here we show that complete loss of IFE-1 by gene disruption causes a temperature-sensitive block to cytokinesis in secondary spermatocytes that results in multinucleated, immature sperm. Lack of IFE-1 does not promote germ cell apoptosis, but induces precocious expression of the pro-apoptotic factor, CED-4, during spermatogenesis. Defects in oogenesis are not overtly apparent; however, we found that oocyte production is modestly reduced. In addition ife-1 oocytes are defective in ways that make them less likely to be fertilized when they encounter wild-type spermatozoa. Our data also indicate that IFE-1 is specifically utilized for efficient translation of maternal mRNAs during oogenesis, since the recruitment of certain mRNAs is compromised in worms lacking IFE-1. Our data indicate that the germline-specific translation factor IFE-1 is required to promote temporal and spatial translational control during critical steps in gametogenesis.
Results
Characterization of the ife-1(bn127) strain
We used trimethylpsoralen and UV mutagenesis followed by PCR to isolate a deletion allele of the C. elegans ife-1 gene. The 590 bp deletion (bn127) starts at nucleotides (nt) 191 in exon 1 and extends through exon 2 and into the 3′ UTR to nt 780 (Fig. 1A). The deletion is detectable by whole worm genomic PCR; flanking primers amplify a 1359 bp PCR product from wild-type worms and a shorter 769 bp PCR product from mutant worms (Fig. 1B). The deletion removes over 70% of the coding region for IFE-1 (149 of the 212 codons), including the α-helices and β-sheets that make up the mRNA platform and a Trp residue essential for m7GTP cap binding, suggesting it is a null mutation. In all experiments described below, a stable mutant worm strain homozygous for ife-1(bn127) was used.
Fig. 1.

Characterization of ife-1(bn127) mutant worms. (A) Depiction of ife-1 gene and location of the 590 bp bn127 deletion. Exons are shown as colored boxes with scale bar indicating 100 bp. (B) Whole worm genomic PCR of three individual ife-1 clones, one heterozygous ife-1/mT1, and one wild-type hermaphrodite using primers –523s and 827a. The wild-type ife-1 gene produces a PCR product of 1359 bp. The bn127 deletion produces a shorter 769 bp product. (C-E) Brood size and stage of offspring after 96 hours at 15°C (C), 20°C (D) or 25°C (E) for ife-1 and wild-type worms. Progeny were scored as oocytes (ooc), unhatched embryos (emb), early (L1/L2) or late (L3/L4) larvae or adults (Ad). Numbers are averages per adult worm (n=9). Error bars indicate standard deviation. At 15°C and 20°C, ife-1 mutant worms produced a small number of viable offspring. At 25°C ife-1 mutants produced only oocytes.
Growth and fertility characteristics of ife-1 worms were similar to the previously published ife-1(RNAi) phenotype (Amiri et al., 2001; Keiper et al., 2000). Worms homozygous for ife-1 completed embryonic, larval and adult development, but had severely compromised fertility. To assess the number and quality of offspring produced, we analyzed brood sizes of wild-type and ife-1 worms over 96 hours. When grown at 15°C or 20°C, ife-1 hermaphrodites produced only 20-30 viable progeny per worm, which represents 10-13% the number of viable progeny produced by the wild type (Fig. 1C,D). ife-1 hermaphrodites also produced a significant population of unfertilized oocytes and arrested embryos. Wild-type hermaphrodites also produced unfertilized oocytes, most probably because of sperm depletion by the end of the egg laying period. However, unfertilized oocytes represented a larger percentage of the overall progeny produced from ife-1 hermaphrodites. Therefore, in the absence of IFE-1 the production of viable offspring at 15°C and 20°C was greatly diminished and may be the result of impaired sperm or oocyte production.
At 25°C, ife-1 mutants failed to produce any viable offspring or arrested embryos (Fig. 1E); instead they laid just a few unfertilized oocytes. Wild-type worms produced a significant number of viable progeny (∼150) and more unfertilized oocytes than ife-1 mutants. The unfertilized oocytes are probably the result of depletion of sperm supply by 96 hours at elevated temperature. These results indicate that loss of IFE-1 causes temperature-sensitive sterility similar to that seen with other mutants of P-granule components (e.g. pgl-1). In general, the total oocytes output (viable and inviable) for wild-type and ife-1 worms at 25°C was similar to their respective outputs at 20°C. The data suggest that sperm and, to a lesser extent, mature oocyte production are limited by loss of IFE-1. One or both of these processes is completely prevented at higher temperature (25°C).
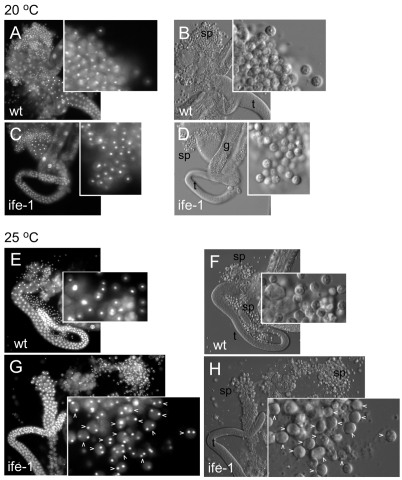
To determine if the sterility at 25°C was caused solely by the absence of mature sperm as previously observed by ife-1(RNAi), we examined the spermatheca of wild-type and ife-1 hermaphrodites by DAPI staining to look for sperm nuclei. For comparison, we also raised fem-2(b245) hermaphrodites that fail to produce sperm at 25°C (Kimble et al., 1984). Wild-type hermaphrodites grown at 25°C successfully produced sperm and stored them in the spermatheca (Fig. 2A). By contrast, ife-1 hermaphrodites, like fem-2, failed to show any mature sperm in the spermatheca (Fig. 2B,C). Their absence suggests that IFE-1 is essential for production of functional spermatozoa at 25°C. Interestingly, when oocyte production is not impaired in worms that lack sperm, oocytes are retained in the ovary and begin to `stack up' (Kimble et al., 1984). We observed dramatic stacking of oocytes in fem-2 young adult hermaphrodites, which accumulated 15 or more late oocytes in the proximal gonad, compared with seven to ten oocytes in wild-type worms (Fig. 2D,F). Wild-type hermaphrodites have an adequate supply of spermatozoa at 25°C, allowing fertilization to occur and preventing the stacking of oocytes. ife-1 oocytes did not stack in young adult hermaphrodites even though spermatozoa were not evident (Fig. 2E). If oocytes were being produced at the same rate as in wild type and fem-2, stacking would occur. Therefore, the rate of oocyte production is decreased in the absence of IFE-1. Our data suggest that sterility as a result of the ife-1 mutation is the consequence of separate defects in both sperm and oocyte production.
Fig. 2.
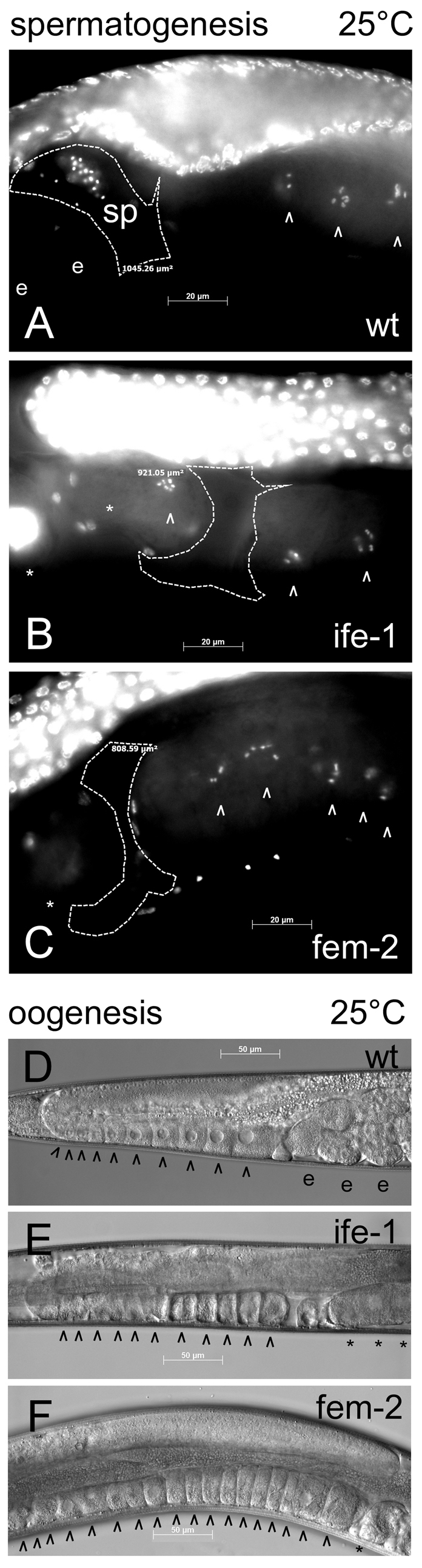
Nuclear and cellular morphology of wild-type, ife-1 and fem-2 hermaphrodite gonads. Whole worms were analyzed during their maximal fertile period at 3 days (wt and fem-2) or 4 days (ife-1; as a result of prolonged L4 stage) post hatching at 25°C. (A) DAPI staining of whole worms indicated the presence of sperm (sp) in wild-type spermatheca. Sperm were not present in the spermatheca of ife-1 (B) and fem-2 (C) hermaphrodites. The spermatheca is outlined by a dashed line. Arrowheads (^) indicate post-pachytene oocytes; e, fertilized embryos in the uterus of wild-type hermaphrodite; asterisks, unfertilized oocytes undergoing endomitosis. (D-F) Wild-type (D), ife-1 (E) and fem-2 (F) gonads with mature oocytes prior to fertilization. fem-2 oocytes stacked up in the absence of sperm for fertilization. Stacking of oocytes was not observed in the ife-1 gonad which also lacks sperm.
Defective steps in oocyte and sperm development
The experiments described above show only that IFE-1 deficiency prevents sperm production and hinders oocyte output, but do not describe the developmental steps affected by the deficiency. We next examined the nature of the defects in oogenesis and spermatogenesis at the cellular level, and considered the effects of temperature on each. The progression of oogenesis was addressed by dissecting gonads of young adult ife-1 mutant hermaphrodites grown at 20°C and 25°C and analyzing cellular and nuclear morphology by microscopy. In wild-type gonads, maturing oocytes in the proximal arm arrest at diakinesis of meiosis I and contain condensed bivalent chromosomes; meiosis is completed after ovulation and fertilization (Yamamoto et al., 2006). ife-1 gonads similarly contained growing oocytes with condensed chromosomes (diakinesis) that appeared normal in cell and nuclear morphology. On close inspection, many ife-1 oocytes were more elongated, flaccid and less well packed than wild-type ones (Fig. 3C,D). However, we also observed one or two post-pachytene oocytes in each ife-1 gonad that had lost integrity of the formed bivalent chromosomes at both 20°C and 25°C (Fig. 3D,H `d'). Unfertilized oocytes were frequently observed in the uterus at both temperatures. To address whether such oocyte defects were merely a consequence of the lack of sperm, we compared ife-1 oocyte morphology to that of fem-2 oocytes at 25°C. The loss of bivalent chromosome integrity was never observed in the feminized strain (i.e. fem-2) and indeed, many more diakinetic oocytes populated the proximal gonad than in ife-1 (compare number of arrowheads in Fig. 3G-J). Oocytes that pass through the spermatheca unfertilized undergo pseudo-activation in the uterus, resulting in chromosome decondensation and endomitosis in a similar fashion for both ife-1 and fem-2. These data demonstrate that the lack of IFE-1 results in less overall oocyte production or progression at both 20°C and 25°C, regardless of sperm availability.
Fig. 3.
Oocyte development in ife-1 hermaphrodite gonads. Whole gonads were dissected from worms grown at 20°C (A-D) and 25°C (E-J). At 20°C, in the wild-type gonad there is normal late oocyte progression in the proximal region (A, DIC image; B, Hoechst stained). There are ten sequential oocytes with condensed chromosomes (post-pachytene) arranged in linear array (arrowheads; ^). ife-1 gonads grown at 20°C showed somewhat elongated, but otherwise normal oocytes in the proximal region (C,D). In contrast to wild type, only seven post-pachytene oocytes were aligned in the ife-1 gonad. Nuclear staining indicated that most ife-1 oocytes contained aligned bivalent chromosomes (^); a few failed to condense (d). Wild-type worms raised at 25°C (E,F) showed similar characteristics to those raised at 20°C with eight post-pachytene oocytes aligned in the proximal region. The gonad from ife-1 worms raised at 25°C (G,H) also showed similar characteristics to the ife-1 gonad at 20°C, with five oocytes with condensed bivalents (^). Fertilization did not occur in ife-1 worms at 25°C, and four unfertilized oocytes were observed in the uterus undergoing endomitosis (*). The feminized fem-2 gonad (I,J), in which no fertilization occurs, showed normal oocyte progression in the proximal region and accumulated 14 post-pachytene oocytes.
In order to discover the stage of sperm development that was aberrant in ife-1 germ cells, we dissected young male gonads and analyzed them by DIC microscopy and Hoechst nuclear staining. ife-1 males were capable of producing spermatocytes, but fewer secondary spermatocytes developed to mature spermatids compared with wild-type males at 20°C (Fig. 4A-D). Mature ife-1 spermatids had normal dimpled morphology, rough cell surface, and single highly condensed chromatin spheres (L'Hernault, 2006). At 25°C, however, ife-1 secondary spermatocytes were completely blocked from progressing to spermatids. ife-1 gonads accumulated a large number of late pachytene-staged spermatocytes. Those spermatocytes that progressed further showed defects in cytokinesis, specifically in spermatid budding from the residual body (Fig. 4E-H). These cells appeared to complete the meiotic nuclear divisions but not cell division. Eventually, large, smooth cells that retained the morphology of secondary spermatocytes accumulated, but these contained two or four haploid nuclei (Fig. 4G,H). Despite their number, these cells were apparently not competent to fertilize either ife-1 or wild-type oocytes (see Fig. 5). Assuming that male spermatogenesis mirrors the process in hermaphrodites, the defect in cytokinesis also prevented hermaphrodite secondary spermatocytes (which are not motile) from maintaining their presence in the spermatheca (Fig. 2). These observations suggest that IFE-1 regulation of protein synthesis is required either during or before the budding event in spermatogenesis.
Fig. 4.
Morphology of ife-1 male gonads, spermatocytes and sperm. Squashed dissected male gonads from wild-type (wt) and ife-1 males grown at 20°C (A-D) and 25°C (E-H) depict progression through spermatogenesis. Wild-type testes stained for DNA with Hoechst (A) or visualized by DIC microscopy (B) showed highly condensed DNA in individual spermatids (sp) at the proximal (mature) end and immature germ cells in pachytene stages in the distal portion of the testis (t). The gut is indicated by g. ife-1 testes show a similar progression but fewer spermatids (C,D). Testes from male worms grown at 25°C were similarly prepared and analyzed (E-H). Wild-type testes (E,F) look similar to those at 20°C, with the highly condensed haploid nuclei in distinct individual spermatids. (Insets) Fourfold magnification of a selected field containing secondary spermatocytes and mature spermatids. The latter were identified by their rough appearance and a central `dimple'. By contrast, ife-1 testes at 25°C (G,H) contained many secondary spermatocytes with multiple nuclei (2-4; ^), but lacked mature spermatids. Terminal multinucleated spermatocytes failed in cytokinesis during residual body formation.
Fig. 5.
Effect of temperature on ife-1 fertility. Brood size from wild-type (A) and ife-1 (B) hermaphrodites shifted from 20°C to 25°C (blue) and 25°C to 20°C (pink) at 12 hour intervals. Wild-type hermaphrodites showed no temperature-induced sterility (A). By the L4 -young adult transition (60-72 hours), temperature downshifts no longer rescued fertility for ife-1 hermaphrodites, and temperature upshifts no longer induced complete sterility (B). Temperature-shifted wild-type hermaphrodites had similar brood sizes (120-220). Fertile ife-1 worms had much smaller broods (5-25). Staging of worms by morphology at time of shift demonstrated the prolonged ife-1 L4 stage (C). Points between stages indicate roughly equal percentages of worms at both stage. (D,E) Mating experiments with three males and one hermaphrodite of each of the indicated genotypes. Total bar height indicates brood size over 96 hours. Male offspring are shown in purple, and hermaphrodites shown in blue at 20°C (D) and 25°C (E). The mean number of offspring (#F1 worms) of triplicate matings were plotted. Error bars indicate the standard deviation from the mean. For clarity, only the `positive error' is displayed for purple bars, and the `negative error' is displayed for blue bars. (F) The number of inviable progeny (unfertilized oocytes and arrested embryos) of wild-type or ife-1 hermaphrodites mated with wild-type males. Percentage values above the bars indicate inviable offspring normalized to respective brood size. Error bars indicate standard deviation.
Temperature sensitivity during gonad development in larvae
C. elegans adult hermaphrodites utilize a temporal switch to produce both sperm and oocytes from the same pool of germ cells (Kimble and Crittenden, 2007). Germ cells start mitotic division in the L1 larval gonad and meiotic divisions in the L3 larval stage. During the L4 stage meiotic divisions lead to the development of mature sperm. At the end of L4, germ cell development switches to oogenesis exclusively, and oocytes become fertilized by sperm that were stored in the spermatheca (Kimble and Crittenden, 2007). To determine which stage in gametogenesis is responsible for ife-1 temperature-sensitive sterility, we conducted temperature-shift experiments between 20°C and 25°C. As previously noted, ife-1 hermaphrodites have low fertility at 20°C and are completely sterile at 25°C. To define the temperature-critical period, we synchronized wild-type and ife-1 L1s and shifted them from 20°C to 25°C and vice versa at 12-hour intervals. Total brood size (viable progeny) was recorded from each temperature-shifted animal at the end of egg laying and hatching (∼108 hours; Fig. 5A,B). Developmental stage at the time of the shift was also recorded. Animals moved to 25°C before 60 hours developed into sterile adults. Conversely, animals shifted from 25°C to 20°C after 60 hours were sterile. These results identify 60 hours post L1 stage as the temperature-critical period (TCP). The TCP correlates temporally with spermatogenesis during the L4 stage. Unexpectedly, the L4 stage was consistently prolonged (nearly twofold) in ife-1 worms as indicated by worm size and the development of the vulva (Fig. 5C). Progression of ife-1 worms from L4 to adults required at least 24 hours at both 20°C and 25°C, whereas the wild type required just 12 hours. Our data demonstrate that defects in spermatogenesis are responsible for temperature-sensitive sterility in strains lacking IFE-1.
Based on the results described above, we suggest that loss of IFE-1 function causes mild defects in oogenesis, and severe defects in spermatogenesis. However, it is possible that reduced oocyte production could stem solely from the absence of viable sperm in ife-1 hermaphrodites. Sperm produce major sperm protein (MSP) that stimulates ovulation (Detwiler et al., 2001; Jud et al., 2007; Miller et al., 2001). In order to test the contributions of IFE-1 function in oogenesis versus spermatogenesis, and the temperature sensitivity of each process, we conducted mating experiments with wild-type and ife-1 worms at 20°C and 25°C. In matings of C. elegans, male sperm efficiently out-compete the hermaphrodite's own sperm and give rise to a 1:1 ratio of male to hermaphrodite offspring. Wild-type hermaphrodite sperm give rise to essentially no males. As expected, sperm from wild-type males were able to dramatically increase (approx. sixfold) brood sizes from ife-1 hermaphrodites at both 20° and 25°C (Fig. 5D,E; wt × ife-1 versus ife-1 × ife-1). Despite the enhancement by wild-type sperm, ife-1 worms produced approximately half the number of offspring of wild-type worms (wt × ife-1 versus wt × wt; Fig. 5D), suggesting that ife-1 causes a reduction in oocyte production. This proportion (half the brood size) was maintained at 25°C, indicating that the viability of IFE-1-deficient oocytes, unlike sperm, is not altered by temperature (Fig. 5E). The gonads of ife-1 hermaphrodites were depleted of late oocytes in diakinesis by mating to wild-type males (data not shown), whereas wild-type and fem-2 hermaphrodites maintained a stream of oocytes ready for fertilization. These observations show that ife-1 worms fail to produce equivalent numbers of mature oocytes relative to wild-type and fem-2 worms, whether or not fertilization occurs.
Temperature-shift experiments indicated spermatogenesis was the primary determinant of temperature-sensitive sterility. Consequently, ife-1 males had little success in fertilizing wild-type or ife-1 oocytes at 20°C, producing ten- to 20-fold fewer male offspring in both cases (Fig. 5D). By contrast, fem-2 males, which produce sperm when grown at 20°C but not at 25°C, were able to produce adequate numbers of male offspring (fem-2 × wt, fem-2 × ife-1, fem-2 × fem-2) at the permissive temperature. Male offspring were completely absent in ife-1 male matings at 25°C, regardless of the recipient, confirming that spermatogenesis is further compromised by high temperature. Similar results were seen with the fem-2 male mating at 25°C (Fig. 5E). Finally, when the ife-1 males were crossed with ife-1 hermaphrodites, the combined effects of defective oocytes and defective sperm led to >tenfold decrease in fertility relative to wild-type counterparts and complete sterility at 25°C (Fig. 5D,E). Further evidence that ife-1 oocytes were less fertile than wild-type oocytes was provided by the number of unfertilized oocytes and arrested embryos produced from ife-1 oocytes and wild-type sperm (Fig. 5F). The availability of wild-type sperm did not prevent ife-1 hermaphrodites from producing unfertilized oocytes (4.7% of total oocyte production versus 0.3% for wt), indicating that a substantial proportion of ife-1 oocytes are not receptive for fertilization. A higher percentage of arrested embryos also resulted from such matings. These results indicate that the role of IFE-1 during spermatogenesis is more critical than during oogenesis, but ife-1 infertility results from a combination of both defects.
Translation efficiency of stored maternal mRNAs
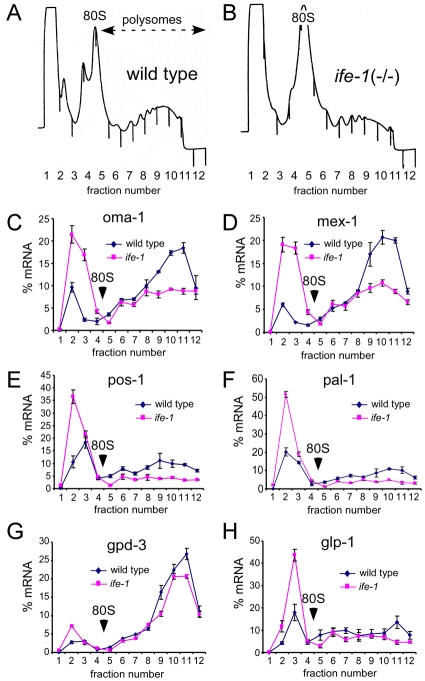
IFE-1 is one of three isoforms of eIF4E in the C. elegans germ line, and the only isoform to associate with the P granules protein PGL-1 (Amiri et al., 2001). As IFE-1 is a translation factor, we hypothesized that its loss might compromise ribosome binding and translation of germ cell mRNAs. To address its role in translational control of maternal mRNAs in these cells, we analyzed polysomes in wild-type and ife-1 worms (Fig. 6). We fractionated mixed-stage, whole worm lysates and analyzed the sedimentation of particular mRNAs by sucrose gradient centrifugation. RNA isolated from gradient fractions was analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) to determine the distribution of specific maternal mRNAs throughout the gradient. mRNAs that sedimented toward the bottom of the gradient (at or below 80S) were loaded in polysomes, indicating a high translation efficiency. By contrast, mRNAs remaining at the top of the gradient (<80S) were not actively translating. Absorbance profiles demonstrated that both content and size of polysomes was similar in wild-type and ife-1 strains (Fig. 6A,B). Only a modest decrease in polysome content was observed in the ife-1 strain compared with the wild type, suggesting that overall protein synthesis is supported in large part by other IFE isoforms. The ife-1 strain also grows and develops more slowly than the wild type (see Fig. 5C), consistent with a slightly lower level of protein synthesis.
Fig. 6.
Analysis of maternal mRNA translation efficiency by polysome fractionation. Absorbance (A254) profiles for continuous gradient fractionation of wild-type (A) and ife-1 (B) mixed staged hermaphrodite worm extracts depict the polysome content of each strain. The highest resolved peak is the monosome (80S) and each peak to the right represents the addition of one ribosome (polysomes). (C-H) Sedimentation of individual maternal mRNAs. mRNA distribution of oma-1 (C), mex-1 (D), pos-1 (E), pal-1 (F), glp-1 (G) and gpd-3 (H) throughout the gradients was quantified by qRT-PCR. mRNA signal was normalized to RNA content in each fraction. Normalization corrects for slight differences in polysome yield, allowing direct comparison of the translational efficiency of each mRNA. Error bars indicate the standard deviation of triplicate qRT-PCR determinations.
Several examples of translational control of specific mRNAs during oogenesis and early embryogenesis in C. elegans have been described (Goodwin and Evans, 1997). Many maternal mRNAs are repressed, then derepressed in temporally and spatially regulated manners. We propose that the cap-binding protein IFE-1 plays an active role in recruiting such mRNAs to the ribosome in late oogenesis and spermatogenesis. In light of the decreased oocyte production observed in ife-1 hermaphrodites (Fig. 5), it was of particular interest to assay the translational efficiency of mRNAs that encode proteins required for developmental functions in late oogenesis and fertilization. oma-1 mRNA is expressed throughout oogenesis, but OMA-1 protein only accumulates in late oocytes and more strongly in newly fertilized embryos (Detwiler et al., 2001; Shimada et al., 2006). Analysis of oma-1 mRNA distribution indicated that its loading to polysomes was markedly decreased in the absence of IFE-1 (fivefold; Fig. 6C). A correspondingly large increase in oma-1 mRNA in the non-translated region indicated that initiation events on this mRNA had become very inefficient. This suggests that IFE-1 facilitates oma-1 mRNA recruitment to ribosomes. Another oocyte-expressed protein, MEX-1, is responsible for proper segregation of P granules during early embryogenesis (Guedes and Priess, 1997). MEX-1 protein must therefore accumulate in oocytes prior to fertilization. Both the MEX-1 protein and mex-1 mRNA associate with P granules suggesting that IFE-1 may play a role in its synthesis. We examined the distribution of mex-1 mRNA and found it had decreased sevenfold in loading to polysomes in the absence of IFE-1 (Fig. 6D). Therefore, mex-1 mRNA, like oma-1 mRNA, is highly dependent on the IFE-1 isoform for translation. As a control for overall translation in both strains, we evaluated the translation efficiency of a general housekeeping mRNA. C. elegans gpd-3 encodes the metabolic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which is present in all cell types. The translation of this general metabolic mRNA continued to be efficient (>87% of gpd-3 mRNA remained on polysomes) and was largely unaffected by the lack of IFE-1 (Fig. 6G).
POS-1 and PAL-1 are proteins involved in early cell fate polarity determination during early embryonic divisions (Hunter and Kenyon, 1996; Tabara et al., 1999). Like MEX-1, both pos-1 mRNA and the POS-1 protein associate with P granules. By contrast, neither the pal-1 mRNA nor PAL-1 protein are found in P granules. Although the localization of the two mRNA differs, their distribution in polysomes is similar (Fig. 6E,F). Interestingly, both pos-1 and pal-1 mRNAs are poorly translated even in wild-type worms. Less than 66% of mRNA for each was associated with any ribosomes. Both mRNAs are subject to translation repression by proteins that bind their 3′UTRs (Hunter and Kenyon, 1996; Tabara et al., 1999). We observed that, in the absence of IFE-1, the poor recruitment of these regulated mRNAs was further compromised (pos-1: fourfold, pal-1: fivefold). As with oma-1 and mex-1 mRNAs, more pos-1 and pal-1 mRNA shifts to non-translating fractions indicating that initiation has become less efficient. But given their already weak efficiency, it is unclear whether IFE-1 plays a substantial role in new synthesis of these proteins in early embryos.
We next examined the translation efficiency of the glp-1 mRNA (Fig. 6H). GLP-1 is a Notch-like receptor involved in early mitotic division during gametogenesis and again during embryonic divisions. Regulation of glp-1 mRNA is complex. glp-1 translation is repressed at various times during gametogenesis and embryogenesis by repressor proteins binding to the 3′ UTR (Kimble and Crittenden, 2007). Similarly to pos-1 and pal-1, we observed moderately reduced loading of glp-1 mRNA in polysomes (fourfold) with the removal of IFE-1, but this appeared to have two components. glp-1 mRNA disappeared from very heavy polysomes (>7-mers) but was maintained in medium polysomes (∼3- to 5-mers). A proportional increase in non-ribosome-bound glp-1 indicated that some of the mRNA requires IFE-1, while another population of glp-1 sustains translation without IFE-1. The independent populations may reflect mRNAs in mitotic versus meiotic germ cells and embryos in our whole-worm lysate, and suggest that IFE-1 recruits glp-1 mRNA in some cells but not others.
We attempted to correlate the accumulation of the protein products of these regulated mRNAs by immunostaining of dissected gonads and embryos. The steady-state level of these proteins may provide insight into the observed phenotype of the ife-1 strain and provide cell-specific information about the IFE-1-mediated translational activity of these mRNAs. In wild-type hermaphrodites, MEX-1 was first detectable in the pachytene stage of meiosis in the germ line of wild-type worms and accumulated progressively through late oogenesis to its highest levels in the oocyte adjacent to the spermatheca (–1 position) prior to fertilization (Fig. 7A). This is consistent with efficient translation of its mRNA. In ife-1 worms, MEX-1 immunostaining was observed beginning at pachytene, as in the wild type (Fig. 7B,C). However, unlike the wild-type, ife-1 mutant gonads showed no increase in MEX-1 accumulation as oocytes grew and matured, consistent with the sevenfold poorer loading of mex-1 mRNA into polysomes (Fig. 6D). This suggests that IFE-1 impacts the spatial and temporal expression of some important developmental proteins such as MEX-1.
Fig. 7.

Accumulation of MEX-1 protein in the absence of IFE-1. Dissected hermaphrodite gonads were fixed and immunostained for MEX-1 protein and costained with DAPI to visualize oocyte nuclei. (A) In the wild type, MEX-1 was first detected in the late pachytene stage of meiosis and progressively accumulates to the –1 oocyte. (B,C) In ife-1 gonads, the MEX-1 protein appeared in late pachytene, but then remained relatively constant (B) or decreased (C) in later oocytes. MEX-1 signal was normalized to exposure time using identical incident light and filter settings to allow direct comparisons.
Production of pro-apoptotic proteins during gametogenesis
The results above show that the lack of IFE-1 is detrimental to both oogenesis and spermatogenesis. To further evaluate the fate of ife-1 germ cells, we examined apoptotic events during oogenesis and spermatogenesis. Apoptosis is a natural occurrence in the C. elegans hermaphrodite gonad for the removal of immature oocytes that act as nurse cells that are destined to die (Gumienny et al., 1999). To assess the extent of germ cell apoptosis during oogenesis, we used a worm strain expressing CED-1::GFP. CED-1 is a scavenger receptor protein expressed in engulfing cells that decorates oocytes undergoing apoptosis (Zhou et al., 2001). Using this sensitive detection method, we previously demonstrated that loss of isoforms of another translation factor (eIF4G/IFG-1 p170) dramatically increases the number of apoptotic corpses (Contreras et al., 2008). In contrast to IFG-1 depletion, the loss of IFE-1 caused no increase in germ cell apoptosis in the hermaphrodite gonad. Approximately two to three apoptotic corpses per gonad arm were observed in ife-1 worms, similar to that observed in wild-type worms (Fig. 8A,B). We also used immunostaining to assay expression of another pro-apoptotic protein, CED-4 (Apaf-1 in mammals), in IFE-1-deficient oocytes. This protein, too, is induced by IFG-1 p170-depletion in oocytes undergoing apoptosis (Contreras et al., 2008). No CED-4 was detected in ife-1 hermaphrodite gonads (data not shown). Thus, there is no evidence that IFE-1 depletion induces apoptosis in oocytes.
Fig. 8.
Expression of pro-apoptotic proteins in hermaphrodite and male gonads. (A,B,E) Germ cell apoptotic corpses were detected by expression of the CED-1::GFP engulfment reporter. Only two germ cell corpses (*) were evident in the wild-type hermaphrodite gonad (A). The ife-1 gonad similarly showed two apoptotic corpses decorated with CED-1:GFP (B). (C,D) DAPI-staining verified the progression of oocyte nuclei through pachytene and diakinesis stages. As a positive control, depletion of IFG-1 p170 by ifg-1(RNAi) significantly increased germline apoptosis in the hermaphrodite gonad (E). Asterisks and brackets indicate the position of a cluster (six to eight) of cell corpses. (F-I) Expression of CED-4 was detected by immunostaining of dissected male testes. (F) Wild-type male gonad shows CED-4 accumulation in apoptosome-like structures only in late spermatocytes and spermatids. (G) Gonad from an ife-1 male showed CED-4 accumulation throughout spermatogenesis that eventually also resided in apoptosome-like structures, suggesting that CED-4 synthesis began earlier in spermatogenesis in the ife-1 male. (H,I) DAPI staining to show the nuclear morphology of early meiotic germ cells, primary spermatocytes (ps) and secondary spermatocytes and spermatids (sp).
The defect in spermatogenesis in the ife-1 male gonads is clearly more severe than that in oogenesis. The lack of IFE-1 in spermatogenesis results in the failure of cytokinesis and production of multinucleated spermatocytes. No evidence for apoptosis in the C. elegans male gonad has been reported. However, the native accumulation of pro-apoptotic proteins such as Apaf-1 and active caspase 3 has been observed during spermatogenesis in flies and mammals (Cagan, 2003). These destructive activities, normally associated with apoptosis, have been found to be required for the removal of cytoplasm in normal, maturing spermatids (Arama et al., 2003; Honarpour et al., 2000; Huh et al., 2004). We evaluated CED-4/Apaf-1expression in young adult C. elegans male testes. Wild-type male germ cells expressed CED-4 late in spermatogenesis during the transition from secondary spermatocyte to mature spermatids (Fig. 8F), consistent with observations in fly and mouse testes (Cagan, 2003). The gonad of ife-1 males grown at 20°C showed extensive CED-4 expression throughout the spermatocyte region (Fig. 8G). CED-4 appeared earlier in spermatogenesis and became more abundant than in wild-type gonads. An increasing number of ife-1 cells displayed apoptosome-like punctate structures associated with nuclei. Despite the difference in CED-4 expression during oogenesis and spermatogenesis, we have no evidence for increased apoptotic corpses in the gonads of either ife-1 males or hermaphrodites. CED-4 accumulation may not cause spermatocyte apoptosis, but may perform a physiological role that becomes enhanced by the lack of IFE-1.
Discussion
IFE-1 is required for efficient translation of maternal mRNAs in C. elegans
Many studies have elucidated the importance of translational control in development from the standpoint of mRNA repression, but few describe mechanisms for subsequent resumption of synthesis from these messages (Curtis et al., 1995; Goodwin and Evans, 1997; Macdonald and Smibert, 1996; Richter, 1991; Wickens et al., 1997). Previous work from our lab has shown that IFE-1 is a unique germline isoform of eIF4E that associates with P granules, which are known to contain numerous translationally repressed maternal mRNAs (Amiri et al., 2001; Pitt et al., 2000; Schisa et al., 2001). In this study we have addressed the specific requirement for IFE-1 in the recruitment of such regulated maternal mRNAs in the C. elegans germ line. Diminished translation efficiency has the potential to change the steady state levels of proteins involved in oocyte and embryo development. In wild-type worms, MEX-1 is first detected in pachytene stages and increases in abundance as oocytes grow, with the highest levels found just after fertilization (Guedes and Priess, 1997). A different pattern of MEX-1 protein accumulation was evident in ife-1 oocytes. The level of MEX-1 attained at pachytene did not continue to increase throughout oogenesis. Based on the poor translation of mex-1 mRNA in the absence of IFE-1, we predict that altered distribution and accumulation of the MEX-1 protein results from disruption of IFE-1-mediated translational control.
Analysis of polysome distribution clearly indicated that general protein synthetic activity was maintained in the absence of IFE-1. The presence of four other eIF4E isoforms, some of which are co-expressed in the germ line (IFE-3 and -5), insures that mRNA cap recognition is sufficient for most protein synthesis. However, the translational efficiencies of oocyte mRNAs known to undergo translational regulation and/or associate with P granules was dramatically reduced by loss of IFE-1. oma-1 and mex-1 mRNAs were translated fivefold and sevenfold less efficiently, respectively. The marked change in polysomal loading suggests that these mRNAs have a specific requirement for the IFE-1 isoform of eIF4E, which IFE-3 and IFE-5 do not sufficiently meet. The robust translation of these mRNAs in the wild type suggests that IFE-1 facilitates the rapid synthesis of proteins which have critical functions at the end of oogenesis. pos-1, pal-1 and glp-1 mRNAs translated four- to fivefold less efficiently in the absence of IFE-1. These mRNAs spend much of their time translationally repressed by 3′ UTR-binding proteins (Evans et al., 1994; Hunter and Kenyon, 1996; Ogura et al., 2003; Tabara et al., 1999). Translational control of glp-1 mRNA is particularly complex. glp-1 mRNA translation is active during germ cell proliferation, then repressed for cells to enter meiosis (Crittenden et al., 2003). Following fertilization, the message is again translated, but only in two blastomeres of the four-cell embryo (Evans et al., 1994). The best evidence that IFE-1 influences mRNA-specific translational control is its range of effects on mex-1 (sevenfold) to glp-1 (fourfold) to gpd-3 (twofold). We previously demonstrated just such discrimination by the muscle and neuronal eIF4E isoform (IFE-4), which recruits a small subset of mRNAs whose translational efficiency similarly decreased in ife-4(ok320) worms (Dinkova et al., 2005). Data from our current study show that IFE-1 is preferentially used for a different set of mRNAs. In conjunction with the observed ife-1 sperm and oocyte defects, these data indicate that IFE-1 plays a specific role in mRNA discrimination in late oocyte and sperm development.
Absence of IFE-1 results in abnormal gametogenesis
There is growing evidence that translation factors play important regulatory roles in germ cell development (Baker and Fuller, 2007; Contreras et al., 2008; Franklin-Dumont et al., 2007; Mendez and Richter, 2001; Yu et al., 2006). We recently demonstrated that two isoforms of eIF4G (IFG-1 p130 and p170) are required to maintain a balance between completion of meiotic development and selective removal during C. elegans oogenesis (Contreras et al., 2008). Oocytes depleted of the large IFG-1 isoform underwent germline apoptosis in far greater numbers. These oocytes induced ectopic expression of the Apaf-1 homolog, CED-4. Here we show that loss of IFE-1, a binding partner to IFG-1 p170 in the cap-binding complex, results in a defect in spermatogenesis and a modest decrease in oocyte production. However, IFE-1-deficiency did not induce germline apoptosis or ectopic expression of CED-4 during oogenesis. Surprisingly, we observed an increase in CED-4 synthesis during late spermatogenesis in wild-type males (Fig. 8). In ife-1 male testes, CED-4 was expressed earlier, became more abundant, and was associated with more nuclei throughout spermatogenesis. Coincidentally, IFE-1 deficiency also inhibited maturation of secondary spermatocytes to spermatids, but we have no evidence to suggest the arrested spermatocytes enter apoptosis. In fact, programmed cell death has not been observed in the C. elegans male germ line (Baum et al., 2005). It is therefore possible that CED-4 expression in male gonads is indicative of a natural maturation step in spermatogenesis such as the loss of cytoplasmic contents. Precisely this function has been ascribed to CED-4/Apaf-1 in both mouse and Drosophila spermatogenesis (Huh et al., 2004). Loss of Apaf-1 expression leads to a degeneration of spermatogonia and the absence of mature sperm (Honarpour et al., 2000). In these species a natural process of bona-fide spermatocyte apoptosis has also been observed (Cagan, 2003). We currently have no evidence that CED-4 accumulation causes apoptosis in C. elegans spermatogenesis. However, the ectopic CED-4 expression in ife-1 testes correlates with the poor viability of ife-1 sperm and may contribute to aberrant development.
These studies underscore the importance of cell-specific translational regulation for sperm development. A defect in spermatogenesis has also been observed with depletion of CBP-1, a protein involved in translational control of stored mRNAs by poly(A) elongation (Luitjens et al., 2000). Here we show that loss of IFE-1-mediated translation disrupted cytokinetic events that led to multinucleated spermatocytes. Germ cell nuclei normally replicate, then enter meiosis I and II during which the segregate to secondary spermatocytes (L'Hernault, 2006). A final round of nuclear division is followed by segregation, this time to small spermatids, leaving behind nearly all cytoplasm in non-nucleated residual bodies. Only factors essential to fertilization are sequestered in the spermatid, but ribosomes, mRNAs and the cytoplasmic translation factor IFE-1 remain in the residual body. It is probable that IFE-1 supports protein synthetic events before budding that are important for nuclear segregation and bud formation. Interestingly, a deficiency in spe-4, spe-5 or spe-39 also causes the accumulation of multinucleated secondary spermatocytes (L'Hernault, 2006). We might then speculate that their mRNAs are targets for IFE-1 binding. Disregulation of translation causes meiotic nuclear divisions to become disconnected from cell divisions, which leads to the arrest of spermatogenesis and male infertility.
Both sperm and oocyte production are dependent on IFE-1. The more severe fertility defect (sperm development) is exacerbated at slightly elevated temperature (25°C), whereas the more moderate (oocyte development) is temperature insensitive. A functional role in oogenesis was expected because IFE-1 is expressed throughout all stages of germ line development. However, no gross defects were observed in oocyte cell and nuclear morphology (maturation) through late oogenesis (diakinesis) in ife-1 hermaphrodites. The lack of oocyte accumulation (stacking) in the absence of sperm, the decreased progeny when mated to normal sperm, and the increased number of unfertilized oocytes when mated to normal sperm all indicate that oocyte production and competence are compromised. Importantly, these defects were not observed in feminized hermaphrodite gonads (fem-2), confirming that ife-1 oocytes were not simply suffering secondary effects from the lack of a sperm-mediated maturation signal. The most plausible explanation is that IFE-1-deficient worms produce oocytes more slowly and of those produced, many cannot be fertilized.
Up to this point, the understanding of translational control mechanisms during C. elegans gametogenesis and embryogenesis has been limited to regulation by 3′ untranslated sequences in controlled mRNAs. Here we demonstrate an added component of translational regulation. C. elegans mRNA cap-binding protein IFE-1 promotes the efficient translation of selected germ cell mRNAs. IFE-1 is necessary for efficient recruitment of oma-1 mRNA to ribosomes. Previous work has established that in the absence OMA-1 and OMA-2, oocytes fail to mature and ovulate, leading to a backup of grown oocytes that cannot complete nuclear envelope breakdown (Detwiler et al., 2001). Although the oma-1/oma-2 phenotype is similar but not identical to that of ife-1, it is clear that the lack of IFE-1 has broader consequence for germ cell maturation. The defect in cytokinesis during spermatogenesis indicates that IFE-1 is also essential for male germ cell mRNA translation. Gamete development is thus dependent on the collective activities of IFE-1 in cells that synergize for fertilization. Our data indicate that IFE-1 is at the center of a highly regulated and parentally derived mRNA network. We propose that this network is governed by the translation initiation mechanism, in which an eIF4E isoform selectively recruits mRNAs to coordinate specific patterns of expression during gametogenesis.
Materials and Methods
Strains
C. elegans wild-type strain was N2 var. Bristol. The strain MD701, {bcIs39 V [Plim-7ced-1::gfp and lin15(+)]} was obtained from Barbara Conradt and used to assay hermaphrodite germ cell apoptosis (Zhou et al., 2001). Strain KX56 {ife-1(bn127) III; bcIs39 V[Plim-7 ced-1::gfp and Lin-15(+)]} was created by crossing strain SS712 [ife-1(bn127)] and strain MD701. F1 offspring were self-mated and F2s were screened to be homozygous for the ife-1(bn127) allele by PCR. Strain DH245 fem-2(b245) III was obtained from the CGC and maintained as described previously (Kimble et al., 1984). Worms were cultured on NGM plates with E. coli strain OP50 or strain HT115 and grown at 20°C unless otherwise described (Brenner, 1974). For polysome analysis, worms were grown at 20°C on chicken egg yolk plates containing OP50 and maintained well fed (Portman, 2006). Mixed-stage hermaphrodites were washed from the plates with M9 buffer, floated on 35% sucrose to remove debris, and frozen by dropping into liquid nitrogen (Portman, 2006).
Isolation of the ife-1(bn127) deletion mutant
Worm libraries mutagenized with trimethylpsoralen and UV irradiation were screened for a deletion mutation in ife-1 (F53A2.6) according to the protocols of M. Koelle et al. (http://info.med.yale.edu/mbb/koelle/protocols/protocol_Gene_knockouts.html) and the C. elegans Knock-Out Consortium (http://celeganskoconsortium.omrf.org/). PCR primers used in the screen were as follows. External primer set: forward 5′-GCACAAATGTTTGGAGGACC-3′ and reverse 5′-AACGACCATGGACTTTGACC-3′. Poison primer: 5′-ACGGAAAACGTTGTAATCGC-3′. Internal primer set: forward 5′-CCAACCGACGATGACTACG-3′ and reverse 5′-TTTGCAGTGTATTTGTGTGG-3′. The latter were used to follow the deletion allele during ten rounds of outcrossing to the wild-type strain. Breakpoints of the deletion were determined by subcloning and sequencing of the genomic deletion PCR product. Subsequently, single worm genomic PCR was used to follow the deletion employing the internal forward primer above and a new reverse primer (5′-TTGGGCCTGGAAACCACAAG-3′) near to the breakpoint. These allowed efficient amplification of both wild-type and deleted alleles during subsequent genetic manipulations, including screening for homozygosity. A stable heterozygous strain was also produced by balancing the ife-1(bn127) allele over mT1 (Edgley et al., 2006).
Characterizing the ife-1(bn127) phenotype
For fertility and brood size assays, three L4 hermaphrodite worms (wt and ife-1) were placed on each of three E. coli OP50-seeded NGM plates. Worms were transferred to new plates every 24 hours for 72 hours. Offspring number and growth were evaluated every 24 hours for 96 hours. The temperature-shift experiment was performed by synchronizing wild-type and ife-1 worms at the L1 stage by incubating isolated embryos in M9 for 6-12 hours at 22°C. At the L1 stage, four to ten worms were transferred to 18 seeded NGM plates and placed at either 20°C or 25°C. Every 12 hours, one plate was shifted from 20°C to 25°C or 25°C to 20°C and developmental stage scored. The brood size of the shifted hermaphrodites was scored at 120 hours post L1 stage. Mating experiments were performed by incubating three males with one L4 hermaphrodite for 48 hours and transferring to new plates over an additional 48 hours at 20°C or 25°C. Males were replenished if the number of surviving males dropped below three. Brood size and hermaphrodite to male ratios were calculated after 96 hours of possible mating.
ifg-1 RNA interference
L2 worms were fed E. coli strain HT115 expressing dsRNA from the plasmid pT72ifgN2 containing 398 bp (nt 24–479) of the ifg-1 cDNA as described previously (Contreras et al., 2008). Offspring were cultured for 96 hours at 25°C and imaged by fluorescence microscopy as described below.
Microscopy and immunostaining
Male and hermaphrodite gonads were prepared for live imaging as follows. Worm heads were removed by cutting just below the pharynx with 30 gauge syringe needles, and where possible, the gonad was teased out of the carcass. Either egg buffer (60 mM NaCl, 32 mM KCl, 3 mM Na2HPO4, 2 mM CaCl2, 5 mM Hepes pH 7.2, 0.2% glucose, 4 mM levamisole) (Edgar, 1995), sperm buffer (45 mM NaCl, 25 mM KCl, 1 mM MgCl2, 0.5 mM CaCl2, 50 mM Hepes pH 7.8, 10 mM dextrose) (Zannoni et al., 2003), or M9 was supplemented with 0.05-1.0 μg/ml Hoechst dye. Gonads were overlaid with a coverslip and immediately imaged on an Axiovert 200M inverted microscope (Carl Zeiss) using DIC, FITC, DAPI and Texas Red filter cubes and analyzers. Image analysis was with Axiovision 4.3 software (Carl Zeiss). Immunostaining methods were those described by Contreras et al. (Contreras et al., 2008) with the following exceptions. The anti-MEX-1(cN-18) goat polyclonal antibody (Santa Cruz Biotechnology) was diluted 1:50. CED-4 localization was determined using the anti-CED-4 (cN-21) goat polyclonal IgG (Santa Cruz Biotechnology) diluted 1:50. Gonads and carcasses were transferred to a poly-L-lysine treated slide, overlaid with a coverslip, and frozen on dry ice for 10 minutes. After removal of the cover slip, samples were fixed by submerging in methanol at –20°C for 10 minutes and acetone at –20°C for 10 minutes. Primary antibody was applied overnight at 4°C. Donkey anti-goat Alexa Fluor 488 (Molecular Probes) at 1:1000 was used as the secondary antibody for MEX-1 and CED-4 staining. Samples were washed in Tris-buffered saline with Tween 20 (TST) containing DAPI and mounted in a 90% glycerol/DABCO (Arcos) medium.
Analysis of polysomes by sucrose gradient fractionation
Lysis and gradient buffers, gradient setup, centrifugation and fractionation were conducted as described by Dinkova et al. (Dinkova et al., 2005) with the following exceptions. Frozen wild-type and ife-1 worms of mixed stages were ground under liquid nitrogen in a mortar and pestle in buffer C. RNA was isolated from sucrose gradient fractions in three volumes TRIzol (Invitrogen) following manufacturer's instructions.
qRT-PCR for maternal mRNAs
Reverse transcripts of isolated RNA was made using iScript cDNA Synthesis Kit (BioRad) from 1 μg of RNA from each fraction. Real-time PCR was performed using an iCycler iQ Real-time PCR machine with iQ SYBR Green supermix (BioRad) in triplicate according to the manufacturer's protocol. Primer sequences included: mex-1 (forward 5′-AACAAACGTTTCTAGGCTTC-3′, reverse 5′-CCAACATTAGGCTTATCCATTT-3′); oma-1 (forward 5′-CAAACTGCTAACTTGATTGCT-3′, reverse 5′-GGATCAAACTGTTGAAATGG-3′); glp-1 (forward 5′-TGGACTTGTGAAGTCTGATG-3′, reverse 5′-ATCGATTTCGTTTCTCTTTG-3′); gpd-3 (forward 5′-GATCTCAGCTGGGTCTCTT-3′, reverse 5′-TCCAGTACGATTCCACTCAC-3′); pos-1 (forward 5′-AAATTGTCGATGGGAATG-3′, reverse 5′-ACGAAGAGTGAATGATTGTG-3′); pal-1 (forward 5′-AAGGATCGAAGATCAAGCA-3′, reverse 5′-AATTCTTTTTCCAGTTCAAGG-3′). It should be noted that real-time quantification of each mRNA (Fig. 5C-H) was normalized to total RNA content (mostly rRNA), which accounts for the slight difference in polysome content and therefore accurately reflects the translational efficiency of each mRNA.
The authors are greatly indebted to Diane Shakes (College of William and Mary, Williamsburg, VA) for advice on microscopy and characterization of spermatogenesis. Jennifer Schisa (University of Central Michigan, Mount Pleasant, MI) and Jennifer Tenlan (University of North Carolina, Chapel Hill, NC) gave helpful advice on immunostaining, and Enhui Hao provided expert technical assistance. This work was supported by grants IRG 5-89812 from the American Cancer Society, RDA Award from ECU Division of Research and Graduate Studies, and MCB-0321017 from the NSF to B.D.K., and GM34059 from the NIH to S.S. Deposited in PMC for release after 12 months.
References
- Amiri, A., Keiper, B. D., Kawasaki, I., Fan, Y., Kohara, Y., Rhoads, R. E. and Strome, S. (2001). An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128, 3899-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama, E., Agapite, J. and Steller, H. (2003). Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4, 687-697. [DOI] [PubMed] [Google Scholar]
- Baker, C. C. and Fuller, M. T. (2007). Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 134, 2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, D. C., Cao, Q. and Richter, J. D. (2005). Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell. Biol. 25, 7605-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, J. S., St George, J. P. and McCall, K. (2005). Programmed cell death in the germline. Semin. Cell Dev. Biol. 16, 245-259. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan, R. L. (2003). Spermatogenesis: borrowing the apoptotic machinery. Curr. Biol. 13, R600-R602. [DOI] [PubMed] [Google Scholar]
- Contreras, V., Richardson, M. A., Hao, E. and Keiper, B. D. (2008). Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ. 15, 1232-1242. [DOI] [PubMed] [Google Scholar]
- Crittenden, S. L., Eckmann, C. R., Wang, L., Bernstein, D. S., Wickens, M. and Kimble, J. (2003). Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, D., Lehmann, R. and Zamore, P. D. (1995). Translational regulation in development. Cell 81, 171-178. [DOI] [PubMed] [Google Scholar]
- Dahanukar, A., Walker, J. A. and Wharton, R. P. (1999). Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, 209-218. [DOI] [PubMed] [Google Scholar]
- Detwiler, M. R., Reuben, M., Li, X., Rogers, E. and Lin, R. (2001). Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1, 187-199. [DOI] [PubMed] [Google Scholar]
- Dinkova, T. D., Keiper, B. D., Korneeva, N. L., Aamodt, E. J. and Rhoads, R. E. (2005). Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 25, 100-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, L. G. (1995). Blastomere culture and analysis. Methods Cell Biol. 48, 303-321. [DOI] [PubMed] [Google Scholar]
- Edgley, M. K., Baillie, D. L., Riddle, D. L. and Rose, A. M. (2006). Genetic balancers. In WormBook (ed. The C. elegans Research Community): WormBook. [DOI] [PMC free article] [PubMed]
- Evans, T. C., Crittenden, S. L., Kodoyianni, V. and Kimble, J. (1994). Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77, 183-194. [DOI] [PubMed] [Google Scholar]
- Evsikov, A. V., Graber, J. H., Brockman, J. M., Hampl, A., Holbrook, A. E., Singh, P., Eppig, J. J., Solter, D. and Knowles, B. B. (2006). Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 20, 2713-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Dumont, T. M., Chatterjee, C., Wasserman, S. A. and Dinardo, S. (2007). A novel eIF4G homolog, Off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development 134, 2851-2861. [DOI] [PubMed] [Google Scholar]
- Gingras, A.-C., Raught, B. and Sonenberg, N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913-963. [DOI] [PubMed] [Google Scholar]
- Goodwin, E. B. and Evans, T. C. (1997). Translational control of development in C. elegans. Semin. Cell. Dev. Biol. 8, 551-559. [DOI] [PubMed] [Google Scholar]
- Guedes, S. and Priess, J. R. (1997). The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development 124, 731-739. [DOI] [PubMed] [Google Scholar]
- Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. and Hengartner, M. O. (1999). Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011-1022. [DOI] [PubMed] [Google Scholar]
- Honarpour, N., Du, C., Richardson, J. A., Hammer, R. E., Wang, X. and Herz, J. (2000). Adult Apaf-1-deficient mice exhibit male infertility. Dev. Biol. 218, 248-258. [DOI] [PubMed] [Google Scholar]
- Huh, J. R., Vernooy, S. Y., Yu, H., Yan, N., Shi, Y., Guo, M. and Hay, B. A. (2004). Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2, E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C. P. and Kenyon, C. (1996). Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87, 217-226. [DOI] [PubMed] [Google Scholar]
- Jud, M., Razelun, J., Bickel, J., Czerwinski, M. and Schisa, J. A. (2007). Conservation of large foci formation in arrested oocytes of Caenorhabditis nematodes. Dev. Genes Evol. 217, 221-226. [DOI] [PubMed] [Google Scholar]
- Jud, M. C., Czerwinski, M. J., Wood, M. P., Young, R. A., Gallo, C. M., Bickel, J. S., Petty, E. L., Mason, J. M., Little, B. A., Padilla, P. A. et al. (2008). Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 318, 38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper, B. D., Lamphear, B. J., Deshpande, A. M., Jankowska-Anyszka, M., Aamodt, E. J., Blumenthal, T. and Rhoads, R. E. (2000). Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275, 10590-10596. [DOI] [PubMed] [Google Scholar]
- Kimble, J. and Crittenden, S. L. (2007). Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23, 405-433. [DOI] [PubMed] [Google Scholar]
- Kimble, J., Edgar, L. and Hirsh, D. (1984). Specification of male development in Caenorhabditis elegans: the fem genes. Dev. Biol. 105, 234-239. [DOI] [PubMed] [Google Scholar]
- L'Hernault, S. W. (2006). Spermatogenesis. In WormBook (ed. The C. elegans Research Community): WormBook. [DOI] [PMC free article] [PubMed]
- Luitjens, C., Gallegos, M., Kraemer, B., Kimble, J. and Wickens, M. (2000). CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P. M. and Smibert, C. A. (1996). Translational regulation of maternal mRNAs. Curr. Opin. Genet. Dev. 6, 403-407. [DOI] [PubMed] [Google Scholar]
- Mendez, R. and Richter, J. D. (2001). Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell. Biol. 2, 521-529. [DOI] [PubMed] [Google Scholar]
- Miller, M. A., Nguyen, V. Q., Lee, M. H., Kosinski, M., Schedl, T., Caprioli, R. M. and Greenstein, D. (2001). A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144-2147. [DOI] [PubMed] [Google Scholar]
- Minshall, N., Reiter, M. H., Weil, D. and Standart, N. (2007). CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 282, 37389-37401. [DOI] [PubMed] [Google Scholar]
- Morales, J., Mulner-Lorillon, O., Cosson, B., Morin, E., Belle, R., Bradham, C. A., Beane, W. S. and Cormier, P. (2006). Translational control genes in the sea urchin genome. Dev. Biol. 300, 293-307. [DOI] [PubMed] [Google Scholar]
- Ogura, K., Kishimoto, N., Mitani, S., Gengyo-Ando, K. and Kohara, Y. (2003). Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 130, 2495-2503. [DOI] [PubMed] [Google Scholar]
- Pitt, J. N., Schisa, J. A. and Priess, J. R. (2000). P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315-333. [DOI] [PubMed] [Google Scholar]
- Portman, D. S. (2006). Profiling C. elegans gene expression with DNA microarrays. In WormBook (ed. The C. elegans Research Community): WormBook. [DOI] [PMC free article] [PubMed]
- Proud, C. G. (2006). Regulation of protein synthesis by insulin. Biochem. Soc. Trans. 34, 213-216. [DOI] [PubMed] [Google Scholar]
- Richter, J. D. (1991). Translational control during early development. BioEssays 13, 179-183. [DOI] [PubMed] [Google Scholar]
- Richter, J. D. (2007). CPEB: a life in translation. Trends Biochem. Sci. 32, 279-285. [DOI] [PubMed] [Google Scholar]
- Robalino, J., Joshi, B., Fahrenkrug, S. C. and Jagus, R. (2004). Two zebrafish eIF4E family members are differentially expressed and functionally divergent. J. Biol. Chem. 279, 10532-10541. [DOI] [PubMed] [Google Scholar]
- Rodriguez, C. M., Freire, M. A., Camilleri, C. and Robaglia, C. (1998). The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13, 465-473. [DOI] [PubMed] [Google Scholar]
- Schisa, J. A., Pitt, J. N. and Priess, J. R. (2001). Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128, 1287-1298. [DOI] [PubMed] [Google Scholar]
- Seydoux, G. and Strome, S. (1999). Launching the germline in Caenorhabditis elegans: regulation of gene expression in early germ cells. Development 126, 3275-3283. [DOI] [PubMed] [Google Scholar]
- Shimada, M., Yokosawa, H. and Kawahara, H. (2006). OMA-1 is a P granules-associated protein that is required for germline specification in Caenorhabditis elegans embryos. Genes Cells 11, 383-396. [DOI] [PubMed] [Google Scholar]
- Syntichaki, P., Troulinaki, K. and Tavernarakis, N. (2007). eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445, 922-926. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Hill, R. J., Mello, C. C., Priess, J. R. and Kohara, Y. (1999). pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1-11. [DOI] [PubMed] [Google Scholar]
- Tadros, W. and Lipshitz, H. D. (2005). Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232, 593-608. [DOI] [PubMed] [Google Scholar]
- Wickens, M., Anderson, P. and Jackson, R. J. (1997). Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 7, 220-232. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J. E. and Smibert, C. A. (2005). Mechanisms of translational regulation in Drosophila. Biol. Cell 97, 235-252. [DOI] [PubMed] [Google Scholar]
- Yamamoto, I., Kosinski, M. E. and Greenstein, D. (2006). Start me up: cell signaling and the journey from oocyte to embryo in C. elegans. Dev. Dyn. 235, 571-585. [DOI] [PubMed] [Google Scholar]
- Yu, X., Vought, V. E., Conradt, B. and Maine, E. M. (2006). Eukaryotic translation initiation factor 5B activity regulates larval growth rate and germline development in Caenorhabditis elegans. Genesis 44, 412-418. [DOI] [PubMed] [Google Scholar]
- Zannoni, S., L'Hernault, S. W. and Singson, A. W. (2003). Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev. Biol. 3, 10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Hartwieg, E. and Horvitz, H. R. (2001). CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43-56. [DOI] [PubMed] [Google Scholar]