Abstract
This paper estimates the accuracy of hippocampal subfield alignment via shape-based normalization. Evaluation takes place in post-mortem MRI dataset acquired at 9.4 Tesla with many averages and approximately 0.01 mm3 voxel resolution. Continuous medial representations (cm-reps) are used to establish geometrical correspondences between hippocampal formations in different images; the extent to which these correspondences match up subfields is evaluated and compared to normalization driven by image forces. Shape-based normalization is shown to perform only slightly worse than image-based normalization; this is encouraging because the former is more applicable to in vivo MRI, which typically lacks features that distinguish hippocampal subfields.
1 Introduction
The hippocampus is arguably one of the most fascinating structures in the brain, and much remains unknown about its precise role in the memory system [13]. Neurons in the hippocampus are uniquely susceptible to neurodegenerative disorders, and, in particular, Alzheimer’s disease (AD) [5]; imaging-based hippocampal volumetry is a reliable early predictor of AD onset [9, 15]. The hippocampus is not a uniform structure: it is formed by two interlocking folded layers of neurons, the cornu Ammonis (CA) and the dentate gyrus (DG), which are further divided into subfields and subserve different functional roles. Non-uniform neuron loss across the hippocampal subfields has been reported in neurodegenerative disorders [3, 4, 18], as has a non-uniform rate of neuroplasticity [8, 10, 14].
Recently, these has been much interest in measuring subfield-level changes associated with neurodegenerative diseases using MRI. However, it is very difficult to identify the boundaries of hippocampal layers in clinical MRI modalities, since the dark band separating these layers, formed by the vestigial hippocampal sulcus (VHS) and strata radiata (SR), is thinner than the typical voxel resolution. In the absence of MRI acquisitions that specifically target the hippocampus (e.g., [22]), the labeling of subfields relies primarily on shape cues, i.e., on the position of the subfields relative to the overall hippocampus shape. The primary instance of this approach is the Washington University hippocampus atlas [6,16], in which the hippocampus template was manually labeled with the help of an anatomical reference [7].
Our group is developing an atlas of the human hippocampus using high-field postmortem MRI [19]. Imaging at 0.01 mm3 voxel resolution, we are able to separate hippocampal layers and label the major subfields of the hippocampus. However, one of the main challenges associated with this atlas is how to effectively apply it to in vivo studies in which there are few reliable intensity features within the hippocampus. A natural approach is to use shape cues to map subfields from the atlas to manually or automatically generated segmentations of the hippocampus from in vivo data; as is typically done for the Washington University atlas [12, 16]. The goal of this paper is to evaluate how accurately shape-based normalization aligns hippocampal subfields between subjects.
A full evaluation of this kind would require a dataset where in vivo and postmortem images for the same subjects would be available. In absence of such data, we restrict our attention to the postmortem dataset, making a somewhat strong assumption that the segmentation of the overall hippocampus in the post-mortem dataset would be sufficiently similar to the segmentation in in vivo data. Clearly, both manual and automatic hippocampus segmentation in in vivo MRI are quite challenging problems in themselves, and whatever subfield alignment errors we report in this paper would be exacerbated by errors in the segmentation of the overall hippocampus shape. Nevertheless, given the recent interest in subfield-level inference in in vivo MRI, we believe that the results reported in the postmortem dataset will be valuable to the field.
The evaluation experiment compares how well shape-based normalization, which does not use intensity cues, performs vis-à-vis diffeomorphic image registration driven by intensity. The latter is treated as a “gold standard”, since it is using all the cues that are available in the postmortem dataset, and which would not be available in typical in vivo data.
2 Materials and Methods
2.1 Specimens and Imaging
Formalin-fixed brain specimens (≥21 days) from three autopsy cases with no abnormal neuropathological findings were studied. Hemispheres were separated from the cerebellum and brain stem, and samples containing the intact hippocampus and not larger than 70mm in diameter were extracted from each hemisphere by making two incisions: the first, orthogonal to the midsagittal plane and parallel to the main axis of the hippocampus, passing through the corpus callosum; the second, parallel to the midsagittal plane, removing the lateral-most third of the sample. In our observation, the deformation caused to the hippocampus while cutting the brain was minimal, as the cuts were made a good distance away from the hippocampus.
Images of five hippocampus samples (3 right, 2 left) were acquired on a 9.4 Tesla Varian 31cm horizontal bore scanner (Varian Inc, Palo Alto, CA) using a 70mm ID TEM transmit/receive volume coil (Insight Neuroimaging Systems, Worchester, MA) and a stock multi-slice spin echo sequence. MRI parameters varied slightly across the samples; typical values are TR/TE=5000/26ms; voxel size: 0:2mm × 0:3mm × 0:2mm; matrix: 300 × 280; 130 slices; 45 averages; acquisition time around 15 hours.
2.2 Manual Segmentation
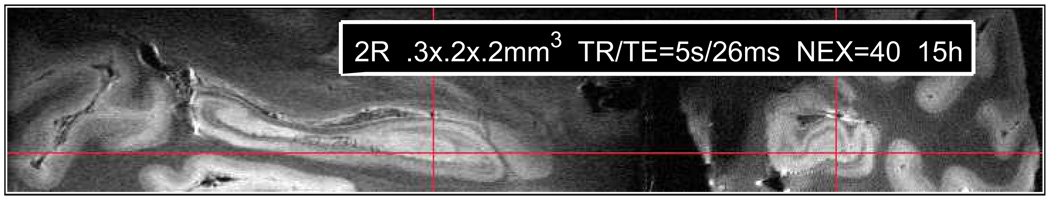
The layered structure of the hippocampus is clearly visible in our images (Fig. 1). One of the authors ([omitted for blind review]) performed the segmentation manually in ITK-SNAP (itksnap.org), using the Duvernoy atlas [7] as the main reference, and taking as much as 80 hours to label each hippocampus. The segmentation included the following labels: CA1, CA2+CA3, DG and VHS+SR. The CA4 subfield was also segmented but, following [1], we treat is a as component of the DG.
Fig. 1.
Sagittal and coronal views of one of the postmortem hippocampus MRIs.
2.3 Shape-Based Normalization
Shape-based normalization uses the continuous medial representation (cm-rep) method [20,21]. A deformable cm-rep model is fitted to each binary segmentation of the hippocampus, so as to maximize the overlap between the model and the binary image, while minimizing distortion across models. As the cm-rep model deforms, the method preserves the branching topology of the its skeleton, making it possible to leverage medial geometry for normalization and shape analysis.
A cm-rep model is defined as follows. A triangle mesh is used to represent the skeleton of the model. Each mesh node consists of a point m ∈ ℝ3 and a positive radius value R. The mesh is constructed by recursive subdivision of a coarse mesh using Loop subdivision rules [11]. The boundary of the model consists of two surface patches b+ and b−, one on each side of the skeleton. These boundary patches are derived from the skeleton mesh using the inverse skeletonization formula:
| (1) |
where ∇mR denotes the Riemannian (surface) gradient of R on m and 𝒩m denotes the unit normal to m. These first-order properties are computed using finite difference approximations. Constraints are in place to ensure that the two boundary patches share a common edge and do not self-intersect, thus forming a boundary of a simple region in ℝ3 [21].
The interior of the cm-rep model is spanned by line segments, called spokes, which extend from points mi on the skeleton to the corresponding points and on the model’s boundary and which are orthogonal to the boundary. As argued in [20] and illustrated in Fig. 2, the spokes form a shape-based coordinate system on the interior of the cm-rep model.
Fig. 2.
A conceptual 2D illustration of shape-based normalization via the cm-rep co-ordinate system. The central curve is the skeleton m; the radial lines from the skeleton to the boundary are the spokes; the shape-based mapping between the two models is given by the locations of corresponding grid vertices.
In order to establish correspondences between cm-rep models fitted to different hippocampi, we minimize the variance in Euclidean distance between corresponding pairs of points in the shape-based coordinate system. This is done in an iterative process, to allow each model to be fitted in parallel. The correspondence energy function for model s at iteration t has the form
| (2) |
where N(i) denotes the grid points adjacent to grid point i, is the Euclidean distance between grid points i and j in model s at iteration t, and is the average of this distance across all models at the previous iteration.
2.4 Deformable Registration
We use a diffeomorphic fluid warping method known as Symmetric Normalization (SyN) [2] to perform pairwise registration between hippocampus images. Given an image match metric Π (in our case, normalized cross-correlation), registration of images I and J seeks a pair of diffeomorphic maps ϕ1; ϕ2 that minimize
| (3) |
Symmetric diffeomorphic maps from I to J and from J to I are given, respectively, by . Binary masks of the hippocampus are incorporated into the registration because the goal of registration in this experiment is to establish an upper bound on the performance of shape-based normalization.
2.5 Evaluation of Subfield Alignment
Quantitative evaluation of the alignment of hippocampal subfields via shape-based and image-based normalization is performed in the leave-one-out frame-work. For a given hippocampus image Is, a deformation field from each other hippocampus image to Is is constructed. In the case of image registration, this field is produced by the registration algorithm; in the case of cm-rep normalization, the field is interpolated from the correspondences assigned by the coordinate system. The manually generated subfields labels for every image except Is are deformed into the space of Is using these warp fields, and a consensus labeling of Is is produced by applying the STAPLE algorithm [17] to the warped labels. This consensus segmentation of Is is compared to the manual segmentation of Is in terms of Dice overlap and root mean square boundary distance.
3 Results
Fig. 3 illustrates cm-rep models fitted to binary hippocampus segmentations. A checkerboard map is used to illustrate the correspondence between different models. The cm-rep models do not fit the binary segmentations with complete accuracy. This is caused by the fact that the cm-rep model is constrained to have a skeleton of predefined branching topology, as well as by the contribution of the correspondence term. The average Dice overlap between the fitted models and the segmentations is 0.93. The greatest mismatch occurs in the head of the hippocampus, which has complex folding features. These features are not typically seen in hippocampus segmentations in in vivo data, for which the accuracy of cm-rep fitting is somewhat higher (0.95) [20].
Fig. 3.
Examples of cm-rep model fitting. The models’ skeletons are shown in the top row, with color representing the radius field R (red: thicker, blue: thinner). The boundary is plotted in the middle row, with the checkerboard pattern showing the correspondence between models. The last row superimposes in semi-transparent blue the boundaries binary segmentations to which these models were fitted.
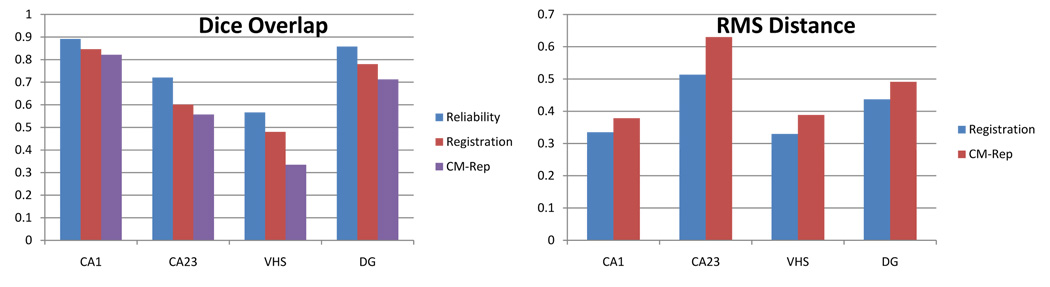
Fig. 4 compares the labeling of hippocampal subfields in some of the cross-validation experiments. For each experiment, the labeling achieved by image-based registration, cm-rep normalization and the target manual segmentation are shown. The average over all five samples of the overlap between the manual segmentation of the subfields in each hippocampus and the segmentation generated by cm-rep normalization is plotted in Fig. 5 for each subfield, compared to the same overlap computed for image-based normalization. These overlaps are also compared to the average overlap between repeated manual segmentations by the same rater (these reliability measurements were computed only on a sub-set of slices). A similar comparison of root mean square distance is also shown in Fig. 5.
Fig. 4.
Comparison of subfield labeling in two leave-one-out experiments (top row shows sample 3L in sagittal plane; second row shows sample 2R in coronal plane). Shown are the manual segmentation of the subfields, the labeling using shape-based normalization (via cm-reps) and the labeling using image registration.
Fig. 5.
Average Dice overlap (left) and average root mean square boundary distance (right) between cm-rep based subfield labeling and the manual segmentation, compared to registration-based labeling. The overlap plot also includes overlap between repeated manual segmentations by the same rater as a reference.
4 Discussion and Conclusions
The results of our evaluation experiment are encouraging but also open to interpretation. The fact that shape-based normalization, which does not use any intensity information, performs only slightly worse than image registration for subfield labeling is significant. It suggests that it may be possible to use subfield maps in the analysis of low resolution in vivo datasets where intensity information inside of the hippocampus is not informative for subfield differentiation. On the other hand, it has to be recognized that the overall error associated with subfield labeling, whether manual, image-based or shape-based, is substantial, even in this high-resolution postmortem dataset. It is likely that additional improvements to the segmentation protocol and the computational techniques used in this paper would result in greater consistency. Nevertheless, subfield mapping in in vivo datasets, which is exacerbated by the difficulty of segmenting the hippocampus itself, will likely be of limited accuracy for years to come.
Limitations of this paper include the fact that evaluation was performed on postmortem data, thus the results we report may not be representative of performance in in vivo datasets. There may be nonuniform shape changes introduced across the hippocampus by the preparation of brain samples after autopsy. Also, the current postmortem dataset is small and does not include pathology. We are working to address these limitations, by acquiring additional data, as well as by incorporating more advanced hippocampus-specific in vivo imaging techniques (e.g. [22]) into our analysis. Despite the limitations, we believe that the proposed technique, as well as the postmortem hippocampus atlas on which is is based, will be of value to morphometric studies of aging and dementia. Perhaps the ‘safest’ way to apply the technique is in a longitudinal study, where fluid registration can be used to derive a subject-specific atlas image, and shape-based subfield labeling would be applied to the atlas.
Acknowledgement
This project is supported by NIH grants AG027785, NS061111, MH068066 and NS045839, and the Pilot Project Award from the Penn Comprehensive Neuro-science Center.
References
- 1.Amaral David G, Scharfman Helen E, Lavenex Pierre. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies) Prog Brain Res. 2007;163:3–22. doi: 10.1016/S0079-6123(07)63001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2007 Jun; doi: 10.1016/j.media.2007.06.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobinski M, Wegiel J, Tarnawski M, Bobinski M, Reisberg B, de Leon MJ, Miller DC, Wisniewski HM. Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of alzheimer disease. J Neuropathol Exp Neurol. 1997 Apr;56(4):414–420. doi: 10.1097/00005072-199704000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Neuropathological stageing of alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 5.Braak H, Braak E. Staging of alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16(3):271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–84. [DOI] [PubMed] [Google Scholar]
- 6.Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer’s disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005 Apr;25(3):783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 7.Duvernoy HM. The human hippocampus, functional anatomy, vascularization and serial sections with MRI. third edition edition. Springer; 2005. [Google Scholar]
- 8.Eriksson PS, Perfilieva E, Bjrk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998 Nov;4(11):1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 9.Jack CR, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998 Oct;51(4):993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996 Mar;16(6):2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loop C, DeRose T. Computer Graphics (ACM SIGGRAPH Proceedings) 1990. Generalized b-spline surfaces of arbitrary topology; pp. 347–356. [Google Scholar]
- 12.Miller Michael I, Trouvé Alain, Younes Laurent. Geodesic shooting for computational anatomy. J. Math. Imaging Vis. 2006;24(2):209–228. doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squire Larry R, Stark Craig E L, Clarkx Robert E. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- 14.van Praag Henriette, Schinder Alejandro F, Christie Brian R, Toni Nicolas, Palmer Theo D, Gage Fred H. Functional neurogenesis in the adult hippocampus. Nature. 2002 Feb;415(6875):1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visser PJ, Verhey FRJ, Hofman PAM, Scheltens P, Jolles J. Medial temporal lobe atrophy predicts Alzheimer’s disease in patients with minor cognitive impairment. J Neurol Neurosurg Psychiatry. 2002 Apr;72(4):491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Lei, Miller J. Philp, Gado Mokhtar H, McKeel Daniel W, Rothermich Marcus, Miller Michael I, Morris John C, Csernansky John G. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. Neuroimage. 2006 Mar;30(1):52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warfield Simon K, Zou Kelly H, Wells William M. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004 Jul;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West Mark J, Kawas Claudia H, Stewart Walter F, Rudow Gay L, Troncoso Juan C. Hippocampal neurons in pre-clinical Alzheimer’s disease. Neurobiol Aging. 2004 Oct;25(9):1205–1212. doi: 10.1016/j.neurobiolaging.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Yushkevich Paul, Avants Brian, Pluta John, Minkoff David, Pickup Stephen, Liu Weixia, Detre John, Grossman Murray, Gee James. Building an atlas of hippocampal subfields using postmortem mri. IEEE International Symposium on Biomedical Imaging: Macro to Nano; 2008. to appear. [Google Scholar]
- 20.Yushkevich Paul A, Zhang Hui, Gee James. Continuous medial representation for anatomical structures. IEEE Trans Med Imaging. 2006 December;25(2):1547–1564. doi: 10.1109/tmi.2006.884634. [DOI] [PubMed] [Google Scholar]
- 21.Yushkevich Paul A, Zhang Hui, Simon Tony J, Gee James C. Structure-specific statistical mapping of white matter tracts. Neuroimage. 2008 Jun;41(2):448–461. doi: 10.1016/j.neuroimage.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeineh Michael M, Engel Stephen A, Thompson Paul M, Bookheimer Susan Y. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science. 2003 Jan;299(5606):577–580. doi: 10.1126/science.1077775. [DOI] [PubMed] [Google Scholar]