SUMMARY
Recurrent epidemics of influenza are observed seasonally around the world with considerable health and economic consequences. A key quantity for the control of infectious diseases is the reproduction number, which measures the transmissibility of a pathogen and determines the magnitude of public health interventions necessary to control epidemics. Here we applied a simple epidemic model to weekly indicators of influenza mortality to estimate the reproduction numbers of seasonal influenza epidemics spanning three decades in the United States, France, and Australia. We found similar distributions of reproduction number estimates in the three countries, with mean value 1·3 and important year-to-year variability (range 0·9–2·1). Estimates derived from two different mortality indicators (pneumonia and influenza excess deaths and influenza-specific deaths) were in close agreement for the United States (correlation=0·61, P<0·001) and France (correlation=0·79, P<0·001), but not Australia. Interestingly, high prevalence of A/H3N2 influenza viruses was associated with high transmission seasons (P=0·006), while B viruses were more prevalent in low transmission seasons (P=0·004). The current vaccination strategy targeted at people at highest risk of severe disease outcome is suboptimal because current vaccines are poorly immunogenic in these population groups. Our results suggest that interrupting transmission of seasonal influenza would require a relatively high vaccination coverage (>60%) in healthy individuals who respond well to vaccine, in addition to periodic re-vaccination due to evolving viral antigens and waning population immunity.
INTRODUCTION
Annual influenza epidemics are observed worldwide with substantial morbidity and mortality impact. In the United States, between 5% and 20% of the population become sick with influenza every year, and 36 000 people on average die from complications of the disease, often following a secondary bacterial infection [1, 2]. The overall economic burden of influenza in the United States alone has been estimated at more than 11 billion dollars annually [3].
Influenza has marked seasonal patterns in temperate areas of the world, where large and intense outbreaks occur once a year in wintertime, followed by fade-out periods in warmer months where no influenza activity is detected. The influenza virus is able to persist in populations through continuous evolution in the form of point mutations in the virus antigenic structure [4]. Major changes in the virus composition can give rise to pandemics, which are major global epidemics that can cause dramatic morbidity and mortality rates. There are currently three influenza (sub)types co-circulating in humans (A/H3N2, A/H1N1 and B) [4].
An important quantity for disease control is the basic reproduction number (R0), which represents the number of secondary cases generated by a primary case during the infectious period, in an entirely susceptible population [5, 6]. This quantity is a measure of the transmissibility of a pathogen and can help determine the intensity of interventions necessary to control an outbreak [5]. If R0>1 then an epidemic may occur, while transmission cannot be sustained when R0<1.
The reproduction number is a central quantity to evaluate whether disease control is possible, in particular when a vaccine is available. Large-scale influenza vaccination programmes have been established in developed countries since the early 1980s, and are now starting in developing countries [7]. Despite widespread vaccine use, there are indications that the current annual vaccination strategy is not optimal for reducing influenza mortality burden [8]. Vaccination is targeted at people at high risk of severe disease outcome, i.e. the elderly and those with chronic conditions, for whom influenza vaccines may be less immunogenic [9]. A key issue today is whether annual immunization of high transmitter groups, in particular children, could achieve herd immunity and interrupt seasonal transmission [10]. Estimates of the transmissibility of seasonal influenza epidemics are necessary to evaluate whether this is feasible and hence refine existing control strategies.
Past studies have estimated the reproduction number of individual influenza seasons, in particular for pandemics [11–19]. However, no study has yet reported estimates of the reproduction number for several countries and consecutive influenza seasons in the inter-pandemic period, where a fraction of the population is immune due to previous influenza exposure or vaccination. Considering multiple epidemics occurring in different years and locations is important to capture the year-to-year variability of influenza epidemics [20], as well as their potential geographical heterogeneities [19, 21, 22]. Here, we apply a simple epidemic model to weekly indicators of influenza-related mortality in the United States, France, and Australia. We estimate the reproduction number of seasonal epidemics spanning three decades in these countries, and explore the relationship between reproduction number and viral prevalence. We then use our reproduction number estimates to discuss whether interrupting transmission of seasonal influenza by vaccination is feasible, given the efficacy of currently available vaccines.
MATERIALS AND METHODS
Demographic and epidemiological data
General approach
Assessment of the transmission patterns in influenza epidemics is not straightforward because of lack of reliable morbidity data, due to non-specific clinical symptoms and infrequent confirmation by laboratory tests [2, 23]. Alternatively, influenza-related mortality is considered a good indicator of disease patterns, albeit with some caveats. The severe complications triggered by influenza infection, such as bacterial pneumonia, are often diagnosed after the virus has been cleared [2, 24]. Many influenza-related deaths are therefore not coded as influenza but rather as underlying respiratory or chronic conditions. The traditional way to assess influenza activity patterns is to calculate ‘excess mortality’ from broad death categories, as the mortality in winter seasons in excess of a non-epidemic baseline. Excess deaths from pneumonia and influenza (P&I) have been shown in several studies to be a reliable and specific endpoint for studying timing and amplitude of influenza-related mortality, both at the local and national scales [2, 18, 23]. Alternatively, the subset of P&I deaths coded specifically as influenza may be even more specific, especially for epidemic timing [2, 18], but these data capture a very small fraction of the overall influenza mortality burden. Both P&I excess mortality and influenza-specific deaths have been used in the past to estimate influenza transmissibility [14–19], and we consider both indicators here.
Mortality data
Weekly P&I mortality time series were computed from death certificates collected by national agencies for vital statistics in the three countries (United States, 1972–2002, National Center for Health Statistics; Australia, 1972–1997, Australian Bureau for Statistics; France, 1972–1997, Institut National de la Santé et de la Recherche Médicale, Service Commun 8). We used codes 470–474 and 480–486 from the International Classification of Diseases (ICD) 8th revision, codes 480–487 from ICD-9 and codes J10·0-J18·9 from ICD-10 to select deaths due to P&I. For US data, we used the standard correction factor given by the National Center for Health Statistics to account for the decrease in pneumonia code use following the transition between ICD-9 and ICD-10 in 1999. To derive death rates, we obtained annual population size estimates from the U.S. Census Bureau [25], the French National Institute for Statistics and Economic Studies [26], and the Australian Bureau of Statistics [27].
P&I excess mortality models
We measured the influenza contribution to weekly P&I mortality series as those P&I deaths in excess of a seasonal baseline, where the baseline mirrors the expected level of mortality in the absence of influenza activity (Fig. 1 and online Supplement). To fit the baseline, we used a classical seasonal regression approach developed by Serfling in 1963 [28], where a linear regression model with harmonic terms is fitted to non-epidemic weeks to produce the baseline. This approach has been applied to mortality data in different countries [8, 21, 22, 29] and at different geographical scales [18], and has also been compared with morbidity data [18].
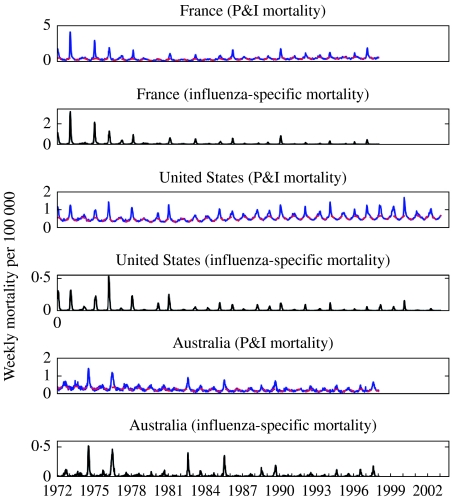
Fig. 1.
Time-series of weekly number of pneumonia and influenza (P&I) and influenza-specific deaths per 100 000 in three countries (France, United States and Australia, blue curve). The red dashed line indicates the baseline mirroring the expected level of P&I mortality in the absence of influenza epidemic activity.
Influenza-specific mortality data
The Serfling approach we applied to P&I data has limitations because it assumes that non-influenza deaths are seasonal and can be modelled by harmonic functions [28]. By contrast, we can use the raw time-series of influenza-specific mortality to estimate transmissibility, without fitting a seasonal baseline, because there are no deaths in non-influenza months.
Transmission model
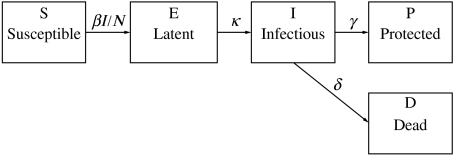
Several studies have used mathematical models to describe the transmission dynamics of influenza within a susceptible population (e.g. see [30, 31]), although none has explored whether interrupting transmission by vaccination was feasible given currently available vaccines. We adapted a mass-action model previously developed for studying the transmissibility of the 1918 influenza pandemic in Geneva, Switzerland [11]. In the ‘SEIR’ model, the population is divided in five categories: Susceptible (S), Exposed (E), Infectious (I), Recovered/Protected (P) and Dead (D) (Fig. 2). In this model, infection is transmitted between infectious and susceptible individuals, and no particular route of transmission is assumed.
Fig. 2.
Compartmental model indicating the transition of individuals among the different epidemiological stages during an influenza outbreak. β=transmission rate; N=total population; 1/κ=latent period; 1/γ=recovery period; δ=mortality rate (see Table for parameter values).
The total population size at time t is given by N(t)=S(t)+E(t)+I(t)+P(t). We assume homogenous mixing, that is, each individual has the same probability of having contact with any other individual in the population. Moreover, for each influenza season, the total population is assumed constant according to the population size estimate for a given country and year. As in a previous models of seasonal influenza (e.g. [30]), susceptible individuals infected with the virus enter the latent period (category E) at the rate βI/N where β is the mean transmission rate per day and I/N is the probability of contacting an infected individual out of the total population size N. Note that β represents a generic transmission rate that combines the effect of direct and indirect contacts (including, droplets, fomites and aerosols [32, 33]). Latent individuals progress to the infectious class at the rate κ (1/κ is the mean latent period). Infectious individuals either recover or die from influenza at the mean rates γ and δ, respectively. Recovered individuals are assumed protected for the duration of the influenza season. The mortality rate is given by δ=γ[CFP/(1−CFP)], where CFP is the mean case fatality proportion. The system of differential equations that describes the above epidemic process is given by:
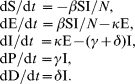 |
The population is assumed completely susceptible at the beginning of each influenza season prior to the first epidemic week, which is defined as the first week with non-zero influenza-related deaths. The initial number of influenza deaths D(0) is set to be the number of influenza deaths in the first epidemic week. Furthermore, using a CFP, we also estimate the number of recovered individuals in the first epidemic week as P(0)=D(0)/CFP – D(0).
The reproduction number
The number of secondary cases generated by a primary infectious case during its period of infectiousness in an entirely susceptible population is known as the basic reproduction number, R0. From our model R0 is given by the product of the transmission rate β and the mean infectious period 1/(γ+δ); that is, R0=β/(γ+δ).
In the case of seasonal influenza epidemics, R0 cannot be estimated due to partial immunity in individuals infected in previous years with antigenically related strains, and annual vaccination of a fraction of the high-risk population. However, we can estimate a different reproduction number, Rp, which measures the transmissibility at the beginning of an epidemic in a partially immune population [5]. For example, if a proportion p of a completely susceptible population is successfully immunized prior to an epidemic, then in a well-mixed population with a constant force of infection Rp=(1 – p)R0. Estimating Rp is equivalent to assuming a fixed R0 for seasonal influenza and estimating a changing susceptibility (1−p) each season [34].
Parameter estimation
Definitions and baseline values for fixed and estimated parameters are given in the Table; the mean latent and recovery periods (respectively 1/κ and 1/γ), and the CFP of influenza were fixed according to previous studies [3, 14, 35]. To estimate the value of unknown parameters, we rely on the general approach of ‘trajectory matching’, where one searches for the combination of model parameters that produces an epidemic curve most statistically similar to the observed one. The transmission rate and the initial numbers of individuals in the exposed E(0) and infectious I(0) categories at the beginning of each influenza season were estimated by least squares fitting to the cumulative number of weekly influenza-related deaths or cases, during the initial take-off of the epidemic. The advantage of using the cumulative over the weekly number of deaths is that the former smoothes out demographic noise and reporting delays [16]. Due to the short latent period for influenza (1·9 days), we assumed E(0)=I(0); this simplification allowed us to estimate only two parameters from the exponential growth phase of the epidemic. We also made the assumption that errors in the data (e.g. underreporting and misdiagnosis of cases) occurred at random and observations were as likely to overestimate, as they were to underestimate the true number of influenza deaths.
Table.
Parameter definitions and baseline values used with the SEIR compartmental epidemic model
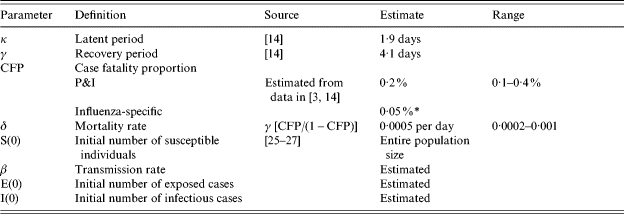
Only two parameters were estimated, the transmission rate and the initial numbers of exposed and infectious cases. That is, for simplicity we assume E(0)=I(0).
Influenza-specific deaths represent ~25% of P&I excess mortality.
The reproduction number (Rp) was estimated using data comprising the four epidemic weeks immediately preceding the epidemic peak. Hence, we only estimated the reproduction number (Rp) for those influenza seasons for which the increasing phase of the epidemic included at least four consecutive weeks. We define the peak week as the week with the maximum death rate; this definition is not ambiguous since there is only major peak of illness or mortality for each influenza season [18]. In addition P&I and influenza-specific mortality peaks are meaningful since they coincide with peaks in influenza laboratory surveillance [18], and occur synchronously in other causes of deaths on which influenza has an impact [23].
Uncertainty of parameter estimates
We estimated the uncertainty of the reproduction number via parametric bootstrap [36]. For each flu season, we simulated 200 alternate realizations of the epidemic trajectory, by perturbation of the best-fit curve of cumulative number of influenza-related deaths. We added to the best-fit curve a simulated error structure computed using the increment in the ‘true’ number of deaths/cases from day j to day j+1 as the Poisson mean for the number of new deaths observed in the j to j+1 interval. The cumulative epidemic curves were then aggregated by week, the temporal scale of the epidemic data. The 95% bootstrap-based confidence intervals for the reproduction number should be interpreted as containing 95% of estimates if the analysis was repeated with the same model assumptions and if observational error was the only source of noise.
Sensitivity analyses
We conducted a number of sensitivity analyses to assess the validity of our assumptions and the robustness of our estimates. In particular, we checked the impact of choosing a Poisson error for bootstrap resampling, our assumptions on the distribution of the latent and infectious periods, the boundaries of confidence intervals, the number of weeks used in the estimations, and the CFP values. These analyses are summarized below (more details are given in the online Supplement).
In our simple SEIR model, we implicitly assumed that the latent and infectious periods were exponentially distributed. But we also simulated more realistic distributions of the latent and infectious periods, such as a gamma distribution, as described in [37], and assessed the potential bias on estimates of the reproduction number (Rp).
Next, we explored the uncertainty of Rp estimates. In the main analysis, we used Poisson distribution to model observational errors, where variance is equal to the mean, but we also tested higher levels of uncertainty, where the variance is 2, 3, or 4 times the mean. Moreover, to check whether the reproduction number estimates were well-constrained, we derived likelihood ratio confidence bounds for Rp.
Last, we assessed the robustness of Rp estimates to a twofold increase or decrease in the predefined CFP values (Table) and changes in the number of weeks used in the computations (4–6).
Relationships between the reproduction number (Rp) and seasonal virus surveillance
We explored the relationships between Rp and the prevalence of the three influenza viral (sub)types circulating each season (A/H3N2, A/H1N1, B). Virus prevalence was defined as the proportion of respiratory samples that tested positive for each influenza (sub)type, as reported by the CDC laboratory surveillance conducted since 1976 in the United States [38–41]. The United States was the only country among the three studied with publicly available virus surveillance data for many years.
RESULTS
Estimates of the reproduction number (Rp)
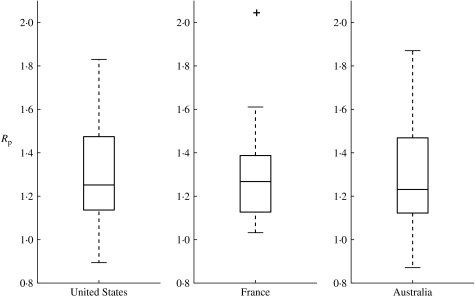
We estimated the reproduction number (Rp) for three decades in the inter-pandemic period using weekly P&I excess mortality curves in the United States, France and Australia. Overall we found similar averages in the three countries: the mean reproduction number (Rp) was 1·3 (s.e.=0·05) in the United States, 1·3 (s.e.=0·05) in France and 1·3 (s.e.=0·07) in Australia (Fig. 3, Wilcoxon test for pairwise comparisons between countries, all P⩾0·87). The reproduction number across influenza seasons and countries lied in the range 0·9–2·0 with an overall mean of 1·3, and 95% confidence interval (CI) 1·2–1·4. Larger variability in the estimates of the reproduction number was observed for Australia than for the United States and France; perhaps explained by the larger demographic noise and spatial heterogeneity in Australia (Fig. 1). Indeed, Australia has the smallest population size of the three countries studied, and most of the population is concentrated in coastal areas of the South-East. Overall, the SEIR transmission model fitted well to the epidemic rise of influenza-related deaths in the three countries, as shown in Fig. 4.
Fig. 3.
Boxplots of the reproduction number (Rp) of influenza seasons (1972–1997) in the United States, France, and Australia. The boxes have lines at 25, 50 and 75 percentiles. The whiskers show the extent of the rest of the data extending to a maximum of 1·5 times the interquartile range. Points outside the ends of the whiskers are indicated with a ‘+’ symbol.
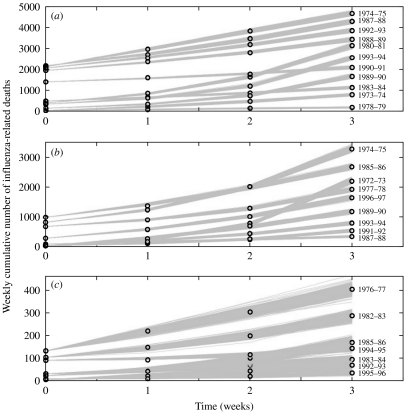
Fig. 4.
Model fits to the epidemic rise of a number of representative influenza seasons in (a) the United States, (b) France, and (c) Australia. The data are the circles and the solid lines are 200 realizations of the model fitted to the data obtained through the parametric bootstrap as explained in the text.
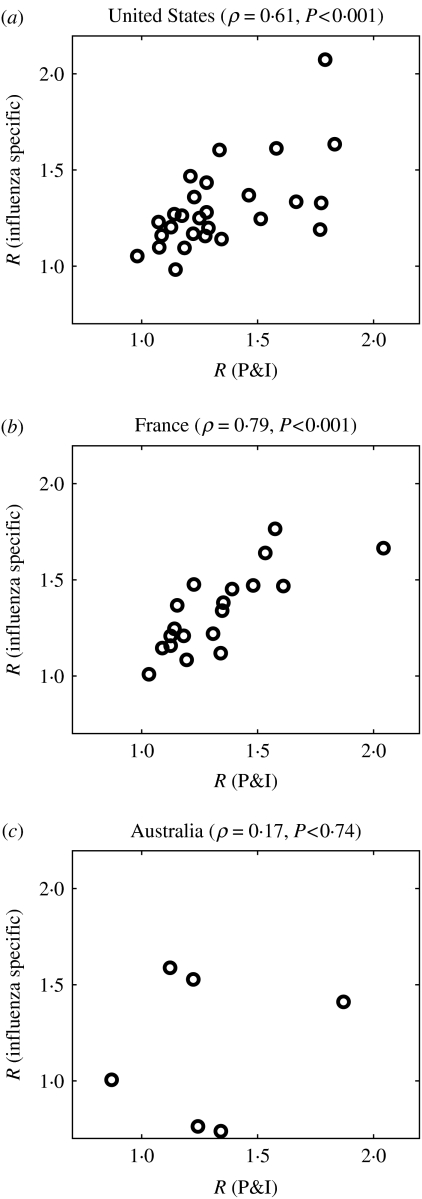
Applying the same methodological approach to influenza-specific deaths, rather than P&I excess deaths, gave similar results in the three countries. The mean reproduction number (Rp) was 1·3 (s.e.=0·04) in the United States, 1·3 (s.e.=0·05) in France and 1·2 (s.e.=0·08) in Australia (Wilcoxon test for pairwise comparisons between countries, all P⩾0·68). Further, estimates of the reproduction number using influenza-specific mortality were strongly correlated with those obtained using P&I excess mortality data in the United States (ρ=0·61, P<0·001, n=27) and France (ρ=0·79, P<0·001, n=19) (Fig. 5a, b). A weaker association was found in Australia (ρ=0·17, P=0·74, n=6) (Fig. 5c), probably due to the few overlapping seasons for which it was possible to estimate the reproduction number for both time series, as well as larger demographic noise and spatial heterogeneity.
Fig. 5.
Correlation of the reproduction number estimates derived from pneumonia and influenza excess mortality and influenza-specific mortality data in three countries (a) United States, (b) France and (c) Australia. While a significant and positive correlation was observed for the United States and France, a weaker correlation was found in Australia, probably due to the few overlapping seasons for which it was possible to estimate the reproduction number (n=6) and the larger demographic noise and spatial heterogeneity.
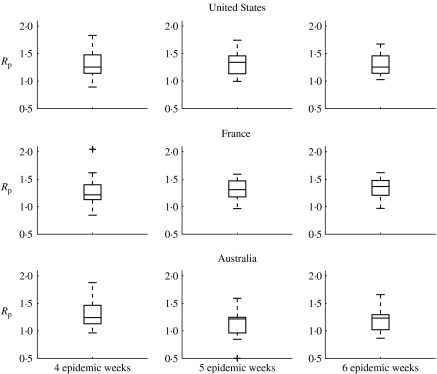
Next, we assessed the sensitivity of Rp estimates to the CFP value and the number of epidemic weeks of mortality data used in the estimation. We found that Rp estimates did not change when we increased or decreased CFP by twofold (respectively 0·1% and 0·4%). Moreover, Rp estimates were robust to increasing the number of epidemic weeks used in the estimation from 4 weeks, to 5 or 6 weeks, for those seasons for which sufficient epidemic data was available (Fig. 6, Wilcoxon test for differences in mean Rp between 4 and 6 weeks, P>0·25 for all three countries).
Fig. 6.
Sensitivity analysis: boxplots of the reproduction number (Rp) as a function of the number of epidemic weeks used in the estimation, for the United States, France and Australia. Our estimates of Rp are robust to increasing the number of epidemic weeks used in the estimation to 5 or 6 epidemic weeks, given a sufficient time period of epidemic take-off (Wilcoxon test for differences in mean Rp between 4 and 6 wks: P>0·66, United States; P>0·34, France; P>0·25, Australia).
Additional sensitivity analyses (see online Supplement) showed that our estimates were robust to model assumptions and observational errors. When the latent and infectious periods were modelled via more realistic distributions than the exponential, the bias incurred in point estimates of Rp rapidly declined with the amount of epidemic data. When using 4 epidemic weeks of data as in our main analysis, we estimated that the bias was <0·13 (9%) on average (Fig. S1, online). Further, increasing the variance of measurement errors changed point estimates of Rp by <0·03 (3·3%) and increased the width of the 95% CI by 0·05 (26·8%) on average [maximum 0·24 (64·4%), Supplementary Table, online]. In addition, profile likelihood, showed that the two parameters estimated in the model, Rp and the initial number of infected individuals, were well-bounded and identifiable (Fig. S2, online).
We then explored the relationship between seasonal estimates of influenza transmissibility and virus surveillance in the United States and found that the reproduction number (Rp) differed by (sub)type of circulating viruses. Influenza A/H3N2 viruses were more frequently isolated in seasons with higher transmission (correlation between Rp and A/H3N2 virus prevalence; Spearman ρ=0·52, P=0·006) whereas by contrast, B viruses were associated with low transmission seasons (correlation between Rp and B virus prevalence, Spearman ρ=−0·55, P=0·004). There was no clear pattern of association for A/H1N1 viruses.
DISCUSSION
We used an epidemic model to estimate the reproduction number (Rp) of inter-pandemic influenza seasons spanning three decades, using data on the weekly cumulative number of influenza-related deaths from the United States, France, and Australia. We found an average Rp of 1·3 (95% CI 1·2–1·4) across influenza seasons and countries, with substantial inter-annual variability. Our model, based on SEIR framework and homogeneous mixing assumption, was kept minimal to estimate two parameters each influenza season (Rp and the initial number of infected individuals). Although we did not consider an age-structured model, previous sensitivity analyses suggest that estimates of the reproduction number are not biased due to differences in age-specific transmission parameters and CFP [14].
Our study is limited from the epidemic data used in the estimation. Excess deaths from P&I capture about 25% of the overall mortality burden of influenza [2]. Consequently, it was not possible to estimate the reproduction number for very mild epidemics (9%), where mortality is not a good proxy indicator for influenza transmission. The use of less sensitive mortality data can explain why Rp estimates and their 95% CI were below 1·0 for two mild influenza seasons (one in France and one in Australia). Encouragingly, recent simulations have shown that estimation approaches relying on the cumulative number of cases are expected to be robust to substantial measurement error and underreporting of cases (such as occurs in mortality data), in particular when the true theoretical reproduction number is below 4·0 [42]. Moreover, we performed a number of sensitivity analyses showing the robustness of our estimates to various assumptions, in line with previous research [14].
Unlike P&I data, influenza-specific death time-series do not require modelling a seasonal baseline prior to fitting a transmission model [15–17, 19]. It is reassuring that mean and individual-season estimates using influenza-specific and P&I mortality data were similar for the two countries where sufficient years were available for comparison (United States, France). Further, a similar modelling approach applied to long-term data on weekly influenza cases from France (41) gave consistent estimates (mean Rp=1·5, s.e.=0·08). Taken together, the consistency of mean and variance estimates of Rp confirms that long-term influenza mortality records can be used to study patterns of disease transmission, even at a refined weekly time-scale [14, 18].
An important limitation of using data aggregated by country is the potential effect of spatial heterogeneity, which could lead to underestimation of the reproduction number. However, recent US studies have shown that estimates of Rp for influenza are surprisingly similar across locations and spatial scales, and do not depend on local geographical or population factors [14, 18]. In addition, the comparison of historical influenza epidemic curves for England and Wales and Greater London concluded that even considerable geographical heterogeneity did not substantially alter Rp estimates [15].
In our model, influenza is transmitted by serial contacts between infected and susceptible individuals. The model does not specify whether transmission occurs by direct or indirect contact, via droplets, fomites or aerosols, since the relative contribution of each mode of transmission is unclear for influenza [32, 33, 43–45]. Unfortunately, there are very few experimental studies of human-to-human transmission; besides, existing studies are inconclusive and predate the discovery of the influenza virus [32, 46, 47]. Indirect evidence for influenza transmission between humans can nevertheless be found in observational studies in close contact settings, such as aeroplanes, hospitals, households, schools, and day-care centres [48–53]. In particular, influenza transmission is more intense in the household than in the community [51, 54], while clinical trials have shown that treating index cases with antivirals reduces secondary transmission to household members [55]. As regards animal studies, recent experiments in guinea pigs suggest that transmission occurs via aerosols in this mammalian host [44] – but whether this also applies to people is unclear. Overall, uncertainty in the source of exposure (household/community) and mode of transmission makes the estimation of the serial interval between infection in two successive cases difficult from observational studies alone [56, 57]. To account for this uncertainty, the different routes of influenza transmission are lumped into a single contact rate parameter (β) in our model.
While this is the first study to systematically estimate the reproduction numbers of influenza for multiple inter-pandemic seasons in different countries, our results are in overall agreement with a previous study reporting an estimate of 1·5 for a single A/H3N2 season in France [12]. An earlier study proposed estimates higher than ours (range 1·4–2·6) for several consecutive influenza seasons in England and Wales [15, 16], however the exact quantity measured in this work remains controversial [14, 31]. A particularly high Rp estimate (Rp>2·0) has also been reported for the 1951 influenza epidemic in England and Canada, however, this epidemic was associated with unusually high mortality and transmissibility locally [19, 22].
By contrast to the scarcity of estimates for the reproduction number of inter-pandemic influenza, several studies have recently proposed estimates for pandemic influenza [11, 13, 14, 17, 19]. The analysis of historical mortality and morbidity curves from past pandemics has revealed that Rp ranged between 1·5 and 3·0 for the 1918 pandemic [11, 13, 14, 17, 19], 1·5–1·7 for the 1957 pandemic [13, 19], and 1·9–2·2 for the 1968 pandemic [13, 19, 58], depending on the pandemic wave studied, geographical location, and estimation method. This is consistent with higher transmissibility in pandemic than inter-pandemic seasons, since the fraction of susceptible individuals is largest when an immunologically naive population is exposed to an entirely novel pandemic virus (and Rp~R0).
While the average inter-pandemic Rp seems rather invariant across geographical locations at around 1·3, there is substantial year-to-year variability around this average (s.e.=0·09, maximum Rp=2·1). Our results suggest that H3N2 viruses are more transmissible than B viruses; A/H3N2 viruses are known to cause more severe epidemics than A/H1N1 and B viruses in terms of overall disease burden on mortality and severe morbidity [20]. This difference in transmissibility could be attributed to underlying differences in the age patterns of infection among influenza (sub)types. Influenza A/H3N2 viruses have the fastest evolutionary rates [59], causing re-infection of a single individual with the same subtype multiple times through life, thereby allowing for a larger pool of susceptibles, and higher transmissibility. By contrast, B viruses have the slowest evolutionary rate [59] and infect mainly children [60], perhaps explaining their lower transmissibility. A/H1N1 viruses have intermediate evolutionary rate [59] and did not display a clear relationship with transmissibility in our study. Our finding of increased transmissibility of A/H3N2 viruses reinforces a recent study showing more rapid dispersal of A/H3N2 epidemics across the United States, as compared with A/H1N1-B epidemics [18].
One could further expect that transmissibility of inter-pandemic influenza is highest when new antigenic clusters of influenza A/H3N2 strains emerge every 2–5 years and escape population immunity to haemagglutinin [61]; yet we could not detect a pattern in our data (not shown). Recent epidemiological studies of pandemic and inter-pandemic influenza suggest that antigenic changes in the haemagglutinin, the most studied component of antibody-mediated immunity, may not be the only determinant of viral fitness and ability to spread in a population [19, 21]. The exact relationship between virus antigenic changes and population dynamics is still unclear, and a key area for future research.
Influenza epidemics display marked seasonal patterns in temperate countries, including the United States, France and Australia [29, 62]. Our estimates of the reproduction number reflect transmissibility in wintertime, and it is likely that transmissibility declines in warmer months, due to changes in the host, the virus, or their interaction. The reasons for the seasonality of influenza are still heavily debated, although the susceptibility of the respiratory mucosa, vitamin D production, human contacts, and virus survival are subject to seasonal variations, which could favour influenza transmission in colder months in high-latitude countries [56, 57, 63, 64]. Influenza displays a pattern of year-round circulation and high disease burden in the Tropics [62, 65] and it would be useful to elucidate the seasonal triggers of influenza epidemics globally and estimate transmissibility in the Tropics. This type of work is contingent upon obtaining reliable data on influenza virus circulation and disease burden in Tropical countries [62].
Overall, our results indicate that the reproduction number Rp for inter-pandemic influenza is below 2·1 in temperate countries, which has important implications for disease control. In a fully mixed population, transmission could be interrupted if about 52% of the population was successfully immunized and we discuss below whether this could be achieved with currently available vaccines.
The influenza vaccination programme in the United States has traditionally targeted individuals who are at highest risk of severe disease outcome and are immunologically impaired relative to the general population, in particular people over 65 years of age and those with chronic conditions [66]. Vaccination prevents 70% of laboratory-confirmed infections in healthy adults and children [67, 68], but the immune response to vaccines varies greatly with age and is particularly weak in the elderly [9] and young children [69]. In particular, there has been no decline in influenza-related mortality among the elderly in the past two decades in the United States, while vaccine coverage increased from 5% to 65% in this age group [8]. Analysis of long-term trends in mortality in Italy revealed similar patterns, suggesting an alarming lack of effectiveness of traditional vaccination programmes targeted at the elderly [70, 71]. In recent years, about 23% of the US population received influenza vaccination [72, 73], but we estimate that only 13% is effectively protected by vaccination at the start of the influenza season (range 11–16%), assuming no mismatch between the vaccine strain and circulating strain.
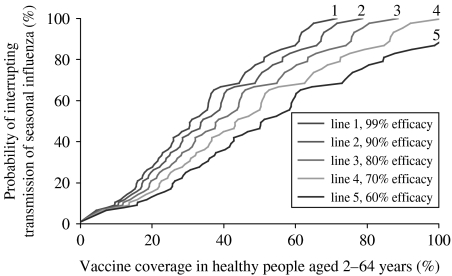
Complementary immunization strategies could target high transmitter groups such as school-age children, who are likely to respond well to influenza vaccination and are prone to transmit infection to their immediate contacts [74]. Fig. 7 illustrates that interrupting transmission in most influenza seasons (>90%) would require vaccination of 60–100% of the population who responds well to vaccine, healthy people aged 2–64 years, depending on vaccine efficacy assumptions (see online Supplement for details on methods). On the one hand, this estimate would probably be lower if we had considered an age-structured transmission model with increased contacts in children [3]; however, data on influenza transmissibility and contact rates between and within different age groups are scarce. On the other hand, our estimate of herd immunity threshold is based on the current background level of natural immunity to influenza, resulting from continuous exposure to viruses circulating in the community. If influenza circulation gradually decreased due to improved vaccination coverage, natural immunity would wane, increasing the level of vaccine coverage necessary to achieve herd immunity.
Fig. 7.
Probability of interrupting transmission of seasonal influenza for various vaccination scenarios. Values are based on the empirical cumulative distribution of reproduction number estimates in the three countries (y axis) and different vaccine coverage in the healthy population aged 2–64 years (x axis), who is supposed to respond well to influenza vaccines (see online Supplement for methods). Different curves represent different assumptions about vaccine efficacy in the healthy population.
In conclusion, given that vaccine efficacy at one dose is suboptimal in population groups currently targeted for vaccination, many are not benefiting from direct protection. Relatively high vaccination coverage would be necessary in the immunocompetent population, healthy people aged 2–64 years, in order to interrupt transmission of seasonal influenza, in addition to periodic re-vaccination due to evolving viral antigens and waning population immunity. An alternative strategy, perhaps less costly in the long run, would be the use of vaccines that are more immunogenic and provide longer-term protection, obviating the need for repeated vaccination.
ACKNOWLEDGEMENTS
This study was funded by the Los Alamos National Laboratory and the Fogarty International Center, National Institutes of Health.
NOTE
Supplementary information accompanies this paper on the Journal's website (http://journals.cambridge.org).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.CDC http://www.cdc.gov/flu/keyfacts.htm. http://www.cdc.gov/flu/keyfacts.htm . Key facts about influenza and the influenza vaccine ( ). Centers for Disease Control and Prevention. Accessed 16 November 2005.
- 2.Simonsen L. The global impact of influenza on morbidity and mortality. Vaccine. 1999;17:S3–10. doi: 10.1016/s0264-410x(99)00099-7. (Suppl 1): [DOI] [PubMed] [Google Scholar]
- 3.Weycker D et al. Population-wide benefits of routine vaccination of children against influenza. Vaccine. 2005;23:1284–1293. doi: 10.1016/j.vaccine.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson K, Hay A. Textbook of Influenza. Oxford; Blackwell: 1998. [Google Scholar]
- 5.Anderson RM, May RM. Infectious Diseases of Humans. New York: Oxford University Press; 1991. [Google Scholar]
- 6.Diekmann O, Heesterbeek J. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. Chichester; Wiley: 2000. [Google Scholar]
- 7.Macroepidemiology of Influenza Vaccination (MIV) Study Group. The macro-epidemiology of influenza vaccination in 56 countries, 1997–2003. Vaccine. 2005;23:5133–5143. doi: 10.1016/j.vaccine.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Simonsen L et al. Impact of influenza vaccination on seasonal mortality in the US elderly population. Archives of Internal Medicine. 2005;165:265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: A quantitative review. Vaccine. 2005;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 10.Halloran ME, Longini IM., Jr. Public health. Community studies for vaccinating schoolchildren against influenza. Science. 2006;311:615–616. doi: 10.1126/science.1122143. [DOI] [PubMed] [Google Scholar]
- 11.Chowell G et al. Transmission dynamics of the great influenza pandemic of 1918 in Geneva, Switzerland: assessing the effects of hypothetical interventions. Journal of Theoretical Biology. 2006;241:193–204. doi: 10.1016/j.jtbi.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 12.Flahault A et al. Modelling the 1985 influenza epidemic in France. Statistics in Medicine. 1988;7:1147–1155. doi: 10.1002/sim.4780071107. [DOI] [PubMed] [Google Scholar]
- 13.Gani R et al. Potential impact of antiviral drug use during influenza pandemic. Emerging Infectious Diseases. 2005;11:1355–1362. doi: 10.3201/eid1109.041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spicer CC. The mathematical modelling of influenza epidemics. British Medical Bulletin. 1979;35:23–28. doi: 10.1093/oxfordjournals.bmb.a071536. [DOI] [PubMed] [Google Scholar]
- 16.Spicer CC, Lawrence CJ. Epidemic influenza in Greater London. Journal of Hygiene (Cambridge) 1984;93:105–112. doi: 10.1017/s0022172400060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson NM et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 18.Viboud C et al. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312:447–451. doi: 10.1126/science.1125237. [DOI] [PubMed] [Google Scholar]
- 19.Viboud C et al. Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine. 2006;24:6701–6707. doi: 10.1016/j.vaccine.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 20.Simonsen L et al. The impact of influenza epidemics on mortality: introducing a severity index. American Journal of Public Health. 1997;87:1944–1950. doi: 10.2105/ajph.87.12.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viboud C et al. Multinational impact of the 1968 Hong Kong influenza pandemic: evidence for a smoldering pandemic. Journal of Infectious Diseases. 2005;192:233–248. doi: 10.1086/431150. [DOI] [PubMed] [Google Scholar]
- 22.Viboud C et al. 1951 influenza epidemic, England and Wales, Canada, and the United States. Emerging Infectious Diseases. 2006;12:661–668. doi: 10.3201/eid1204.050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichert TA et al. Influenza and the winter increase in mortality in the United States, 1959–1999. American Journal of Epidemiology. 2004;160:492–502. doi: 10.1093/aje/kwh227. [DOI] [PubMed] [Google Scholar]
- 24.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annual Review of Medicine. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 25.U.S. Census Bureau http://www.census.gov/popest/estimates.php. http://www.census.gov/popest/estimates.php . Population estimates ( ). Accessed 3 August 2005.
- 26.Institut National de la Statistique et des Etudes Economiques http://www.insee.fr/fr/recensement/page_accueil_rp.htm. http://www.insee.fr/fr/recensement/page_accueil_rp.htm . Census ( ). Accessed 3 August 2005.
- 27.Australian Bureau of Statistics. http://www.abs.gov.au. 2005. http://www.abs.gov.au ). Accessed 3 August .
- 28.Serfling R. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Reports. 1963;78:494–506. [PMC free article] [PubMed] [Google Scholar]
- 29.Viboud C et al. Influenza epidemics in the United States, France, and Australia, 1972–1997. Emerging Infectious Diseases. 2004;10:32–39. doi: 10.3201/eid1001.020705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreasen V. Dynamics of annual influenza A epidemics with immuno-selection. Journal of Mathematical Biology. 2003;46:504–536. doi: 10.1007/s00285-002-0186-2. [DOI] [PubMed] [Google Scholar]
- 31.Gog JR et al. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proceedings of the National Academy of Sciences USA. 2003;100:11143–11147. doi: 10.1073/pnas.1830296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridges CB, Kuehnert MJ, Hall CB. Transmission of influenza: implications for control in health care settings. Clinical Infectious Diseases. 2003;37:1094–1101. doi: 10.1086/378292. [DOI] [PubMed] [Google Scholar]
- 33.Tellier R. Review of Aerosol Transmission of Influenza A Virus. Emerging Infectious Diseases. 2006;12:1657–1662. doi: 10.3201/eid1211.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Earn DJ et al. A simple model for complex dynamical transitions in epidemics. Science. 2000;287:667–670. doi: 10.1126/science.287.5453.667. [DOI] [PubMed] [Google Scholar]
- 35.Longini IM, Jr. et al. Containing pandemic influenza with antiviral agents. American Journal of Epidemiology. 2004;159:623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 36.Efron B, Tibshirani RJ. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Statistical Science. 1986;1:54–75. [Google Scholar]
- 37.Wearing HJ, Rohani P, Keeling MJ. Appropriate models for the management of infectious diseases. PLoS Medicine. 2005;2:e174. doi: 10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson WW et al. Mortality associated with influenza and respiratory syncytial virus in the United States. Journal of the American Medial Association. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 39.CDC. Update: Influenza activity – United States and Worldwide, 1999–2000 season, and composition of the 2000–01 influenza vaccine. Morbidity and Mortality Weekly Report. 2000;49:375–381. [PubMed] [Google Scholar]
- 40.CDC. Update: Influenza activity – United States and Worldwide, 2000–2001 season, and composition of the 2001–02 influenza vaccine. Morbidity and Mortality Weekly Report. 2001;50:466–470. [PubMed] [Google Scholar]
- 41.CDC. Update: Influenza activity – United States and Worldwide, 2001–2002 season, and composition of the 2002–03 influenza vaccine. Morbidity and Mortality Weekly Report. 2002;51:503–506. [PubMed] [Google Scholar]
- 42.Ferrari MJ, Bjornstad ON, Dobson AP. Estimation and inference of R0 of an infectious pathogen by a removal method. Mathematical Biosciences. 2005;198:14–26. doi: 10.1016/j.mbs.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Alford RH et al. Human influenza resulting from aerosol inhalation. Proceedings of the Society for Experimental Biology Medicine. 1966;122:800–804. doi: 10.3181/00379727-122-31255. [DOI] [PubMed] [Google Scholar]
- 44.Lowen AC et al. The guinea pig as a transmission model for human influenza viruses. Proceedings of the National Academy of Sciences USA. 2006;103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bean B et al. Survival of influenza viruses on environmental surfaces. Journal of Infectious Diseases. 1982;146:47–51. doi: 10.1093/infdis/146.1.47. [DOI] [PubMed] [Google Scholar]
- 46.Leake JP. The transmission of influenza. Boston Medical and Surgical Journal. 1919;181:675. [Google Scholar]
- 47.Rosenau JM. Experiments to determine mode of spread of influenza. Journal of the American Medical Association. 1919;73:311. [Google Scholar]
- 48.Moser MR et al. An outbreak of influenza aboard a commercial airliner. American Journal of Epidemiology. 1979;110:1–6. doi: 10.1093/oxfordjournals.aje.a112781. [DOI] [PubMed] [Google Scholar]
- 49.Anon. Influenza in a boarding school. British Medical Journal. 1978;1:587. [Google Scholar]
- 50.Blumenfeld HL et al. Studies on influenza in the pandemic of 1957–1958. I. An epidemiologic, clinical and serologic investigation of an intrahospital epidemic, with a note on vaccination efficacy. Journal of Clinical Investigation. 1959;38:199–212. doi: 10.1172/JCI103789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longini IM, Jr. et al. Estimating household and community transmission parameters for influenza. American Journal of Epidemiology. 1982;115:736–751. doi: 10.1093/oxfordjournals.aje.a113356. [DOI] [PubMed] [Google Scholar]
- 52.Riley RL. Airborne infection. American Journal of Medicine. 1974;57:466–475. doi: 10.1016/0002-9343(74)90140-5. [DOI] [PubMed] [Google Scholar]
- 53.Viboud C et al. Risk factors of influenza transmission in households. British Journal of General Practice. 2004;54:684–689. [PMC free article] [PubMed] [Google Scholar]
- 54.Cauchemez S et al. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Statistics in Medicine. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- 55.Hayden FG et al. Inhaled zanamivir for the prevention of influenza in families. Zanamivir Family Study Group. New England Journal of Medicine. 2000;343:1282–1289. doi: 10.1056/NEJM200011023431801. [DOI] [PubMed] [Google Scholar]
- 56.Hope-Simpson RE. The Transmission of Eepidemic Influenza. New York: Plenum Press; 1992. [Google Scholar]
- 57.Viboud C et al. Association of influenza epidemics with global climate variability. European Journal of Epidemiology. 2004;19:1055–1059. doi: 10.1007/s10654-004-2450-9. [DOI] [PubMed] [Google Scholar]
- 58.Longini M, Jr., Fine PE, Thacker SB. Predicting the global spread of new infectious agents. American Journal of Epidemiology. 1986;123:383–391. doi: 10.1093/oxfordjournals.aje.a114253. [DOI] [PubMed] [Google Scholar]
- 59.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 60.Glezen WP. Serious morbidity and mortality associated with influenza epidemics. Epidemiological Reviews. 1982;4:25–44. doi: 10.1093/oxfordjournals.epirev.a036250. [DOI] [PubMed] [Google Scholar]
- 61.Smith DJ et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 62.Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions. PLoS Medicine. 2006;3:e89. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cannell JJ et al. Epidemic influenza and vitamin D. Epidemiology and Infection. 2006;134:1129–1140. doi: 10.1017/S0950268806007175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dushoff J et al. Dynamical resonance can account for seasonality of influenza epidemics. Proceedings of the National Academy of Sciences USA. 2004;101:16915–16916. doi: 10.1073/pnas.0407293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiu SS et al. Influenza-related hospitalizations among children in Hong Kong. New England Journal of Medicine. 2002;347:2097–2103. doi: 10.1056/NEJMoa020546. [DOI] [PubMed] [Google Scholar]
- 66.Harper SA et al. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP) Morbidity and Mortality Weekly Report. Recommended Reports. 2004;53:1–40. [PubMed] [Google Scholar]
- 67.Demicheli V et al. Vaccines for preventing influenza in healthy adults. The Cochrane Database of Systematic Reviews. 2004 doi: 10.1002/14651858.CD001269.pub2. ; Art. No.: CD001269. [DOI] [PubMed] [Google Scholar]
- 68.Jefferson T et al. Assessment of the efficacy and effectiveness of influenza vaccines in healthy children: systematic review. Lancet. 2005;365:773–780. doi: 10.1016/S0140-6736(05)17984-7. [DOI] [PubMed] [Google Scholar]
- 69.Smith S et al. Vaccines for preventing influenza in healthy children. The Cochrane Database of Systematic Reviews. 2006 doi: 10.1002/14651858.CD004879.pub2. ; Art. No.: CD004879. [DOI] [PubMed] [Google Scholar]
- 70.Glezen WP, Simonsen L. Commentary: benefits of influenza vaccine in US elderly – new studies raise questions. International Journal of Epidemiology. 2006;35:352–353. doi: 10.1093/ije/dyi293. [DOI] [PubMed] [Google Scholar]
- 71.Rizzo C et al. Influenza-related mortality in the Italian elderly: no decline associated with increasing vaccination coverage. Vaccine. 2006;24:6468–6475. doi: 10.1016/j.vaccine.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 72.CDC. Influenza vaccination levels among persons aged >or=65 years and among persons aged 18–64 years with high-risk conditions – United States, 2003. Morbidity and Mortality Weekly Report. 2005;54:1045–1049. [PubMed] [Google Scholar]
- 73.Morbidity and Mortality Weekly Report. 2005;54:304–307. Estimated influenza vaccination coverage among adults and children – United, September 1, 2004–January 31, 2005. [PubMed] [Google Scholar]
- 74.Longini IM, Jr., Halloran ME. Strategy for distribution of influenza vaccine to high-risk groups and children. American Journal of Epidemiology. 2005;161:303–306. doi: 10.1093/aje/kwi053. [DOI] [PubMed] [Google Scholar]