Abstract
Actin carries out many of its cellular functions through its filamentous form, so understanding the detailed structure of actin filaments is an essential step in achieving a mechanistic understanding of actin function. The acromsomal bundle in the Limulus sperm has been shown to be a quasi-crystalline array with an asymmetric unit composed of a filament with 14 actin-scruin pairs. The bundle in its true discharge state penetrates the jelly coat of the egg. Our previous electron crystallographic reconstruction demonstrated that the actin filament cross-linked by scruin in this acrosomal bundle state deviates significantly from a perfect F-actin helix. In that study, the tertiary structure of each of the 14 actin protomers in the asymmetric unit of the bundle filament was assumed to be constant. In the current study, an actin filament atomic model in the acrosomal bundle has been refined by combining rigid body docking with multiple actin crystal structures from the Protein Data Bank (PDB) and constrained energy minimization. Our observation demonstrates that actin protomers adopt different tertiary conformations when they form an actin filament in the bundle. The scruin and bundle packing forces appear to influence the tertiary and quarternary conformations of actin in the filament of this biologically active bundle.
During the Limulus acrosomal reaction, a bundle of actin filaments cross-linked by scruin uncoils and supports an extension of the plasma membrane that mechanically penetrates through the jelly layer surrounding the egg membrane1. Biomechanical studies show the extended bundle has a Young’s modulus in the GPa range2. The coiled bundle stores approximately 10-13J of elastic energy2.
The extended form of the bundle, which is also known as the true discharge state, is a 3-D crystal obeying the symmetry of space group P21, with 14 actin and scruin protomers in the asymmetric unit3. The 9.5 Å cryoEM map of the bundle4 showed that its actin filament is not strictly helical as in the Holmes F-actin filament model5. A single model for an individual F-actin protomer (courtesy of K. Holmes) was fitted as a rigid body into each of the 14 actin positions in the filament in our cryoEM density map. We will refer this model as the homogeneous starting model. The average deviation between the orientation of our model and the Holmes F-actin model5 is 11.3° 4. These orientation deviations resulted in severe steric clashes between subunits in the homogeneous starting filament model. No refinement was carried out to relieve such clashes in our previous study4.
Each actin subunit has 4 subdomains (Fig 1a)6; subdomains 2 and 4 of one molecule interact with subdomains 1 and 3 of the molecule above it in the same strand. The most severe steric clashes in the homogeneous starting model occur at these contacts (supplemental movie 1). Other clashes occur between the hydrophobic plug5 of one subunit and subunits in the opposite strand. This is because the fit of the plug to the opposite strand is quite snug, and only a slight deviation from the canonical F-actin structure5 will cause a steric clash. To resolve all these clashes, an energy minimization on this starting model was carried out using X-PLOR7 in combination with Situs8 to constrain the orientations that were determined above with respect to our cryoEM density fit (Supplementary Methods). Fig. 1b illustrates the refined model based on the Holmes coordinates after minimization. Red color highlights the largest movements required by the energy minimization, involving residues for which the Cα needed to be moved by more than 2.5 Å during the minimization. The total number of such residues for this model is 103.
Figure 1.
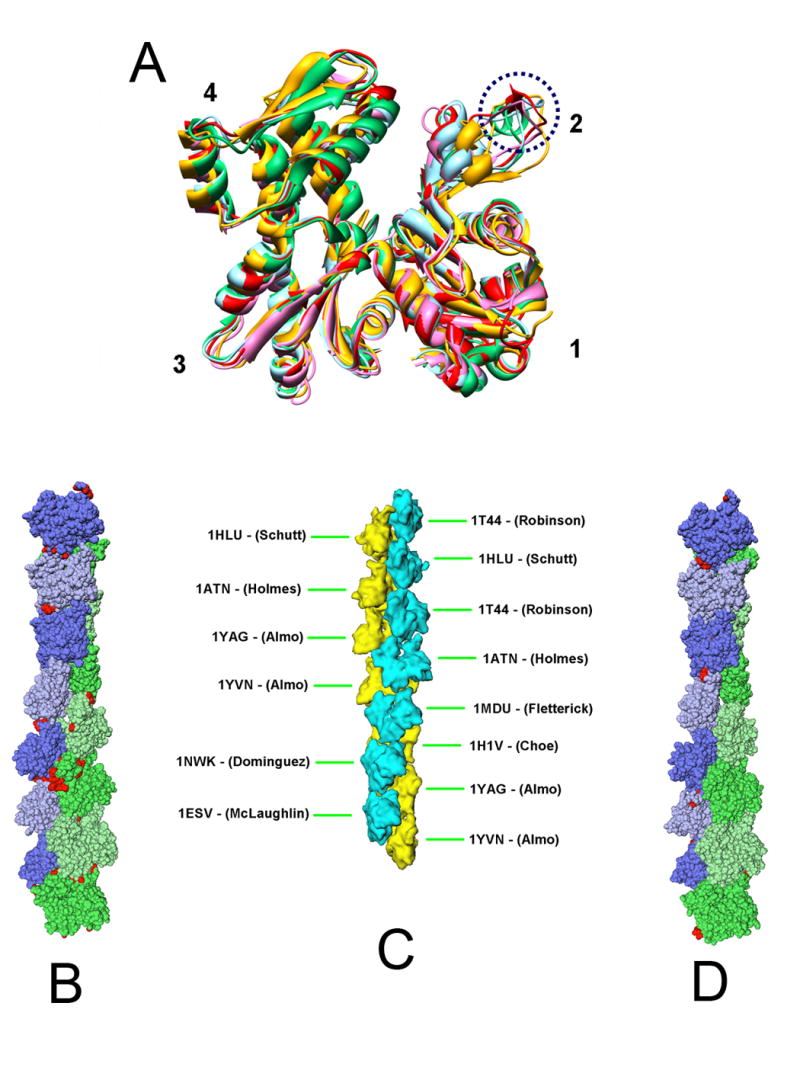
Actin crystal structure conformations and representation of fourteen protomers in the asymmetric unit of the acrosomal actin bundle as a space filling model. (a) Front view of several representative actin crystal structures illustrating the arrangement of the four actin subdomains as well as the difference in the secondary structures in subdomain 2, as discussed in the text (and circled by blue dash-line here). All the structures were aligned to subdomain 1 of 1ATN. The color scheme is as follows: 1ATN (red), 1HLU (gold), 1J6Z (green), 1YAG (ice blue), 2BTF (pink). 1HLU (gold) is obviously in a more open conformation comparing to other structures. (b) Cα representation of the asymmetric unit of the acrosomal bundle modeled with Holmes’ coordinates5 after energy minimization in the context of the bundle. The actin subunits are shown in a slightly different color and shade. Each amino-acid residue is represented by a sphere. Residues colored in red represent those whose Cα moved by more than 2.5Å from their original coordinates after energy minimization. (c) PDB codes and positions of the nine different structure coordinate sets (heterogeneous starting model) assigned to the 14 actins in the asymmetric unit. (d) Cα of the heterogeneous starting model for the acrosomal filament after energy minimization.
In addition to Holmes’ actin filament model, there exist a wealth of monomeric actin (G-actin) crystal structures that vary in their states of the bound nucleotide, crystallization buffers, co-crystallized proteins, and species of origin. These structures share a similar 4 subdomain organization but differ in details (Fig 1a). The spatial relationship of the subdomains in actin is characterized by twist and scissors angle differences with respect to the original Holmes crystal structure (Supplemental movie 2 and Fig 1a), but the deposited crystal structures, varied as they are, may still not express the entire range of possible conformations available to actin9. Based on these differences, we grouped actin structures into four classes: open and closed conformation between subdomains 1 plus 2 and subdomains 3 plus 4 (Fig 1a)10; in the closed conformation group, they can be further subdivided into twisted right (of subdomains 3 and 4 viewed from the left side of Fig 1a), twisted left, and untwisted with respect to the original Holmes structure (Supplemental movie 2). Also, as illustrated in Fig 1a, the backbone secondary structure of the same residues in subdomain 2 of some actin crystal structures is α-helical (eg. res 40-47 in 1J6Z), in others, β-sheet (eg. 42-44 in 1ATN), and in yet others, random coil (eg. 38-51 in 2BTF and 1HLU, 39-55 in 1YAG), or even disordered and unobserved.
To investigate whether the other actin crystal coordinates might provide more suitable models for the actin protomers in the asymmetric unit of the acrosomal bundle in our 9.5 Å cryoEM map4, we replaced Holmes coordinates with each of these other actin coordinates at each of the 14 actin positions in the asymmetric unit. These other coordinates were directly superimposed onto subdomain 1 of Holmes’ coordinates, which corresponds to the highest density in the averaged cryoEM map. The trial model was accepted if it had minimum clashes with the subunit located above it in the same strand. The identity of the upper subunit in testing these alternatives was of secondary importance, because subdomains 1 and 3 of most actin molecules are quite similar to each other (Fig 1a); subdomains 2 and 4 of the lower subunit are most critical to i) the differences in structure, ii) the causes of steric clashes with the molecule above it in the filament, and iii) the means of relieving these clashes by choosing a different model. Given the resolution of our cryoEM map (9.5Å), several actin structures from the same class (see above) fit the map equally well, and thus any of them is equally acceptable, so the reliability of a particular model in a particular position in the filament is limited to the class to which it belongs.
Using nine different actin coordinate sets for the 14 actin positions in the asymmetric unit (Figure 1c) resulted in significantly fewer steric clashes between neighboring subunits. We will refer to this model as the heterogeneous starting model. The improvements include fewer clashes between the neighboring subunits in the same strand, as well as between the hydrophobic plug of one subunit and the adjacent subunit in the opposite strand. These hydrophobic plug clashes were generally alleviated because the corresponding loops in these crystal structures do not protrude as far from the surface. There are still some remaining clashes between neighboring subunits belonging to the same strand (notably subunit 4 and 6; subunit 12 and 14); as well as between adjacent subunits in different strands (subunits 9 and 10). To resolve these clashes, the same energy minimization procedure was carried out as described above for the homogeneous starting model. Figure 1d shows our atomic model based on the heterogeneous starting model after energy minimization. The total number of residues with Cα movement larger than 2.5 Å after minimization for this heterogeneous starting model is 29, many fewer than for the homogeneous starting model (103 residues).
Figure 1b and 1d demonstrate visually that fewer residues need to be moved a large distance for the heterogeneous vs. the homogeneous starting model. In Figure 2 we show this quantitatively in histogram distributions of Cα atoms’ movement during energy minimization for both sets of models. Figure 2 illustrates that the movement of Cα atoms in the heterogeneous starting model has a smaller, tighter distribution compared to the homogeneous starting model. The average Cα movement in the heterogeneous starting model is 0.66 Å, while it is 0.87 Å for the homogeneous starting model.
Figure 2.
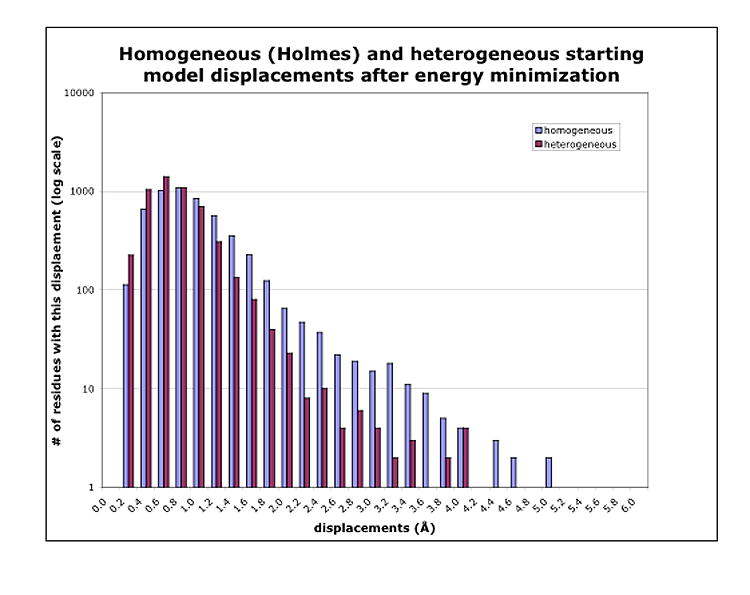
Histograms of the movement of the Cα coordinates in the acrosomal bundle filament after energy minimization using the homogeneous starting model for every actin subunit vs. using the heterogeneous starting model. X-coordinates are the shift-size bins in Å, frequency along the y-axis is shown on a log scale, to display the large range of numbers of Cα coordinates in the bins.
Our acrosomal actin filament model building process, including rigid-body docking and model refinement by constrained energy minimization, clearly demonstrates that the heterogeneous model is a more appropriate actin filament model, because it produces fewer clashes and smaller shifts than a single model when fitted to our experimental density. Our cryoEM map is consistent with multiple actin atomic models based on all four conformation groups classified above (and even deviating slightly further from these, as postulated by Klenchin9, to properly accommodate the acrosomal actin filament), indicating actin can adopt different conformations when it forms a filament in the acrosomal bundle. This observation is consistent with biochemical evidence which reveals that multiple conformations of actin protomers in a filament likely exist at any time11. It has been postulated that the hydrophobic plug of actin swings out and inserts into the opposite strand in the filament, stabilizing the filament structure5. However, in our heterogeneous model built on multiple actin crystal structures, most of the hydrophobic plugs are not in the extended form. It is noteworthy that recent biochemistry experiments11,12 support this observation; they demonstrated that rather than being in the extended position, the hydrophobic plug resides predominantly in a “parked” position within the filament, but is able to dynamically populate other conformational states. This represents a modification of previous conclusions that favored the hydrophobic plug extended conformation when it forms a filament13. Figure 3 shows the average density map of the 14 actin protomers in the acrosomal bundle asymmetric unit we originally reported4. Along with it are the Holmes F-actin model with the extended hydrophobic plug, and two representative x-ray coordinate sets of monomeric G-actin. Our density map shows that the average density in this hydrophobic plug region physically lies between the two kinds of conformation for this loop. These biochemical and structural observations support our heterogeneous model.
Figure 3.
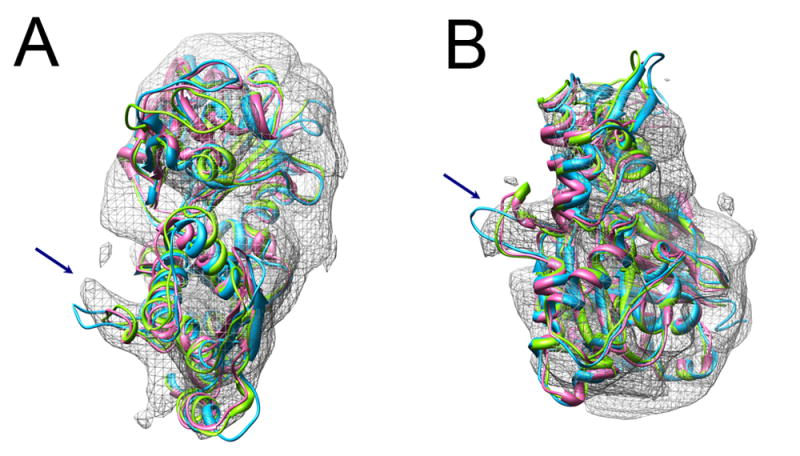
Average density map of the 14 actin protomers in the acrosomal bundle asymmetric unit4, along with the Holmes F-actin model (in cyan) and two representative x-ray structures of G-actin monomers (1MDU in light green, 1ESV in pink). Arrows indicate the hydrophobic loop that is extended toward the opposite strand in Holmes’ structure and “parked” against its own monomer in the x-ray structures. Our average density is intermediate between these two extremes. (a) Top view, looking approximately down the filament axis, (b) Side view.
We earlier established that the twist of the actin filament was affected at the subunit-to-subunit level by the interactions of actin with its binding protein, scruin4. We also showed that the orientation of each protomer in the asymmetric unit was unique, again presumably influenced by the environment in the bundle. Our observations here suggest that this influence may even extend to the tertiary structure of the actin subunit because of the better fit of the multiple crystal structures (Figure 1d). This observation implies that the environment of the actin (i.e. actin-actin, actin-scruin and scruin-scruin packing interactions in this bundle) has a great influence on the conformation of the individual actin subunits. Such conformational variation has also been observed in G-actin monomer crystals with different co-crystallization partners and crystallization conditions10. Furthermore, the average twist in F-actin filaments has been seen to vary in the presence of actin-binding proteins14 or during the process of actin polymerization15. All of these studies support the notion that each and every actin protomer in a filament can have a different tertiary and quaternary structure depending on its cellular environment. This is further emphasized in Supplementary Figure 1, which shows the variance map based on the 14-protomer average. The variance of the density of subdomains 2,3 and 4, and the relatively lower variance within subdomain 1, is a model-independent measure reinforcing the notion that each of the subdomains in the protomers of the actin filament may adopt a different conformation. In fact, the lower density of subdomains 2 and 4 in our published 14-protomer average map4 was one of the clues that led us to try different conformers of actin (some of which vary substantially in subdomains 2 and 4) in the first place. The accompanying PDB entry is thus the first one for which F-actin density has been used to fit atomic models into a biological filament without the assumption of perfect helical symmetry. It should be noted that these coordinates are different from the original ones for each of these molecules, since the molecular dynamics calculation shifted their coordinates to relieve the steric clashes in the acrosomal filament. Conformational flexibility of actin allows the filament to withstand strain and local distortions and still remain at least partly intact11. Since no crystal structure of scruin is available, a full pseudoatomic model of the actin filament with scruin in the context of the acrosomal bundle crystal cannot yet be established. This bundle is a natural system for demonstrating actin structural flexibility without bias because it is both biologically active and experimentally tractable.
Supplementary Material
Note: Supplementary information is available on the website.
Acknowledgments
We acknowledge support from National Institutes of Health (P41RR02250, DK35306, and 2PN2EY016525 through the NIH Roadmap for Medical Research) and National Science Foundation (EIA-0325004 and 1IIS-0705474).
Footnotes
Accession codes. Protein Data Bank: Coordinates have been deposited with accession code ***.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mahadevan L, Matsudaira P. Motility powered by supramolecular springs and ratchets. Science. 2000;288:95–100. doi: 10.1126/science.288.5463.95. [DOI] [PubMed] [Google Scholar]
- 2.Shin JH, Mahadevan L, Waller GS, Langsetmo K, Matsudaira P. Stored elastic energy powers the 60-microm extension of the Limulus polyphemus sperm actin bundle. J Cell Biol. 2003;162:1183–8. doi: 10.1083/jcb.200304006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid MF, Matsudaira P, Jeng TW, Jakana J, Towns-Andrews E, Bordas J, Chiu W. Crystallographic analysis of acrosomal bundle from Limulus sperm. J Mol Biol. 1991;221:711–25. doi: 10.1016/0022-2836(91)80082-6. [DOI] [PubMed] [Google Scholar]
- 4.Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–7. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- 5.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–9. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 6.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 7.Brunger A. X-PLOR (version 3.1): A System for X-ray crystallography and NMR. The Howard Hughes Medical Insttitute and Department of Molecular Biophysics and Biochemistry, Yale University; 1992. [Google Scholar]
- 8.Wriggers W, Birmanns S. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J Struct Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- 9.Klenchin VA, Khaitlina SY, Rayment I. Crystal structure of polymerization-competent actin. J Mol Biol. 2006;362:140–50. doi: 10.1016/j.jmb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Sablin EP, Dawson JF, VanLoock MS, Spudich JA, Egelman EH, Fletterick RJ. How does ATP hydrolysis control actin’s associations? Proc Natl Acad Sci U S A. 2002;99:10945–7. doi: 10.1073/pnas.152329899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shvetsov A, Stamm JD, Phillips M, Warshaviak D, Altenbach C, Rubenstein PA, Hideg K, Hubbell WL, Reisler E. Conformational dynamics of loop 262-274 in G- and F-actin. Biochemistry. 2006;45:6541–9. doi: 10.1021/bi052558v. [DOI] [PubMed] [Google Scholar]
- 12.Scoville D, Stamm JD, Toledo-Warshaviak D, Altenbach C, Phillips M, Shvetsov A, Rubenstein PA, Hubbell WL, Reisler E. Hydrophobic loop dynamics and actin filament stability. Biochemistry. 2006;45:13576–84. doi: 10.1021/bi061229f. [DOI] [PubMed] [Google Scholar]
- 13.Shvetsov A, Musib R, Phillips M, Rubenstein PA, Reisler E. Locking the hydrophobic loop 262-274 to G-actin surface by a disulfide bridge prevents filament formation. Biochemistry. 2002;41:10787–93. doi: 10.1021/bi020205f. [DOI] [PubMed] [Google Scholar]
- 14.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–81. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, Reisler E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci U S A. 2004;101:17664–8. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the website.


