Abstract
We have genotyped 14,436 nsSNPs and 897 MHC tagSNPs in 1000 independent cases of Ankylosing Spondylitis (AS), Autoimmune Thyroid Disease (AITD), Multiple Sclerosis and Breast Cancer. Comparing each of these diseases against a common control set of 1500 unselected healthy British individuals, we report initial association and independent replication of two new loci for AS, ARTS1 and IL23R, and confirmation of the previously reported AITD association with TSHR and FCRL3. These findings, enabled in part by expanding the control reference group with individuals from the other disease groups to increase statistical power, highlight important new possibilities for autoimmune regulation and suggest that IL23R may be a common susceptibility factor for the major ‘seronegative’ diseases.
Genome-wide association scans are currently revealing a number of new genetic variants for common diseases; eg1-11. We have recently completed the largest and most comprehensive scan conducted to date, involving genome-wide association studies of 2000 individuals from each of seven common disease cohorts and 3000 common control individuals using a dense panel of >500k markers12. In parallel with this scan, we conducted a study of 5,500 independent individuals with a genome-wide set of non-synonymous coding variants, an approach which has recently yielded new findings for Type 1 diabetes and Crohn’s disease and which has been proposed as an efficient complementary approach to whole genome scans13-15. Here we report several new replicated associations in our scan of nsSNPs in 1500 shared controls and 1000 individuals from each of 4 different diseases: Ankylosing Spondylitis (AS), Autoimmune Thyroid Disease/Graves’ Disease (AITD), Breast Cancer (BC) and Multiple Sclerosis (MS).
RESULTS
Initial genotyping was performed with a custom-made Infinium array (Illumina) and involved 14,436 nsSNPs (assays were synthesized for 16,078 nsSNPs). At the time of study inception, this comprised the complete set of experimentally validated nsSNPs with MAF > 1% in Caucasian samples. In addition, because three of the diseases were of autoimmune aetiology, we also typed a dense set of 897 SNPs throughout the major histocompatibility complex (MHC) which together with 348 nsSNPs in this region provided comprehensive tag SNP coverage (r2 ≥ 0.8 with all SNPs in ref16). Finally, 103 SNPs were typed in pigmentation genes specifically designed to differentiate between population groups. Similar to previous studies, our data revealed that detailed assessment of initial data is critically important to the process of association inference, as biases in genotype calling lead to clear inflation of false positive rates12,17. This inflation is exaggerated in nsSNP data because they tend to have lower allele frequencies than otherwise anonymous genomic SNPs and genotype calling is often most difficult for rare alleles. If only cursory filtering had been applied in the present case, a large number of striking false-positives would have emerged (Supplementary Figures 1-4). Table 1 displays the total number of SNPs and individuals remaining after genotype and sample quality control procedures (see Methods).
Table 1.
Number of individuals and SNPs tested in each cohort
| Cohort |
|||||
|---|---|---|---|---|---|
| AS | AITD | BC | MS | 58C | |
| Males | 610 | 138 | 0 | 271 | 732 |
| Females | 312 | 762 | 1004 | 704 | 734 |
| Number of SNPs genotyped | 15,436 | 15,436 | 15,436 | 15,436 | 15,436 |
| SNPs with Low GC score | 783 | 816 | 771 | 802 | 796 |
| SNPs with Low Genotyping | 133 | 206 | 124 | 218 | 186 |
| Monomorphic SNPs | 1,842 | 1,829 | 1,854 | 1,810 | 1,687 |
| SNPs with HW p < 10-7* | 129 | 74 | 104 | 97 | 132 |
| Differences in missing rate p < 10-4 | 51 | 101 | 172 | 309 | n/a |
| “Manual” Exclusions | 33 | 33 | 33 | 33 | 33 |
| Total Number of SNPs tested | 12,701 | 12,572 | 12,577 | 12,374 | |
Only SNPs with HW p < 10-7 in the 58C control group were excluded from analyses
Association with the MHC
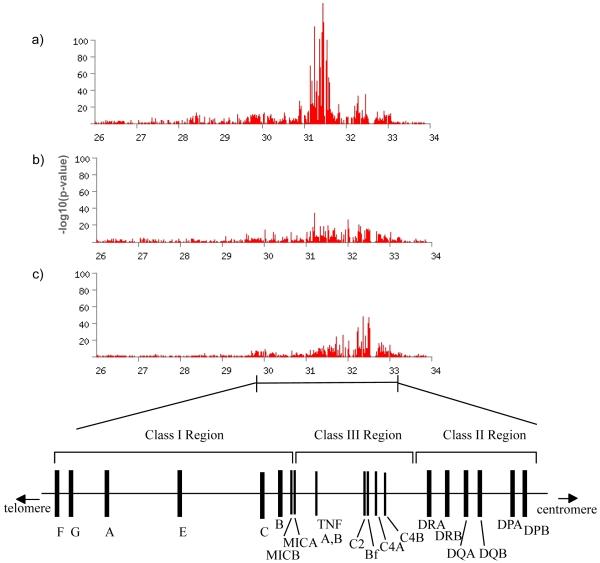
The strongest associations observed in the study were between SNPs in the MHC region and the three auto-immune diseases studied, AS, AITD and MS, with p-values of <10-20 for each disease (Figure 1). No association of the MHC was seen with BC (p > 10-4 across the region). For each of the autoimmune diseases, the maximum signal is centered around the known HLA-associated genes (ie, HLA-B in AS, HLA-DRB1 in MS and the MHC Class I-II for AITD), but in all cases it extends far beyond the specific associated haplotype(s). For example, in AS, association was observed at p < 10-20 across ~ 1.5 Mb. Given the well-known large effect of B27 with AS (odds ratio 100-200 in most populations), the extent of this association signal reflects the fact that with such large effects, even very distant SNPs in modest LD will reveal indirect evidence for association. Strong signals like these may also cloud the evidence for additional HLA loci18. Disentangling similar patterns of association within the MHC has proven extremely challenging in the past and will be addressed in future studies of these data. Here we focus specifically on the nsSNP results.
Figure 1.
Minus log10 p values for the Armitage test of trend for MHC association with Ankylosing Spondylitis (top panel), Auto-Immune Thyroid Disease (middle panel), and Multiple Sclerosis (bottom panel). Note in particular how evidence for association extends along very long regions of the MHC, reflecting statistical power to detect association even when linkage disequilibrium amongst SNPs is relatively low and/or the possibility of multiple disease-predisposing loci.
Association with nsSNPs
A major advantage of the WTCCC design is the availability of multiple disease cohorts which are similar in terms of ancestry and which have been typed on the same genetic markers12,17. Assuming that each disease has at least some genetic loci that differ between diseases, it should be possible to increase power to detect association by combining the other three case groups with the 58C controls19. For each disease we therefore conducted two primary analyses: (1) testing nsSNP associations for each disease against the 1958 Birth Cohort controls (58C); and (2) testing the same associations for each disease against an expanded reference group comprising the combined cases from the other three disease groups plus individuals from the 58C. A similar set of analyses was conducted for each of the autoimmune disorders against a reference group comprising 58C+BC, but the results were very similar to those for the fully expanded groups so here we describe the larger sample (see Supplementary Table 1). In addition, since it is possible that different autoimmune diseases share similar genetic etiologies, we also compared a combined AS, AITD and MS group against the combined set of BC patients and 58C controls. All of our analyses are reported without regard to specific treatment of population structure, as the degree of structure in our final genotype data is not severe (Genomic Control20 λ = 1.07 to 1.13 in the 58C-only datasets; λ = 1.03 to 1.06 in the expanded reference group comparisons, see Table 2), consistent with our recent findings from 17,000 UK individuals involving the same controls12.
Table 2.
Estimates of λ for Single and Combined Cohorts
| λ | ||
|---|---|---|
| Single Cohort | ||
| AS cases vs 1958 | 1.07 | |
| AITD cases vs 1958 | 1.12 | |
| BC cases vs 1958 | 1.13 | |
| MS cases vs 1958 | 1.12 | |
| Mixed Cohorts | ||
| AS cases vs All Others | 1.03 | |
| AITD cases vs All Others | 1.05 | |
| BC cases vs All Others | 1.04 | |
| MS cases vs All Others | 1.06 | |
| IMMUNE cases vs BC and 58C | 1.04 |
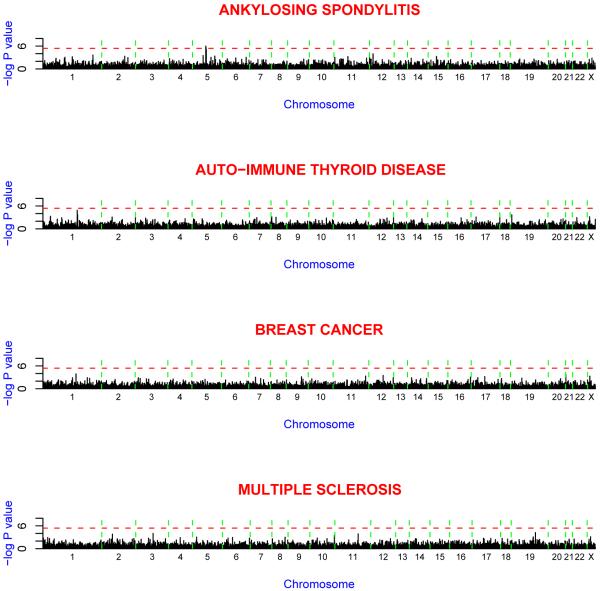
nsSNP association results (i.e. excluding the MHC region) for each of the four disease groups against the 58C controls are shown in Figure 2 and Table 3. Two SNPs on chromosome 5 reached a high-level of statistical significance for AS (rs27044: p = 1.0 × 10-6; rs30187: p = 3.0 × 10-6). This level of significance exceeds the 10-5-10-6 thresholds advocated for gene-based scans21, as well as the oft-used Bonferroni correction at p< 0.05 (see refs12,21 for a discussion of genome-wide association significance). Both of these markers reside in the gene for ARTS1 (ERAAP, ERAP1), a type II integral transmembrane aminopeptidase with diverse immunological functions. Four additional SNPs display significance at p< 10-4, with an increasing number of possible associations at more modest significance levels. Several of the more strongly associated SNPs or others in the same genes have previously been associated with the particular disease, and for others there exists functional evidence for their involvement in the particular condition. These include SNPs in the genes FCRL3 and FCRL5 in the case of AITD, IL23R in the case of AS, MEL-18 in the case of BC, and IL7R for MS. The complete list of single-marker association results is provided in Supplementary Table 1.
Figure 2.
Minus log10 p values for the Armitage test of trend for genome-wide association scans of ankylosing spondylitis, auto-immune thyroid disease, breast cancer and multiple sclerosis. The spacing between SNPs on the plot is uniform and does not reflect distances between the SNPs. The vertical dashed lines reflect chromosomal boundaries. The horizontal dashed lines display the cutoff of p = 10-6. Note that SNPs within the MHC are not included in this diagram.
Table 3.
nsSNPs outside the MHC which meet a point-wise significance level of p < 10-3 for the Cochran-Armitage test for trend
| Disease | SNP | Chromosome | Position (bp) | MAF | OR | χ 2 | p value | Gene |
|---|---|---|---|---|---|---|---|---|
| AS | ||||||||
| rs696698 | 1 | 74777462 | .04 | 1.84 | 11.13 | 8.5 × 10-4 | C1orf173 | |
| rs10494217 | 1 | 119181230 | .17 | 0.77 | 11.62 | 6.5 × 10-4 | TBX15 | |
| rs2294851 | 1 | 206966279 | .13 | 0.73 | 13.55 | 2.3 × 10-4 | HHAT | |
| rs8192556 | 2 | 182368504 | .01 | 0.45 | 12.24 | 4.7 × 10-4 | NEUROD1 | |
| rs16876657 | 5 | 78645930 | .02 | 3.10 | 13.05 | 3.0 × 10-4 | JMY | |
| rs27044 | 5 | 96144608 | .34 | 1.40 | 23.90 | 1.0 × 10-6 | ARTS-1 | |
| rs17482078 | 5 | 96144622 | .17 | 0.76 | 13.55 | 2.3 × 10-4 | ARTS-1 | |
| rs10050860 | 5 | 96147966 | .18 | 0.75 | 14.87 | 1.1 × 10-4 | ARTS-1 | |
| rs30187 | 5 | 96150086 | .40 | 1.33 | 21.82 | 3.0 × 10-6 | ARTS-1 | |
| rs2287987 | 5 | 96155291 | .18 | 0.75 | 14.31 | 1.6 × 10-4 | ARTS-1 | |
| rs2303138 | 5 | 96376466 | .10 | 1.58 | 19.41 | 1.1 × 10-5 | LNPEP | |
| rs11750814 | 5 | 137528564 | .16 | 0.77 | 10.99 | 9.1 × 10-4 | BRD8 | |
| rs11959820 | 5 | 149192703 | .02 | 0.49 | 12.41 | 4.3 × 10-4 | PPARGC1B | |
| rs907609 | 11 | 1813846 | .13 | 0.76 | 10.91 | 9.5 × 10-4 | SYT8 | |
| rs3740691 | 11 | 47144987 | .29 | 0.80 | 11.86 | 5.7 × 10-4 | ZNF289 | |
| rs11062385 | 12 | 297836 | .24 | 0.79 | 11.82 | 5.9 × 10-4 | JARID1A | |
| rs7302230 | 12 | 7179699 | .08 | 1.57 | 14.97 | 1.1 × 10-4 | CLSTN3 | |
| AITD | ||||||||
| rs10916769 | 1 | 20408244 | .17 | 0.76 | 12.10 | 5.0 × 10-4 | FLJ32784 | |
| rs6427384 | 1 | 154321955 | .18 | 1.43 | 18.97 | 1.3 × 10-5 | FCRL5 | |
| rs2012199 | 1 | 154322098 | .17 | 1.35 | 13.18 | 2.8 × 10-4 | FCRL5 | |
| rs6679793 | 1 | 154327170 | .22 | 1.33 | 14.69 | 1.3 × 10-4 | FCRL5 | |
| rs7522061 | 1 | 154481463 | .47 | 1.25 | 13.78 | 2.1 × 10-4 | FCRL3 | |
| rs1047911 | 2 | 74611433 | .15 | 1.34 | 11.24 | 8.0 × 10-4 | MRPL53 | |
| rs7578199 | 2 | 241912838 | .26 | 1.26 | 11.53 | 6.9 × 10-4 | HDLBP | |
| rs3748140 | 8 | 9036429 | .00 | 0.28 | 11.44 | 7.2 × 10-4 | PPP1R3B | |
| rs1048101 | 8 | 26683945 | .42 | 0.82 | 10.98 | 9.2 × 10-4 | ADRA1A | |
| rs7975069 | 12 | 132389146 | .30 | 0.80 | 12.06 | 5.2 × 10-4 | ZNF268 | |
| rs2271233 | 17 | 6644845 | .07 | 0.94 | 11.32 | 7.7 × 10-4 | TEKT1 | |
| rs2856966 | 18 | 897710 | .19 | 0.76 | 14.00 | 1.8 × 10-4 | ADCYAP1 | |
| rs7250822 | 19 | 2206311 | .04 | 1.97 | 13.83 | 2.0 × 10-4 | AMH | |
| rs2230018 | 23 | 44685331 | .14 | 1.41 | 11.55 | 6.8 × 10-4 | UTX | |
| BC | ||||||||
| rs4255378 | 1 | 151919300 | .48 | 1.25 | 14.70 | 1.3 × 10-4 | MUC1 | |
| rs2107732 | 7 | 44851218 | .10 | 1.40 | 10.96 | 9.3 × 10-4 | CCM2 | |
| rs4986790 | 9 | 117554856 | .07 | 1.54 | 11.46 | 7.1 × 10-4 | TLR4 | |
| rs2285374 | 11 | 118457383 | .38 | 0.82 | 12.25 | 4.7 × 10-4 | VPS11 | |
| rs7313899 | 12 | 54231386 | .03 | 2.10 | 13.02 | 3.1 × 10-4 | OR6C4 | |
| rs2879097 | 17 | 34143085 | .20 | 0.78 | 11.73 | 6.1 × 10-4 | MEL18 | |
| rs2822558 | 21 | 14593715 | .13 | 0.73 | 13.87 | 2.0 × 10-4 | ABCC13 | |
| rs2230018 | 23 | 44685331 | .14 | 1.40 | 12.14 | 4.9 × 10-4 | UTX | |
| MS | ||||||||
| rs17009792 | 2 | 74400978 | .02 | 0.44 | 14.41 | 1.5 × 10-4 | SLC4A5 | |
| rs1132200 | 3 | 120633526 | .15 | 0.73 | 15.22 | 9.6 × 10-5 | FLJ10902 | |
| rs6897932 | 5 | 35910332 | .23 | 0.80 | 11.04 | 8.9 × 10-4 | IL7R | |
| rs6470147 | 8 | 124517985 | .36 | 1.23 | 10.92 | 9.5 × 10-4 | FLJ10204 | |
| rs3818511 | 10 | 134309378 | .24 | 1.28 | 12.84 | 3.4 × 10-4 | INPP5A | |
| rs11574422 | 11 | 67970565 | .02 | 2.82 | 14.64 | 1.3 × 10-4 | LRP5 | |
| rs388706 | 19 | 49110533 | .48 | 1.22 | 11.19 | 8.2 × 10-4 | ZNF45 | |
| rs1800437 | 19 | 50873232 | .17 | 0.74 | 16.11 | 6.0 × 10-5 | GIPR | |
| rs2281868 | 23 | 69451484 | .50 | 1.26 | 11.38 | 7.4 × 10-4 | SAP102 |
The results for analyses involving the expanded reference group are presented in Supplementary Figure 5 and Supplementary Table 1. Many of the SNPs that showed moderate to strong evidence for association in the initial analysis revealed substantially increased significance with the larger reference group. Notably, these included the SNPs rs27044 (p = 4.0 × 10-8) and rs30187 (p = 2.1 × 10-7) in ARTS1, as well as several other variants in this gene. A second SNP, rs7302230 in the Calsyntenin-3 gene on chromosome 12, showed substantially stronger evidence for association in the expanded reference group analysis (p = 5.3 × 10-7) relative to the 58C-only results (p = 1.1 × 10-4). Results of the expanded group also showed elevated results for several SNPs which did not appear exceptional in the original (non-combined) analyses, including SNPs in several candidate genes such as sialoadhesin22 and complement receptor 1 for AS, PIK3R2 for MS, and C8B, IL17R and TYK2 in the combined autoimmune disease analysis. SNP rs3783941 in the thyroid stimulating hormone receptor (TSHR) gene emerged as amongst the most significant in the expanded reference group analyses of AITD (p = 2.1 × 10-5). Several polymorphisms in the TSHR have previously been associated with Graves’ disease23,24. This known association did not reach even the modest significance level of 10-3 in the original analyses, but adding an additional 3000 further reference samples delineated it from the background noise and further supports the original independent report.
ARTS1 association confirmed in an independent cohort
In order to validate the most exceptional findings from the initial study, we genotyped the ARTS1, CLSTN3 and LNPEP SNPs in 471 independent AS cases and 625 new controls (all North American Caucasian). Table 4 shows the results of the genes examined for AS. The data strongly suggest that the ARTS1 association is genuine. All ARTS1 nsSNPs reveal independent replication in the same direction of effect, with replication significance levels ranging from 4.7 × 10-4 to 5.1 × 10-5. When combined with the original samples, the results reveal striking evidence for association with AS (p = 1.2 × 10-8 to 3.4 × 10-10). The population attributable risk25 contributed by the most strongly associated marker in the North American dataset (rs2287987) is 26%.
Table 4.
Ankylosing Spondylitis Replication Results
| Gene | SNP | UK Cases | US Cases | All Cases | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case MAF | Control MAF | OR | p val | Case MAF | Control MAF | OR | p val | Case MAF | Control MAF | OR | p val | ||
|
| |||||||||||||
| ARTS1 | rs27044 | 0.34 | 0.27 | 1.40 | 1.0×10-6 | - | - | - | - | - | - | - | - |
| ARTS1 | rs17482078 | 0.17 | 0.22 | 0.76 | 2.3×10-4 | 0.15 | 0.21 | 0.65 | 5.1×10-5 | 0.16 | 0.22 | 0.70 | 1.2×10-8 |
| ARTS1 | rs10050860 | 0.18 | 0.23 | 0.75 | 1.2×10-4 | 0.15 | 0.22 | 0.66 | 8.8×10-5 | 0.17 | 0.22 | 0.71 | 7.6×10-9 |
| ARTS1 | rs30187 | 0.40 | 0.33 | 1.33 | 3.0×10-6 | 0.41 | 0.35 | 1.30 | 0.00047 | 0.41 | 0.34 | 1.40 | 3.4×10-10 |
| ARTS1 | rs2287987 | 0.18 | 0.22 | 0.75 | 1.6×10-4 | 0.15 | 0.21 | 0.66 | 8.4×10-5 | 0.17 | 0.22 | 0.71 | 1.0×10-8 |
| LNPEP | rs2303138 | 0.10 | 0.07 | 1.58 | 1.1×10-5 | 0.11 | 0.09 | 1.40 | 0.018 | 0.11 | 0.07 | 1.48 | 1.1×10-6 |
| CLSTN3 | rs7302230 | 0.08 | 0.05 | 1.57 | 1.1×10-4 | 0.06 | 0.05 | 1.10 | 0.56 | 0.07 | 0.05 | 1.30 | 0.0039 |
| IL23R | rs11209026 | 0.04 | 0.06 | 0.63 | 0.0017 | 0.038 | 0.06 | 0.63 | 0.014 | 0.04 | 0.06 | 0.63 | 4.0×10-6 |
| IL23R | rs1004819 | 0.35 | 0.30 | 1.20 | 0.0013 | 0.35 | 0.30 | 1.30 | 0.0045 | 0.35 | 0.30 | 1.20 | 1.1×10-5 |
| IL23R | rs10489629 | 0.43 | 0.45 | 0.90 | 0.062 | 0.39 | 0.47 | 0.72 | 4.2×10-5 | 0.41 | 0.46 | 0.83 | 0.00011 |
| IL23R | rs11465804 | 0.04 | 0.06 | 0.67 | 0.0019 | 0.049 | 0.06 | 0.68 | 0.04 | 0.04 | 0.06 | 0.68 | 0.0002 |
| IL23R | rs1343151 | 0.30 | 0.34 | 0.85 | 0.0077 | 0.29 | 0.36 | 0.71 | 6.7×10-5 | 0.30 | 0.34 | 0.80 | 1.0×10-5 |
| IL23R | rs10889677 | 0.36 | 0.31 | 1.20 | 0.00066 | 0.37 | 0.29 | 1.40 | 4.7×10-5 | 0.36 | 0.31 | 1.30 | 1.3×10-6 |
| IL23R | rs11209032 | 0.38 | 0.32 | 1.30 | 2.0×10-6 | 0.38 | 0.32 | 1.30 | 0.0013 | 0.38 | 0.32 | 1.30 | 7.5×10-9 |
| IL23R | rs1495965 | 0.49 | 0.44 | 1.20 | 0.0021 | 0.50 | 0.43 | 1.40 | 0.00019 | 0.49 | 0.44 | 1.20 | 3.1×10-6 |
Association was also confirmed with marker rs2303138 in the LNPEP gene, which lies 127kb 3′ of ARTS1. This marker is in strong LD with ARTS1 markers (D’ = 1, rs27044 - rs2303138). We tested the interdependence of the ARTS1 and LNPEP associations using conditional logistic regression. The remaining association at LNPEP is weak after controlling for ARTS1 (p = 0.01), whereas the association at ARTS1 remains strong after controlling for LNPEP (p = 2.7 × 10-6), suggesting that the LNPEP association may only be secondary to LD with a true association at ARTS1.
No association was seen with CLSTN3 in the confirmation set. The US controls exhibited the same allele frequency as the UK controls (5%) but the allele frequency in the US cases was less than that of the UK cases (6% vs 8%), suggesting no association in the US samples and substantially reducing the significance of the combined data. Calystenin-3 is a post-synaptic neuronal membrane protein, and is an unlikely candidate gene for involvement in inflammatory arthritis. The failure to replicate this association suggests that our replication sample size is insufficient to detect the modest effect or it was a false positive in the initial scan.
IL23R variants confer risk of AS
The IL23R variant rs11209026, whilst not striking in the initial nsSNP scan (p = 1.7 × 10-3), is of particular interest as it was recently associated with both Crohn’s Disease26,27 and psoriasis28, conditions which commonly co-occur with AS. To better define this association, 7 additional SNPs in IL23R were genotyped in the same 1000 British AS cases and 1500 58C controls as well as the North American Caucasian replication samples (Table 4). In the WTCCC dataset, strong association was seen in 7 of 8 genotyped SNPs (p ≤ 0.008, including the original nsSNP rs11209026), with the strongest association seen at rs11209032 (p = 2.0 × 10-6). In the replication dataset, association was observed with all genotyped SNPs (p ≤ 0.04), with peak association observed with marker rs10489629 (p = 4.2 × 10-5). In the combined dataset, the strongest association observed was with SNP rs11209032 (odds ratio 1.3, 95% CI 1.2 - 1.4, p = 7.5 × 10-9). The attributable risk for this marker in the replication cohort is 9%. Conditional logistic regression analyses did not reveal a single primary disease-associated marker, with residual association remaining after having controlled for association at the remaining SNPs. Considering only AS cases who self-reported as not having inflammatory bowel disease (n = 1066) the association remained strong and was still strongest at rs11209032 (p = 6.9 × 10-7), indicating that there is a primary association with AS and that the observed association was not due to coexistent clinical inflammatory bowel disease.
In contrast to the pleiotropic effects of IL23R, the ARTS1 association evidence appears confined to AS. We genotyped the five AS-associated SNPs in 755 British Crohn’s disease and 1011 ulcerative colitis cases, and 633 healthy controls. No association was seen with either UC or CD (Armitage trend p > 0.4 for all markers).
FCRL3 confirmed in AITD pathogenesis
In addition to the AS replications, we attempted to confirm and extend the FCRL3 association in AITD. The SNP rs7522061 in the FCRL3 gene was recently reported to be associated with AITD29 and two other autoimmune diseases, rheumatoid arthritis and systemic lupus erythematosus30. Our initial association evidence (p = 2.1 × 10-4) likely reflects the signal of the originally detected polymorphism since the level of linkage disequilibrium (LD) is high across this gene. In fact, the entire 1q21-q23 region (which includes another gene, FCRL5, flagged in our scan) has also been implicated in several autoimmune diseases including psoriasis and multiple sclerosis31,32.
On the basis of the original findings on 1q21-q23, the original cohort was increased from 1,000 to 2,500 Graves Disease (GD) cases and we used 2,500 different 1958 cohort controls. Eight SNPs that tagged the FCRL3 and FCRL5 gene regions were selected and typed in all 5,000 samples using an alternative genotyping platform. SNP rs3761959, which tags rs7522061 and rs7528684 (previously associated with RA and GD), was associated with GD in this extended cohort (Table 5), therefore, confirming the original result. In total, three of the seven FCRL3 SNPs showed some evidence for association (p < .05) with SNP rs11264798 being the most associated of the tag SNPs, p = 4.0 × 10-3. SNP rs6667109 in FCRL5, which tagged SNPs rs6427384, rs2012199 and rs6679793, all found to be weakly associated in the original study, showed little evidence of association in this extended cohort.
Table 5.
Auto-immune Thyroid Disease Replication Results
| Gene | SNP | Replication Cohort | Combined Cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Case MAF | Control MAF | OR | p val | Case MAF | Control MAF | OR | p val | ||
|
| |||||||||
| FCRL3 | rs3761959* | 0.48 | 0.45 | 0.90 | 0.029 | 0.49 | 0.45 | 0.87 | 9.4×10-3 |
| FCRL3 | rs11264794 | 0.42 | 0.46 | 1.10 | 0.079 | 0.42 | 0.46 | 1.12 | 0.013 |
| FCRL3 | rs11264793 | 0.27 | 0.24 | 0.87 | 0.020 | 0.26 | 0.24 | 0.90 | 0.044 |
| FCRL3 | rs11264798 | 0.44 | 0.49 | 1.21 | 4.0×10-3 | 0.44 | 0.49 | 1.22 | 1.6×10-5 |
| FCRL3 | rs10489678 | 0.19 | 0.20 | 1.07 | 0.30 | 0.20 | 0.20 | 1.04 | 0.43 |
| FCRL3 | rs6691569 | 0.28 | 0.28 | 1.03 | 0.60 | 0.29 | 0.29 | 1.00 | 0.93 |
| FCRL3 | rs2282284 | 0.062 | 0.058 | 0.92 | 0.013 | 0.062 | 0.058 | 0.93 | 0.47 |
| FCRL5 | rs6667109 | 0.17 | 0.15 | 0.91 | 0.18 | 0.18 | 0.15 | 0.85 | 7.7×10-2 |
This SNP tags the SNP rs7522061 which was flagged as associated with AITD in the WTCCC screen (p = 2.1 × 10-4)
DISCUSSION
Our scan of nsSNPs has identified and validated two new genes for AS (ARTS1 and IL23R), confirmed and extended markers in the TSHR and FCRL3 genes which have previously been associated with AITD, and provided a dense set of association data for AITD, AS and MS across the MHC region. The challenge now is to design functional studies that will reveal how variation in these genes translates into physiological processes that influence disease risk.
From a functional perspective, the ARTS1 and IL23R genes represent excellent biological candidates. ARTS1 has two known functions, either of which may explain its association with AS. Within the endoplasmic reticulum, ARTS1 is involved in trimming peptides to the optimal length for MHC Class I presentation33,34. AS is primarily an HLA Class I mediated autoimmune disease35, with >90% of cases carrying the HLA-B27 allele. How B27 increases risk of AS is unknown, but if the mechanism of association of ARTS1 with the disease is through effects on peptide presentation, this would inform research into the mechanism explaining the association of B27 with AS. The second known function of ARTS1 is that it cleaves cell surface receptors for the pro-inflammatory cytokines IL-1 (IL-1R2)36, IL-6 (IL-6Rα)37 and TNF (TNFR1)38, thereby downregulating their signaling. Genetic variants that alter the functioning of ARTS1 could therefore have pro-inflammatory effects through this mechanism.
As well as AS, polymorphisms in IL23R have been recently documented in Crohn’s Disease 26,27 and psoriasis 28, suggesting that this gene is a common susceptibility factor for the major ‘seronegative’ diseases, at least partially explaining their co-occurrence. IL-23R is a key factor in the regulation of a newly defined effector T-cell subset, TH17 cells. TH17 cells were originally identified as a distinct subset of T-cells expressing high levels of the pro-inflammatory cytokine IL-17 in response to stimulation, in addition to IL-1, IL-6, TNFa, IL-22 and IL-25 (IL-17E). IL-23 has been shown to be important in the mouse models experimental autoimmune encephalomyelitis39, collagen-induced arthritis40 and mouse models of inflammatory bowel disease41, but has not been studied in AS, either in humans or animal models of disease. These studies show that blocking IL-23 reduces inflammation in these models, suggesting that the IL23R variants associated with disease are pro-inflammatory. Successful treatment of Crohn’s disease has been reported with anti-IL-12p40 antibodies, which block both IL-12 and IL-23, as these cytokines share the IL-12p40 chain42. No functional studies of IL23R variants have been reported to date, and it is unclear to what extent findings in studies targeting IL-23 can be generalised to mechanisms by which IL23R variation affects disease susceptibility. Our genetic findings provide a major novel insight into the aetiopathogenesis of AS, and suggest that treatments targeting IL-23 may prove effective in this condition, but clearly much more needs to be understood about the mechanism underlying the observed association.
Despite the successful identification of the ARTS1 and IL23R genes, it is likely that additional real associations are either present in our data but with modest effect sizes, or that our focus on non-synonymous coding changes led us to miss real genes. For example, we found no evidence for association at even a nominal p < 0.05 in or within 2 Mb of the recently reported and validated breast cancer gene FGFR33,43 (2 nsSNPs in FGFR3 were included in our panel, rs1078816 and rs755793; the former yielded p = 0.12 and the latter was monomorphic in these samples), nor in or near any of the other suggestive breast cancer genes reported in refs3,43. Lack of statistical power in 1000 cases and 1500 controls is a likely contributor to this lack of replication, but some of these loci, notably FGFR3, appear to be intronic and thus would likely have been missed even with larger samples.
The issue of statistical power is emphasized in studies of non-synonymous coding changes, which have a greater number of rare variants than other genetic variants and thus will require even larger sample sizes unless the effect sizes are larger. Other analytical approaches, such as assessing evidence for association between clusters of rare variants rather than individual loci may prove highly informative in this regard44, but most of the nsSNPs available in this study exist either by themselves in each gene or with 1-2 others, which precludes these assessments (Supplementary Figure 6). In our analyses, ARTS1 was the only locus showing exceptional statistical significance in the scan of 1000 cases and 1500 controls, thus emphasizing the need for greater statistical power. We increased power by expanding the controls, or ‘reference set’, to include some or all of the other disease samples. In doing so, ARTS1 showed strong association evidence, the IL23R SNPs increased to a level that began to delineate them from background noise, and the AITD/TSHR confirmation emerged. This demonstration of increased statistical power by combining multiple datasets is timely given the international impetus to make genotype data available to the scientific community. Future investigations will be needed to assess the power vs confounding effects and the statistical corrections needed to combine more heterogeneous samples from broader sampling regions.
These results also highlight the question of how much information may be missed by focusing on coding SNPs rather than searching more broadly over the genome at large. This question is relevant because the trade-off between SNP panel selection and sample size to genotype is a salient factor in every genome-wide study design. In the HapMap data45, a substantial portion of the common non-synonymous variation in our nsSNP set is captured by available genome-wide panels (about 65% of common (MAF > 5%) nsSNPs in the Illumina Human NS-12 Beadchip are tagged with an r2 > .8 using the Affymetrix 500K chip, rising to 90% for the Illumina HumanHap300 which includes almost all of the nsSNPs from the NS-12 Beadchip). The four primary associated variants flagged in our study (i.e., in ARTS1, IL23R, TSHR and FCRL3) would have been detected using any of the genome-wide panels, since either the markers themselves or a SNP in high LD with them (r2 ≥ .78), are present on the genome-wide chips. This LD relationship also emphasizes the fact that observing an association with a nsSNP does not necessarily imply that the nsSNP is causal, as it may be indirectly associated with other genetic variants in or outside the gene. Given this high degree of overlap, the continuously increasing coverage of many available genotyping products and concomitant pressures to decrease assay costs, these data suggest that future gene-centric scans will be efficiently subsumed by the more encompassing and less hypothesis-driven genome-wide SNP panels.
METHODS
Subjects
Individuals included in the study were self-identified as white Europeans and came from mainland UK (England, Scotland and Wales, but not Northern Ireland). The 1500 control samples were from the British 1958 Birth Cohort (58C, also known as the National Child Development Study), which included all births in England, Wales and Scotland during one week in 1958. Recruitment details and diagnostic criteria for each of the four case groups and the 58C are further described in the Supplementary Methods online.
Sample QA/QC
Genome-wide Identity by State (IBS) sharing was calculated for each pair of individuals in the combined sample of cohorts in order to identify first and second degree relatives that might contaminate the study. One subject from any pair of individuals who shared < 400 genotypes IBS = 0 and/or > 80% alleles IBS was removed from all subsequent analyses (i.e. the individual with the most missing genotypes). In order to identify individuals who might have ancestries other than Western European, we merged each of our cohorts with the 60 CEU founder, 60 YRI founder, and 90 JPT and CHB individuals from the International HapMap Project45. We calculated genome-wide identity by state distances for each pair of individuals (i.e. one minus average IBS sharing) on those markers shared between HapMap and our non-synonymous panel, and then used the multidimensional scaling option in R to generate a two dimensional plot based upon individuals’ scores on the first two principal coordinates from this analysis (Supplementary Figure 2). Any WTCCC sample that was not present in the main cluster with the CEU individuals was excluded from subsequent analyses. Finally, any individual with >10% of genotypes missing was removed from the analysis. The number of individuals remaining after these quality control measures is displayed in Table 1.
Genotyping
We genotyped a total of 14,436 nsSNPs across the genome on all case and control samples. Because three of the diseases were of autoimmune etiology, we also typed an additional 897 SNPs within the MHC region, as well as 103 SNPs in pigmentation genes specifically designed to differentiate between population groups. SNP genotyping was performed with the Infinium I assay (Illumina) which is based on Allele Specific Primer Extension (ASPE) and the use of a single fluorochrome. The assay requires ~250 ng of genomic DNA which is first subjected to a round of isothermal amplification generating a “high complexity” representation of the genome with most loci represented at usable amounts. There are two allele specific probes (50mers) per SNP each on a different bead type; each bead type is present on the array 30 times on average (minimum 5), allowing for multiple independent measurements. We processed six samples per array. Clustering was performed with the GenCall software version 6.2.0.4 which assigns a quality score to each locus and an individual genotype confidence score (GC score) which is based on the distance of a genotype from the centre of the nearest cluster. First, we removed samples with more than 50% of loci having a score below 0.7 and then all loci with a quality score below 0.2. Post clustering we applied two additional filtering criteria: (i) omit individual genotypes with a GC score < 0.15 and (ii) remove any SNP which had more than 20% of its samples with GC scores below 0.15. The above criteria were designed so as to optimize genotype accuracy whilst minimizing uncalled genotypes.
Statistical Analysis
Markers that were monomorphic in both case and control samples, SNPs with > 10% missing genotypes, and SNPs with differences in the amount of missing data between cases and controls (p < 10-4 as assessed by χ2 test) were excluded from all analyses involving that case group only. In addition any marker which failed an exact test of Hardy-Weinberg equilibrium in controls (p < 10-7) was excluded from all analyses46.
Cochran-Armitage Tests for trend47 were conducted using the PLINK program48. For the present analyses, we used the significance thresholds of p < 10-4 – 10-6, as suggested for gene-based scans with stronger prior probabilities than scans of anonymous markers21. In the present context, the lower thresholds are similar to Bonferroni significance levels (Bonferroni-corrected p = .05 corresponds to nominal p = 3 × 10-6). The conditional logistic regression analyses involving the LNPEP and ARTS1 SNPs were performed using Purcell’s WHAP program49.
We manually rechecked the genotype calls of every nsSNP with an asymptotic significance level of p < 10-3 by inspecting raw signal intensity values and their corresponding automated genotype calls. Interestingly, this flagged an additional 33 markers with clear problems in genotype calling, which were subsequently excluded from all analyses (see Supplementary Figure 4 for an example). These results indicate that this genotyping platform generally yields highly accurate genotypes, but errors do occur and they can be distributed non-randomly between cases and controls despite stringent QC procedures. It is imperative to check the clustering of the most significant SNPs to ensure that evidence for associations are not a result of genotyping error.
Whilst great lengths were taken to ensure our samples were as homogenous as possible in terms of genetic ancestry, even subtle population substructure can substantially influence tests of association in large genome-wide analyses involving thousands of individuals50. We therefore calculated the genomic-control inflation factor, λ20, for each case-control sample as well as in the analyses where we combined the other case groups with the control individuals (Table 2). In general, values for λ were small (~1.1) indicating a small degree of substructure in UK samples which induces only a slight inflation of the test statistic under the null hypothesis, consistent with the results from our companion paper12. We, therefore, present uncorrected results in all analyses reported.
Supplementary Material
Acknowledgements
We would like to thank all the patients and controls who participated in this study.
AITD: We wish to thank the collection coordinators, Jackie Carr-Smith and all contributors to the AITD national DNA collection of index cases and family members from centres including Birmingham, Bournemouth, Cambridge, Cardiff, Exeter, Leeds, Newcastle and Sheffield. Principle leads for the AITD UK national collection are: Simon HS Pearce (Newcastle), Bijay Vaidya (Exeter), John H Lazarus (Cardiff), Amit Allahabadia (Sheffield), Mary Armitage (Bournemouth), Peter J Grant (Leeds), VK Chatterjee (Cambridge).
AS: We wish to thank the Arthritis Research Campaign (UK). MAB is funded by the National Health and Medical Research Council (Australia). TASC is funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases grants 1PO1-052915-01,RO1 AR046208, and RO1-AR048465,as well as by University of Texas at Houston CTSA grant UL1RR024148, Cedars-Sinai GCRC grant MO1-RR00425, The Rosalind Russell Center for Arthritis Research at The University of California San Francisco, and the Intramural Research Program, National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH. We thank Rui Jin for technical assistance and Laura Diekman, Lori Guthrie, Felice Lin and Stephanie Morgan for their study coordination.
BC: The Breast Cancer samples were clinically and molecularly curated with the assistance of Anthony Renwick, Anita Hall, Anna Elliot, Hiran Jayatilake, Tasnim Chagtai, Rita Barfoot, Patrick Kelly and Katarina Spanova. Our research is supported by US Army Medical Research and Material Command grant #W81XWH-05-1-0204, The Institute of Cancer Research and Cancer Research UK.
MS: Our work has been supported by the Wellcome Trust (Grant Ref 057097), the Medical Research Council (UK) (Grant Ref G0000648), the Multiple Sclerosis Society of Great Britain and Northern Ireland (Grant Ref 730/02) and the National Institutes of Health (USA) (Grant Ref 049477). AG is a Postdoctoral Fellow of the Research Foundation - Flanders (FWO - Vlaanderen).
Luana Galver and Paulina Ng at Illumina and Jonathan Morrison at the Sanger Institute contributed in the design of the nsSNP array. The DNA team of the JDRF/WT DIL. Thomas Dibling, Cliff Hind, and Douglas Simpkin at the Sanger Institute for carrying out the genotyping.
We also with to thank Sheila Bingham and the WTCCC Inflammatory Bowel Disease group for genotyping the ARTS1 markers in their replication samples.
*. Wellcome Trust Case Control Consortium
Management Committee: Paul R Burton1, David G Clayton2, Lon R Cardon3,55, Nick Craddock4, Panos Deloukas5, Audrey Duncanson6, Dominic P Kwiatkowski3,5, Mark I McCarthy3,7, Willem H Ouwehand8,9, Nilesh J Samani10, John A Todd2, Peter Donnelly (Chair)11
Analysis Committee: Jeffrey C Barrett3, Paul R Burton1, Dan Davison11, Peter Donnelly11, Doug Easton12, David M Evans3, Hin-Tak Leung2, Jonathan L Marchini11, Andrew P Morris3, Chris CA Spencer11, Martin D Tobin1, Lon R Cardon (Co-chair)3,55, David G Clayton (Co-chair)2
UK Blood Services & University of Cambridge Controls: Antony P Attwood5,8, James P Boorman8,9, Barbara Cant8, Ursula Everson13, Judith M Hussey14, Jennifer D Jolley8, Alexandra S Knight8, Kerstin Koch8, Elizabeth Meech15, Sarah Nutland2, Christopher V Prowse16, Helen E Stevens2, Niall C Taylor8, Graham R Walters17, Neil M Walker2, Nicholas A Watkins8,9, Thilo Winzer8, John A Todd2, Willem H Ouwehand8,9
1958 Birth Cohort Controls: Richard W Jones18, Wendy L McArdle18, Susan M Ring18, David P Strachan19, Marcus Pembrey18,20
Bipolar Disorder (Aberdeen): Gerome Breen21, David St Clair21; (Birmingham): Sian Caesar22, Katharine Gordon-Smith22,23, Lisa Jones22; (Cardiff): Christine Fraser23, Elaine K Green23, Detelina Grozeva23, Marian L Hamshere23, Peter A Holmans23, Ian R Jones23, George Kirov23, Valentina, Moskivina23, Ivan Nikolov23, Michael C O'Donovan23, Michael J Owen23, Nick Craddock23; (London): David A Collier24, Amanda Elkin24, Anne Farmer24, Richard Williamson24, Peter McGuffin24; (Newcastle): Allan H Young25, I Nicol Ferrier25
Coronary Artery Disease (Leeds): Stephen G Ball26, Anthony J Balmforth26, Jennifer H Barrett26, Timothy D Bishop26, Mark M Iles26, Azhar Maqbool26, Nadira Yuldasheva26, Alistair S Hall26; (Leicester): Peter S Braund10, Paul R Burton1, Richard J Dixon10, Massimo Mangino10, Suzanne Stevens10, Martin D Tobin1, John R Thompson1, Nilesh J Samani10
Crohn's Disease (Cambridge): Francesca Bredin27, Mark Tremelling27, Miles Parkes27; (Edinburgh): Hazel Drummond28, Charles W Lees28, Elaine R Nimmo28, Jack Satsangi28; (London): Sheila A Fisher29, Alastair Forbes30, Cathryn M Lewis29, Clive M Onnie29, Natalie J Prescott29, Jeremy Sanderson31, Christopher G Matthew29; (Newcastle): Jamie Barbour32, M Khalid Mohiuddin32, Catherine E Todhunter32, John C Mansfield32; (Oxford): Tariq Ahmad33, Fraser R Cummings33, Derek P Jewell33
Hypertension (Aberdeen): John Webster34; (Cambridge): Morris J Brown35, David G Clayton2; (Evry, France): Mark G Lathrop36; (Glasgow): John Connell37, Anna Dominiczak37; (Leicester): Nilesh J Samani10; (London): Carolina A Braga Marcano38, Beverley Burke38, Richard Dobson38, Johannie Gungadoo38, Kate L Lee38, Patricia B Munroe38, Stephen J Newhouse38, Abiodun Onipinla38, Chris Wallace38, Mingzhan Xue38, Mark Caulfield38; (Oxford): Martin Farrall39
Rheumatoid Arthritis: Anne Barton40, The Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee*, Ian N Bruce40, Hannah Donovan40, Steve Eyre40, Paul D Gilbert40, Samantha L Hilder40, Anne M Hinks40, Sally L John40, Catherine Potter40, Alan J Silman40, Deborah PM Symmons40, Wendy Thomson40, Jane Worthington40
Type 1 Diabetes: David G Clayton2, David B Dunger2,41, Sarah Nutland2, Helen E Stevens2, Neil M Walker2, Barry Widmer2,41, John A Todd2
Type 2 Diabetes (Exeter): Timothy M Frayling42,43, Rachel M Freathy42,43, Hana Lango42,43, John R B Perry42,43, Beverley M Shields43, Michael N Weedon 42,43, Andrew T Hattersley42,43; (London): Graham A Hitman44; (Newcastle): Mark Walker45; (Oxford): Kate S Elliott3,7, Christopher J Groves7, Cecilia M Lindgren3,7, Nigel W Rayner3,7, Nicolas J Timpson3,46, Eleftheria Zeggini3,7, Mark I McCarthy3,7
Tuberculosis (Gambia): Melanie Newport47, Giorgio Sirugo47; (Oxford): Emily Lyons3, Fredrik Vannberg3, Adrian VS Hill3
Ankylosing Spondylitis: Linda A Bradbury48, Claire Farrar49, Jennifer J Pointon49, Paul Wordsworth49, Matthew A Brown48,49
Autoimmune Thyroid Disease: Jayne A Franklyn50, Joanne M Heward50, Matthew J Simmonds50, Stephen CL Gough50
Breast Cancer: Sheila Seal51, Breast Cancer Susceptibility Collaboration (UK)*, Michael R Stratton51,52, Nazneen Rahman51
Multiple Sclerosis: Maria Ban53, An Goris53, Stephen J Sawcer53, Alastair Compston53
Gambian Controls (Gambia): David Conway47, Muminatou Jallow47, Melanie Newport47, Giorgio Sirugo47; (Oxford): Kirk A Rockett3, Dominic P Kwiatkowski3,5
DNA, Genotyping, Data QC and Informatics (Wellcome Trust Sanger Institute, Hinxton): Claire Bryan5, Suzannah J Bumpstead5, Amy Chaney5, Kate Downes2,5, Jilur Ghori5, Rhian Gwilliam5, Sarah E Hunt5, Michael Inouye5, Andrew Keniry5, Emma King5, Ralph McGinnis5, Simon Potter5, Rathi Ravindrarajah5, Pamela Whittaker5, David Withers5, Panos Deloukas5; (Cambridge): Hin-Tak Leung2, Sarah Nutland2, Helen E Stevens2, Neil M Walker2, John A Todd2
Statistics (Cambridge): Doug Easton12, David G Clayton2; (Leicester): Paul R Burton1, Martin D Tobin1; (Oxford): Jeffrey C Barrett3, David M Evans3, Andrew P Morris3, Lon R Cardon3,55; (Oxford): Niall J Cardin11, Dan Davison11, Teresa Ferreira11, Joanne Pereira-Gale11, Ingeleif B Hallgrimsdóttir11, Bryan N Howie11, Jonathan L Marchini11, Chris CA Spencer11, Zhan Su11, Yik Ying Teo3,11, Damjan Vukcevic11, Peter Donnelly11
PIs: David Bentley5,54, Matthew A Brown48,49, Lon R Cardon3,55, Mark Caulfield38, David G Clayton2, Alastair Compston53, Nick Craddock23, Panos Deloukas5, Peter Donnelly11, Martin Farrall39, Stephen CL Gough50, Alistair S Hall26, Andrew T Hattersley42,43, Adrian VS Hill3, Dominic P Kwiatkowski3,5, Christopher G Matthew29, Mark I McCarthy3,7, Willem H Ouwehand8,9, Miles Parkes27, Marcus Pembrey18,20, Nazneen Rahman51, Nilesh J Samani10, Michael R Stratton51,52, John A Todd2, Jane Worthington40
AITD Replication Group: Sarah L Mitchell50, Paul R Newby50, Oliver J Brand50, Jackie Carr-Smith50, Simon HS Pearce50 and Stephen CL Gough50
Genetic Epidemiology Group, Department of Health Sciences, University of Leicester, Adrian Building, University Road, Leicester, LE1 7RH, UK;
Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Department of Medical Genetics, Cambridge Institute for Medical Research, University of Cambridge, Wellcome Trust/MRC Building, Cambridge, CB2 0XY, UK;
Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK;
Department of Psychological Medicine, Henry Welcome Building, School of Medicine, Cardiff University, Heath Park, Cardiff CF14 4XN, UK;
The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK;
The Wellcome Trust, Gibbs Building, 215 Euston Road, London NW1 2BE, UK;
Oxford Centre for Diabetes, Endocrinology and Medicine, University of Oxford, Churchill Hospital, Oxford, OX3 7LJ, UK;
Department of Haematology, University of Cambridge, Long Road, Cambridge, CB2 2PT, UK;
National Health Service Blood and Transplant, Cambridge Centre, Long Road, Cambridge, CB2 2PT, UK;
Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Groby Road, Leicester, LE3 9QP, UK;
Department of Statistics, University of Oxford, 1 South Parks Road, Oxford OX1 3TG, UK;
Cancer Research UK Genetic Epidemiology Unit, Strangeways Research Laboratory, Worts Causeway, Cambridge CB1 8RN, UK;
National Health Service Blood and Transplant, Sheffield Centre, Longley Lane, Sheffield S5 7JN, UK;
National Health Service Blood and Transplant, Brentwood Centre, Crescent Drive, Brentwood, CM15 8DP, UK;
The Welsh Blood Service, Ely Valley Road, Talbot Green, Pontyclun, CF72 9WB, UK;
The Scottish National Blood Transfusion Service, Ellen's Glen Road, Edinburgh, EH17 7QT, UK;
National Health Service Blood and Transplant, Southampton centre, Coxford Road, Southampton, SO16 5AF, UK;
Avon Longitudinal Study of Parents and Children, University of Bristol, 24 Tyndall Avenue, Bristol, BS8 1TQ, UK;
Division of Community Health Services, St George's University of London, Crammer Terrace, London SW17 0RE, UK;
Institute of Child Health, University College London, 30 Guilford St, London WC1N 1EH, UK;
University of Aberdeen, Institute of Medical Sciences, Foresterhill, Aberdeen, AB25 2ZD, UK;
Department of Psychiatry, Division of Neuroscience, Birmingham University, Birmingham, B15 2QZ, UK;
Department of Psychological Medicine, Henry Wellcome Building, School of Medicine, Cardiff Univeristy, Heath Park, Cardiff CF14 4XN, UK;
SGDP, The Institute of Psychiatry, King's College London, De Crespigny Park Denmark Hill London SE5 8AF, UK;
School of Neurology, Neurobiology and Psychiatry, Royal Victoria Infirmary, Queen Victoria Road, Newcastle upon Tyne, NE1 4LP, UK;
LIGHT and LIMM Research Institutes, Faculty of Medicine and Health, University of Leeds, Leeds, LS1 3EX, UK;
IBD Research Group, Addenbrooke's Hospital, University of Cambridge, Cambridge, CB2 2QQ, UK;
Gastrointestinal Unit, School of Molecular and Clinical Medicine, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU UK;
Department of Medical & Molecular Genetics, King's College London School of Medicine, 8th Floor Guy's Hospital, London, SE1 9RT, UK;
Institute for Digestive Diseases, University College London Hospitals Trust, London NW1 2BU, UK;
Department of Gastroenterology, Guy's and St Thomas' NHS Foundation Trust, London, SE1 7EH, UK;
Department of Gastroenterology & Hepatology, University of Newcastle upon Tyne, Royal Victoria Infirmary, Newcastle upon Tyne, NE1 4LP, UK;
Gastroenterology Unit, Radcliffe Infirmary, University of Oxford, Oxford, OX2 6HE, UK;
Medicine and Therapeutics, Aberdeen Royal Infirmary, Foresterhill, Aberdeen, Grampian AB9 2ZB, UK;
Clinical Pharmacology Unit and the Diabetes and Inflammation Laboratory, University of Cambridge, Addenbrookes Hospital, Hills Road, Cambridge CB2 2QQ, UK;
Centre National de Genotypage, 2 Rue Gaston Cremieux, Evry, Paris 91057;
BHF Glasgow Cardiovascular Research Centre, University of 28 Glasgow, 126 University Place, Glasgow, G12 8TA, UK;
Clinical Pharmacology and Barts and The London Genome Centre, William Harvey Research Institute, Barts and The London, Queen Mary's School of Medicine, Charterhouse Square, London EC1M 6BQ, UK;
Cardiovascular Medicine, University of Oxford, Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK;
arc Epidemiology Research Unit, University of Manchester, Stopford Building, Oxford Rd, Manchester, M13 9PT, UK;
Department of Paediatrics, University of Cambridge, Addenbrooke's Hospital, Cambridge, CB2 2QQ, UK;
Genetics of Complex Traits, Institute of Biomedical and Clinical Science, Peninsula Medical School, Magdalen Road, Exeter EX1 2LU, UK;
Diabetes Genetics, Institute of Biomedical and Clinical Science, Peninsula Medical School, Barrack Road, Exeter EX2 5DU UK;
Centre for Diabetes and Metabolic Medicine, Barts and The London, Royal London Hospital, Whitechapel, London, E1 1BB , UK;
Diabetes Research Group, School of Clinical Medical Sciences, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, UK;
The MRC Centre for Causal Analyses in Translational Epidemiology, Bristol University, Canynge Hall, Whiteladies Rd, Bristol BS2 8PR, UK;
MRC Laboratories, Fajara, The Gambia;
Diamantina Institute for Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Woolloongabba, Qld 4102, Australia;
Botnar Research Centre, University of Oxford, Headington, Oxford OX3 7BN, UK;
Department of Medicine, Division of Medical Sciences, Institute of Biomedical Research, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK;
Section of Cancer Genetics, Institute of Cancer Research, 15 Cotswold Road, Sutton, SM2 5NG, UK;
Cancer Genome Project, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK;
Department of Clinical Neurosciences, University of Cambridge, Addenbrooke's Hospital, Hills Road, Cambridge CB2 2QQ, UK;
PRESENT ADDRESS: Illumina Cambridge, Chesterford Research Park, Little Chesterford, NR Saffron Walden, Essex, CB10 1XL, UK;
PRESENT ADDRESS: Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, Washington 98109 USA.
+. The Australo-Anglo-American Spondylitis Consortium (TASC)
John D. Reveille1, Xiaodong Zhou1, Linda A Bradbury2, Anne-Marie Sims2, Alison Dowling2, Jacqueline Taylor2, Tracy Doan2, Lon R. Cardon3,55, John C Davis4, Jennifer J Pointon5, Laurie Savage6, Michael M Ward7, Thomas L Learch8, Michael H Weisman9, Paul Wordsworth5, Matthew A Brown2,5
Rheumatology and Clinical Immunogenetics, University of Texas-Houston Medical School, Houston, United States;
Diamantina Institute for Cancer, Immunology and Metabolic Medicine, University of Queensland, Brisbane, Australia;
Statistical Genetics, Wellcome Trust Centre for Human Genetics, Oxford, United Kingdom;
Rheumatology, University of California, San Francisco, San Francisco, United States;
Botnar Research Centre, University of Oxford, Oxford, United Kingdom;
The Spondylitis Association of America, Sherman, Oaks, CA;
National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, United States;
Department of Radiology, Cedars-Sinai Medical Centre, Los Angeles, United States;
Department of Medicine/Rheumatology, Cedars-Sinai Medical Centre, Los Angeles, United States.
Footnotes
Membership of the BRAGGS and Breast Cancer Susceptibility Collaboration (UK) is listed in the Supplementary Information.
References
- 1.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 3.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 6.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 8.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 9.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 12.WTCCC Genome-wide association studies of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–683. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 14.Jorgenson E, Witte JS. Coverage and power in genomewide association studies. Am J Hum Genet. 2006;78:884–8. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–9. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 16.Miretti MM, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–46. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 18.Sims AM, et al. Non-B27 MHC associations of ankylosing spondylitis. Genes Immun. 2007;8:115–23. doi: 10.1038/sj.gene.6364362. [DOI] [PubMed] [Google Scholar]
- 19.McGinnis R, Shifman S, Darvasi A. Power and efficiency of the TDT and case-control design for association scans. Behav Genet. 2002;32:135–44. doi: 10.1023/a:1015205924326. [DOI] [PubMed] [Google Scholar]
- 20.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 21.Thomas DC, Clayton DG. Betting odds and genetic associations. J Natl Cancer Inst. 2004;96:421–3. doi: 10.1093/jnci/djh094. [DOI] [PubMed] [Google Scholar]
- 22.Jiang HR, et al. Sialoadhesin promotes the inflammatory response in experimental autoimmune uveoretinitis. J Immunol. 2006;177:2258–64. doi: 10.4049/jimmunol.177.4.2258. [DOI] [PubMed] [Google Scholar]
- 23.Dechairo BM, et al. Association of the TSHR gene with Graves’ disease: the first disease specific locus. Eur J Hum Genet. 2005;13:1223–30. doi: 10.1038/sj.ejhg.5201485. [DOI] [PubMed] [Google Scholar]
- 24.Hiratani H, et al. Multiple SNPs in intron 7 of thyrotropin receptor are associated with Graves’ disease. J Clin Endocrinol Metab. 2005;90:2898–903. doi: 10.1210/jc.2004-2148. [DOI] [PubMed] [Google Scholar]
- 25.Miettinen O. Proportion of disease caused or prevented by a given exposure, trait or intervention. Am J Epidemiol. 1974;99:325–332. doi: 10.1093/oxfordjournals.aje.a121617. [DOI] [PubMed] [Google Scholar]
- 26.Duerr RH, et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science. 2006 doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tremelling M, et al. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology. 2007;132:1657–64. doi: 10.1053/j.gastro.2007.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmonds MJ, et al. Contribution of single nucleotide polymorphisms within FCRL3 and MAP3K7IP2 to the pathogenesis of Graves’ disease. J Clin Endocrinol Metab. 2006;91:1056–61. doi: 10.1210/jc.2005-1634. [DOI] [PubMed] [Google Scholar]
- 30.Kochi Y, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–85. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capon F, et al. Fine mapping of the PSORS4 psoriasis susceptibility region on chromosome 1q21. J Invest Dermatol. 2001;116:728–30. doi: 10.1046/j.1523-1747.2001.01311.x. [DOI] [PubMed] [Google Scholar]
- 32.Dai KZ, et al. The T cell regulator gene SH2D2A contributes to the genetic susceptibility of multiple sclerosis. Genes Immun. 2001;2:263–8. doi: 10.1038/sj.gene.6363774. [DOI] [PubMed] [Google Scholar]
- 33.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–12. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saveanu L, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–97. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 35.Brown MA, et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis. 1996;55:268–70. doi: 10.1136/ard.55.4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui X, Rouhani FN, Hawari F, Levine SJ. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J Immunol. 2003;171:6814–9. doi: 10.4049/jimmunol.171.12.6814. [DOI] [PubMed] [Google Scholar]
- 37.Cui X, Rouhani FN, Hawari F, Levine SJ. An aminopeptidase, ARTS-1, is required for interleukin-6 receptor shedding. J Biol Chem. 2003;278:28677–85. doi: 10.1074/jbc.M300456200. [DOI] [PubMed] [Google Scholar]
- 38.Cui X, et al. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J Clin Invest. 2002;110:515–26. doi: 10.1172/JCI13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–8. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 40.Murphy CA, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–7. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannon PJ, et al. Anti-interleukin-12 antibody for active Crohn’s disease. N Engl J Med. 2004;351:2069–79. doi: 10.1056/NEJMoa033402. [DOI] [PubMed] [Google Scholar]
- 43.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 45.A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armitage P. Test for linear trend in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 48.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 50.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004 doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.