Abstract
Originally designed to block the prenylation of oncogenic Ras, inhibitors of protein farnesyltransferase currently in pre-clinical and clinical trials are showing efficacy in cancers with normal Ras. Blocking protein prenylation has also shown promise in the treatment of malaria, Chagas disease, and progeria syndrome. A better understanding of the mechanism, targets and in vivo consequences of protein prenylation are needed to elucidate the mode of action of current PFTase inhibitors and to create more potent and selective compounds. Caged enzyme substrates are useful tools for understanding enzyme mechanism and biological function. Reported here is the synthesis and characterization of caged substrates of PFTase. The caged isoprenoid diphosphates are poor substrates prior to photolysis. The caged CAAX peptide is a true catalytically caged substrate of PFTase in that it is to not a substrate, yet is able to bind to the enzyme as established by inhibition studies and x-ray crystallography. Irradiation of the caged molecules with 350 nm light readily releases their cognate substrate, and their photolysis products are benign. These properties highlight the utility of those analogues towards a variety of in vitro and in vivo applications.
Keywords: protein prenylation, prenyltransferase, caged substrate, CAAX peptide, isoprenoid diphosphate, farnesyl, photorelease
INTRODUCTION
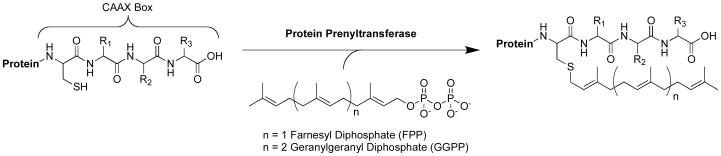
Protein prenylation is a post-translational modification involving the attachment of a C15 farnesyl or C20 geranylgeranyl group to a C-terminal cysteine residue through a thioether linkage[1] (Figure 1). Initially, this cysteine residue is located within a CAAX consensus sequence where C is the cysteine, A is typically aliphatic, and X is serine, methionine, or glutamine, which is prenylated by PFTase; or leucine, isoleucine, or valine; which is prenylated by PGGTase[2]. Protein prenyltransferases function through an ordered sequential mechanism, where the isoprenoid diphosphate binds first, creating a portion of the binding pocket for the CAAX peptide[3, 4]. The cysteine's thiol group ionizes upon coordination with the active site Zn2+, and reaction with the C1 carbon of the isoprenoid occurs[5]. Whether this reaction occurs as an associative or dissociative type mechanism is currently being investigated[6-9].
Figure 1.
Reaction catalyzed by protein prenyltransferase.
Farnesylation of Ras, a protein implicated in 30% of all human cancers, with the highest frequencies being mutated K-Ras in pancreatic (90%), colorectal (50%), and lung (30%) cancer[10], is essential for communication with its downstream effectors and hence, its oncogenic properties[11]. Following this discovery, a massive research effort focused on developing inhibitors of PFTase in an attempt to thwart Ras-induced cancer. To date, there are two PFTase inhibitors in phase II clinical trials and one in phase III, which show modest results[12]. Part of this is due to the fact that K-Ras is an alternative substrate for PGGTase. Upon blocking the farnesylation of K-Ras, it is alternatively geranylgeranylated thus rescuing its oncogenic activity. While inhibition of farnesylation is fairly non-toxic, dual inhibition of PGGTase and PFTase is quite toxic[13]. Despite being designed for cancers expressing mutated Ras, it was discovered that PFTase inhibitors are also effective in cancers expressing normal Ras[14]. In order to comprehend this interesting phenomenon, a greater understanding of the prenylation reaction as well as the in vivo consequences of protein prenylation is needed. Such knowledge should facilitate the creation of more potent and selective inhibitors.
Photoactivatable enzymes and enzyme substrates are well-established tools for the investigation of biological function and catalytic mechanism[15-20]. Using a photocleavable yeast peptide pheromone (α-factor, WHWLQLKPGQPMY), Parker et al. recently demonstrated that the arrest of MATa cells at the G1 stage of division is possible with brief exposure to light[21]. Such control allows for synchronizing yeast cell cultures and investigating signaling processes related to morphogenesis and transcription. Rather and coworkers were able to reversibly inactivate the bacterial Sssl DNA methyltransferase by incorporating a dimethoxynitrobenzyl photolabile group onto an active site cysteine residue[22]. This caged enzyme may be useful for controlling DNA methylation patterns as well as time-resolved structure analysis. One of the earliest examples of the use of a caged enzyme substrate for time-resolved x-ray crystallography is from the work of Schlichting and coworkers[23]. Crystals of Ha-Ras p21 with nitrophenethyl-caged GTP were used to investigate the conformational changes of p21 upon GTP hydrolysis. Using Laue diffraction methods, they were able to observe the short-lived p21:GTP complex as well as structural changes that occurred in the regions of the enzyme believed to interact with the GTPase-activating protein.
To achieve temporal control of prenyltransferase activity, similar to the examples reported above, we have designed a series of photoactivatable substrate analogues. Here we describe the synthesis and characterization of several caged substrates for protein farnesyltransferase. The first generation of compounds contains a nitrobenzyl-based photolabile group incorporated at the distal phosphate of the isoprenoid diphosphate substrate. The second generation has a nitrobenzyl-based photolabile group linked to the cysteine thiol of the CAAX peptide substrate. The kinetics of uncaging these substrates were investigated and their interactions with PFTase were studied by a variety of enzymatic assays and x-ray crystallography.
MATERIALS AND METHODS
General
All synthetic reactions were conducted under air atmosphere and stirred magnetically, unless otherwise indicated. Dowex 50W-X8 ion-exchange resin was obtained from BioRad. Analytical TLC was performed on precoated (250 μm) silica gel 60 F-254 plates from Merck. Visualization was done under UV irradiation or by subjecting the plates to aqueous potassium permanganate (KMnO4) or ethanolic phosphomolybdic acid (PMA) solutions followed by heating. Octylfunctionalized silica gel was purchased from Aldrich. Flash chromatography (silica gel, 60-120 mesh) was obtained from Mallinckrodt Inc. Anhydrous CH2Cl2 and DMF were purchased from Aldrich. Preparative HPLC separations were performed using a Beckman model 127/166 instrument, equipped with a UV detector and a Phenomenex C18 column (Luna, 10 μm, 10 × 250 mm) with a 5 cm guard column. MS spectra for evaluations of caged compounds and their precursors were obtained on a Bruker BioTOF II instrument. Deuterated NMR solvents were used as obtained from Cambridge Isotope Laboratories, Inc. All other chemical reagents were purchased from Sigma Aldrich Chemical Co. unless otherwise noted. 1H NMR spectra were obtained at 300 or 500 MHz. 13C NMR spectra were obtained at 75 MHz. 31P NMR spectra were obtained at 121 MHz. All NMR spectra were obtained on Varian instruments at 25°C. Chemical shifts are reported in ppm and J values are given in Hz. Fluorescence measurements were performed with a Varian Cary Eclipse Fluorescence Spectrophotometer. N-Dansyl-GCVIA and N-Dansyl-GCVLS were prepared as described by Gaon et al[24]. Analytical HPLC was performed on a Beckman model 125/166 instrument, equipped with a UV detector, fluorescence detector, and a Varian C18 column (Microsorb-MV, 5 μm, 4.6 × 250 mm). Bandwidth filters of 305-395 nm and 495-550 nm were utilized for fluorescence excitation and emission, respectively.
β-(2-nitrobenzyl)farnesyl diphosphate (5)
2-nitrobenzylphosphate (4) (93 mg, 0.40 mmol) and 1,1'-carbonyldiimidazole (78 mg, 0.48 mmol) were dissolved in anhydrous DMF (2.0 mL) and stirred at rt for 2 h. 2 (130 mg, 0.40 mmol) was then added and the mixture was stirred at rt for 4 days upon which DMSO (2.0 mL) was added. The resulting solution continued to stir at rt for another 3 days. 8.0 mL of isopropyl alcohol:25 mM NH4HCO3 (1:49) was then added, and the reaction mixture was passed through an ion exchange resin (Dowex W-X8 NH +4 form), and eluted with 25 mM NH4HCO3. The eluent was lyophilized to give a white solid. RP-HPLC of the solid (solvent A: 25 mM NH4HCO3, solvent B: CH3CN) afforded 5 (60 mg, 27% yield). 1H-NMR (D2O, 300 MHz): δ 1.28 (3H, s), 1.33 (6H, s), 1.42 (3H, s), 1.71 (8H, m), 1.16 (2H, t, J = 5.7 Hz), 4.82, (2H, dd, J = 6.6 Hz, 6.0 Hz), 5.00 (1H, t, J = 6.6 Hz), 5.12 (2H, d, J = 6.6 Hz), 7.12 (1H, t, J = 7.8 Hz), 7.51 (1H, t, J = 7.5 Hz), 7.69 (2H, d, J = 8.4 Hz). 31P-NMR (D2O, 121.4 MHz): δ -7.88 (1P, d, J = 20 Hz), -8.27 (1P, d, J = 20 Hz); HRMS calcd for C22H32NO9P2 [M-H]- 516.1552; found 516.1555.
Di-t-butyl 1-(4,5-dimethoxy-2-nitrophenyl)ethyl phosphite (7)
Di-t-butyl diisopropylphosphoramidite (430 mg, 1.5 mmol) was added dropwise to a mixture of 4,5-dimethoxy-α-methyl-2-nitrobenzyl alcohol (6, 230 mg, 1.0 mmol) and tetrazole (140 mg, 2.0 mmol) in 10 mL of dry CH2Cl2. The resulting mixture was stirred at rt for 3 h in the dark, after which 30 mL of a 10% NaHCO3 in H2O was added. Extraction with EtOAc (2 × 20 mL), drying over MgSO4 and concentration in vacuo gave a clear residue. Flash chromatography (hexanes/EtOAc, 5/1) yielded 7 (280 mg, 70% yield). 1H-NMR (CDCl3, 200 MHz): δ 7.49 (1H, s), 7.28 (1H, s), 6.03 (1H, m), 1.51 (3H, d, J = 6.4 Hz), 1.32 (9H, s), 1.15 (9H, s). 13C-NMR (CDCl3, 50 MHz): 25.27 (d, J = 2.8 Hz), 30.74 (d, J = 8.5 Hz), 31.03 (d, J = 9.1 Hz), 56.30 (d, J = 5.6 Hz), 64.47 (d, J = 5.0 Hz), 76.04 (d, J = 9.1 Hz), 106.89, 109.88, 137.10, 138.91, 147.30, 153.28. 31P-NMR (CDCl3, 81 MHz): δ 136.
1-(4,5-dimethoxy-2-nitrophenyl)ethyl dihydrogen phosphate (8)
0.15 M HCl in decane (0.30 mL) was added to 7 (280 mg, 0.69 mmol) in 10 mL dry CH2Cl2 and stirred at rt for 1 h in the dark. Solvent and excess of t-Bu-OOH were removed by vacuum to afford an oily residue. 1.0 M HCl in dioxane (8.0 mL) was added and the mixture was stirred at rt for 4 h in the dark and concentrated in vacuo. 50 mL of H2O was added and extracted with Et2O. Lyophilization resulted in a yellow solid, 8 (200 mg, 94% yield). 1H-NMR (d6-acetone, 200 MHz,): δ 1.63 (3H, d), 3.92 (3H, s), 3.99 (3H, s, 3H), 6.10 (1H, m), 7.33 (1H, s), 7.60 (1H, s); 13C-NMR (d6-acetone, 50 MHz): δ 23.63 (d, J = 4.9 Hz), 55.76 (d, J = 8.5 Hz), 71.40 (d, J = 4.2 Hz), 107.42, 108.99, 133.32 (d, J = 4.2 Hz), 138.98, 148.34, 154.11; 31P-NMR (d6-acetone, 81 MHz): δ -0.20.
β-1-(4,5-dimethoxy-nitrophenyl)ethyl farnesyl diphosphate (9)
A mixture of 8 (56 mg, 0.18 mmol) and 1,1'-carbonyldimidazole (36 mg, 0.22 mmol) in 2.0 mL anhydrous DMF were stirred at rt for 2 h in the dark after which farnesyl monophosphate (2, cyclohexylammomium salt, 60 mg, 0.18 mmol) was added and the resulting mixture was stirred at rt in the dark for 7 days. 10 mL of isopropyl alcohol:25 mM NH4HCO3 (1:49) was added to the mixture, and the solution was passed through an ion exchange column (Dowex W-X8 NH +4 form), and washed with 25 mM NH4HCO3. The eluent was lyophilized to an off-white solid. RP-HPLC (solvent A: 25 mM NH4HCO3, solvent B: CH3CN, linear gradient) afforded 9 as a light yellow solid (40 mg, 27% yield). 1H-NMR (D2O, 500 MHz): δ 1.29 (3H, s), 1.30 (3H, s), 1.37 (3H, s), 1.42 (6H, m), 1.75 (8H, m), 3.65 (3H, s), 3.83 (3H, s), 4.11 (2H, t, J = 5.5 Hz), 4.79 (1H, d, J = 6.5 Hz), 4.84 (1H, d, J = 6.5 Hz), 5.08 (1H, t, J = 6.0 Hz), 5.88 (1H, m), 7.26 (1H, s), 7.37 (1H, s). 31P-NMR (D2O, 121 MHz): δ -9.88 (1P, d, J = 14.0 Hz), -10.90 (1P, d, J = 14.0Hz). HRMS calcd for C25H38NO11P2 [M-H]- 590.1920; found 590.1904.
Purification of yPFTase
yPFTase was purified as described by Mayer et al.[25] and published in our earlier work[24].
Construction of r FPTase E. coli expression vector, pRFT529
We have designed a new E. coli expression construct for production of recombinant rPFTase using the pCDF-Duet1 vector (Novagen). pCDF-Duet1 is ideal for robust co-expression of heterodimeric proteins or protein complexes, as it contains two multiple cloning sites (MCSI and II) under the control of separate IPTG-inducible T7 promoters. The gene encoding the alpha subunit of rPFTase was excised from a pUC13-rPFTalpha construct (American Type Culture Collection) using EcoRI and HindIII restriction enzymes (all restriction enzymes were obtained from New England Biolabs) and subcloned into MCSI of pCDF-Duet1 at the same restriction sites. The vector encodes an N-terminal His6 tag at this MCS, and the Stratagene Quikchange site-directed mutagenesis kit was used to bring the ligated insert in-frame with the His6 tag. Mutagenesis primer sequences were 5'-ATCACCATCATCACCACAAGCCAGGATCCGAATTC-3' and 5'-GAATTCGGATCCTGGCTTGTGGTGATGATGGTGAT-3'. The resulting affinity tag and linker amino acid sequence N-terminal to the wild-type rPFTase alpha subunit sequence is MGSSHHHHHHKPGSEFELAPADGGE. The beta subunit of rPFTase was amplified using the high fidelity Platinum Pfx DNA polymerase (Invitrogen) according to the manufacturer's suggested protocol from a pAlter-Ex2 construct containing the gene that was a kind gift of Dr. Patrick Casey's laboratory at Duke University[26]. The rPFT beta forward primer sequence including an NdeI restriction site (underlined) was 5'-GACCTGATACATATGGCTTCTTCGAGTTCCTTCACCTATTATTGTCC-3', and the reverse primer sequence including the AvrII restriction site (underlined) 5'-GTAAGCCCTAGGGCTAGTCAGTGGCAGGATCTGAGGTCACC-3'. The insert was then subcloned into MCSII of pCDF-Duet1 using the NdeI and AvrII restriction sites. All DNA sequencing to confirm the error-free construction of the rPFTase expression vector was performed by the Duke University Medical Center DNA Analysis Facility.
Purification ofrPFTase
Competent BL21(DE3) E. coli. (Novabiochem) were transformed with the pRFT529 according to manufacturers protocol, using Streptomycin (50 μg/mL) as the selection antibiotic. A single colony was used to inoculate 100 mL LB media containing 50 μg/mL streptomycin. Cells were grown at 37°C for 18 h, shaking at 250 rpm. 10 mL of the overnight culture was used to inoculate 1 L of LB media containing 50 μg/mL streptomycin. Cells were grown at 37°C (250 rpm shaking) until an OD600 of 1.3 was reached. IPTG at a final concentration of 1.0 mM and 0.5 mM ZnSO4 were added to induce rPFTase expression. Cells were grown for an additional 4 h at 37°C (250 rpm shaking) and harvested by centrifugation at 5,000 g (4°C) for 10 min. The cell pellet from 1 L of growth (~ 15 g) was resuspended in 100 mL of lysis buffer containing 50 mM Tris pH 7.7, 200 mM NaCl, 5.0 μM ZnCl2, 5.0 mM MgCl2, 20 mM imidazole, 1.0 mM β-mercaptoethanol, and Sigma protease inhibitor cocktail (EDTA free). Cells were lysed by sonication for 5 min (10 sec on, 10 sec off). Cell debris was removed by centrifugation at 26,000 g (4°C) for 1 h. Supernatant was loaded onto a Ni-NTA chelating column (15 mL resin bed) pre-equilibrated in 50 mM Tris pH 7.7, 200 mM NaCl, 5.0 μM ZnCl2, 5.0 mM MgCl2, 20 mM imidazole and 1.0 mM β-mercaptoethanol. The column was washed with a solution of 50 mM Tris pH 7.7, 200 mM NaCl, 5.0 μM ZnCl2, 5.0 μM MgCl2, 20 mM imidazole and 1.0 mM β-mercaptoethanol and monitored by absorbance at 280 nm. Bound proteins were then eluted with 50 mM Tris pH 7.7, 200 mM NaCl, 5.0 μM ZnCl2, 5.0 mM MgCl2, 250 mM imidazole and 1.0 mM β-mercaptoethanol. Fractions containing active enzyme (determined with a continuous fluorescence assay[27, 28]) were pooled and the salt concentration was adjusted by adding an equivalent volume of 20 mM Tris-HCl pH 7.7, 1.0 M (NH4)2SO4, 5.0 μM ZnCl2, and 1.0 mM DTT slowly (over 5 minutes) while stirring. The solution was left to incubate at 4°C for 15 min without stirring. The solution was then filtered (0.45 μM filter) and applied to a Phenyl Sepharose column (15 mL resin bed) pre-equilibrated in 20 mM Tris-HCl pH 7.7, 500 mM (NH4)2SO4, 5.0 μM ZnCl2, and 1.0 mM DTT (Buffer A). Enzyme was eluted from the column using a linear gradient of 0-100% Buffer B (20 mM Tris-HCl pH 7.7, 5.0 uM ZnCl2, and 1.0 mM DTT). Active enzyme eluted at 100% B. Active fractions (based on continuous fluorescence assay[27, 28]) were then pooled and concentrated used Millipore Centricon centrifuge tubes (10 kDa MWCO) and centrifugation at 4,000 g for 70 min. Glycerol was added to a final concentration of 40% (v/v) to stabilize the enzyme for long term storage at -80°C.
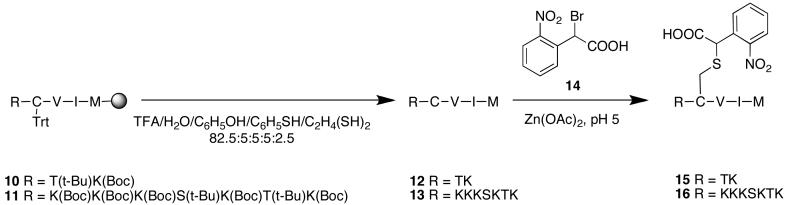
General procedure for peptide synthesis
The linear sequences were assembled by automated Fmoc SPPS, starting with Wang resin, using an ABI 430 synthesizer. DIC and HOBt were used for coupling to avoid racemization of cysteine[29]. The peptide was cleaved with Reagent K[30] (TFA, phenol, thioanisole, water, ethanedithiol 82.5:5:5:5:2.5) for 2 h, precipitated with Et2O, and centrifuged to form a pellet, which was washed with Et2O. The crude peptide was purified by RP-HPLC.
TKCVIM (12)
Yield 200 mg. Purity >95% by RP-HPLC. Deconvoluted ESI-MS calc 694.4, found 694.4.
KKKSKTKCVIM (13)
Yield 130 mg. Purity >95% by RP-HPLC. Deconvoluted ESI-MS calc 1293.8, found 1293.7.
General procedure for cysteine alkylation
Peptide (1.0 eq.) and Zn(OAc)2·2H2O (5.0 eq.) were dissolved in MeOH/0.1 M NaOAc pH 5 (4:1, v/v) (2.0 mL solvent per 1.0 mg peptide). Carboxy-nitrobenzyl bromide[31] (5.0 eq.) was then added. The reaction was monitored by RPHPLC, and once judged complete (typically 24 h), was filtered, and purified by RP-HPLC.
TKC(CNB)VIM (15)
Reaction scale: 100 mg peptide. Yield: ~7 mg. Purity >95% by RPHPLC. Deconvoluted ESI-MS calc 873.4, found 873.3.
KKKSKTKC(CNB)VIM (16)
Reaction scale: 100 mg peptide. Yield ~9 mg. Purity >90% by RP-HPLC. Deconvoluted ESI-MS calc 1472.8, found 1472.6.
Photolysis rate and quantum efficiency of 5
25 mM NH4HCO3 buffered solutions (1.5 mL) of the 5 (0.1 mM) in siliconized quartz tubes (10 × 75 mm) were irradiated with 350 nm light from a UV Rayonet mini-reactor (8 × 4 watt bulbs with spectral distributions from 300-400 nm, λmax 350nm) fitted with a spinning platform, holding samples 1 cm from bulbs. The duration of irradiation ranged from 0.25 to 16 min. After each period of irradiation, a 100 μL aliquot of the solution was removed for analysis by RP-HPLC. Aliquots were flash frozen in liquid N2 and stored at -20°C until analyzed. The compound was eluted with a gradient of 25mM NH4HCO3 in H2O and CH3CN (gradient 0.3%/mL, flow rate 1 mL/min). Absorbance was detected at 248 nm. Photodecomposition curves were plotted in Kaleidagraph 3.0 and analyzed by non-linear regressional analysis (first-order kinetics). Quantum efficiency, Qu1 were determined from the relationship Qu1 = (Iσt90%)-1 [32], where I is the irradiation intensity in ein•cm-2•s-1, σ is the decadic extinction coefficient (103 times ε, molar extinction coefficient) in cm2•mol-1, and t90% is the irradiation time in seconds for 90% conversion to product[33]. The UV intensity of the lamps in the mini-reactor was measured using potassium ferrioxalate actinometry using the same apparatus[34]. The same procedure was used for 9.
Photolysis rate and quantum efficiency of 16
20 mM NH4HCO3, 10 mM EDTA buffered solutions (3 mL) of 16 (1 mM) in siliconized quartz tubes (10 × 75 mm) were irradiated with 350 nm light from a UV Rayonet reactor (16 × 14 watt bulbs with spectral distributions from 300-400 nm, λmax 350nm) fitted with a spinning platform, holding samples 1 cm from bulbs. The duration of irradiation ranged from 0.5 to 20 min. After each period of irradiation, a 150 μL aliquot of the solution was removed for analysis by RP-HPLC. Aliquots were flash frozen in N2(l) and stored at -20°C until analyzed. The compound was eluted with a gradient of 0.1% TFA in H2O and 0.1% TFA in CH3CN (gradient 1%/mL, flow rate 1 mL/min). Absorbance was detected at 220 nm. Photodecomposition curves were plotted in Kaleidagraph 3.0 and analyzed by non-linear regressional analysis (first-order kinetics). Quantum efficiency, Qu1, was determined as described for 5.
yPFTase substrate studies with 5
Photolysis of a 100 μL solution of 5 (230 μM) and yPFTase (550 nM) in a siliconized quartz test tube (10 × 75mm) was conducted for 12 min (~3 times the half-life, t1/2 = 3.60 min) at 4°C in a UV Rayonet mini-reactor fitted with six RMR-3500Å lamps and a circulating platform. The sample was spun to ensure equal distribution of irradiation. yPFTase catalysis rates were determined using a continuous fluorescence assay[27, 28]. A solution containing N-Dansyl-GCVIA was prepared by dissolving solid peptide in buffer (20 mM Tris-HCl, pH 7.0 and 10 mM EDTA) and its concentration was determined by UV absorbance at 340 nm using N-Dansyl glycine as a standard. FPP was prepared either by diluting a purchased solution (Aldrich) in 7:3 25mM NH4OH:MeOH or obtained from photolysis reactions. yPFTase was prepared by diluting purified enzyme with buffer (52mM Tris-HCl, pH 7.0, 5.8 mM DTT, 12 mM MgCl2, 12 μM ZnCl2) containing 1 mg/mL bovine serum albumin. Assay solutions contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 μM ZnCl2, 5.0 mM DTT, 0.04% (w/v) n-dodecyl-β-D-maltoside, 2.4 μM N-Dansyl-GCVIA, and FPP (10 μM) or 5 (12.7 μL of 0.29 mM stock solution or 22.7 μL of irradiated 5 + yPFTase solution). The assay solution was placed in a quartz cuvette, equilibrated at 30°C for 5 min, initiated by the addition of yPFTase or irradiated 5 + yPFTase, and monitored spectrofluorimetrically (340 nm excitation and 505 nm emission, 10 nm slit widths). Enzymatic rates were obtained from linear regression analysis of the time-dependent fluorescence emission data using the fluorimeter software.
y PFTase substrate studies with 9
A similar procedure as described for 5 was employed, except for the following: Photolysis of a 100 μL solution of 9 (190 μM) and yPFTase (550 nM) in a siliconized quartz test tube (10 × 75mm) was conducted for 15 min (~3 times the half-life, t1/2 = 5.82 min). Assay solutions contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 μM ZnCl2, 5.0 mM DTT, 0.04% (w/v) n-dodecyl-β-D-maltoside, 2.4 μM N-Dansyl-GCVIA, and FPP (10 μM) or 9 (17.5 μL of 0.40 mM stock solution or 27.5 μL of irradiated 9 + yPFTase solution).
rPFTase substrate studies with 16
A solution containing 16 or KKKSKTKCVIM was prepared by dissolving solid peptide in buffer (20 mM Tris-HCl, pH 7.0 and 10 mM EDTA) and its concentration was estimated by mass. Photolysis of a 20 μL solution of 16 (10 μM) in a siliconized quartz test tube (10 × 75mm) was conducted for 15 min (~3 times the half-life, t1/2 = 4.9 min) at 4°C in a UV Rayonet mini-reactor fitted with sixteen RPR-3500Å lamps and a circulating platform. The sample was spun to ensure equal distribution of irradiation. 3H-FPP was used as purchased from American Radiolabeled Chemicals (25 μM in 70% EtOH/25mM aqueous NH4HCO3, 20 Ci/mmol). Assay solutions contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 μM ZnCl2, 5.0 mM DTT, 4 μM3H-FPP, and 16 (2.0 μM), irradiated 16 (2.0 μM), or 13 (2.0 μM). The reaction was initiated by addition of rPFTase (4 mg/mL) and incubated at 35°C for 60 min. Eppendorf tubes containing reaction mixture were kept on ice for 2 min, centrifuged (10 s at 1,000 RPM), and the total reaction mixture was spotted onto p81 phosphocellulose paper (Whatman). After 10 min of air drying, each paper was placed on a Millipore 1225 Sampling Manifold and washed first with 1.0% aqueous trichloroacetic acid solution (2 × 5 mL) then distilled water (3 × 5 mL). The wet papers were put into scintillation vials containing 5.0 mL scintillation fluid (Cyto-scint) and the radioactivity associated with the 3H-farnesylated peptide was determined by scintillation counting. Each reaction was performed in triplicate using a solution containing no peptide as a control.
Inhibition of r PFTase by 16
Inhibition of rPFTase was evaluated using the same previously described continuous fluorescence assay (see substrate studies with 5). Assay solutions contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 μM ZnCl2, 5.0 mM DTT, 0.2% (w/v) octylglucopyranoside, 1.0 μM N-Dansyl-GCVLS, FPP (10 μM), and 16 (0, 2, 4, 6, 10, 20, 30 μM). The assay solution was placed in a quartz cuvette, equilibrated at 30°C for 5 min, initiated by the addition of rPFTase (5 ng/mL) and monitored spectrofluorimetrically (340 nm excitation and 505 nm emission, 10 nm slit widths). Enzymatic rates were obtained from linear regression analysis of the time-dependent fluorescence emission data using the fluorimeter software.
X-ray crystallography studies with 15
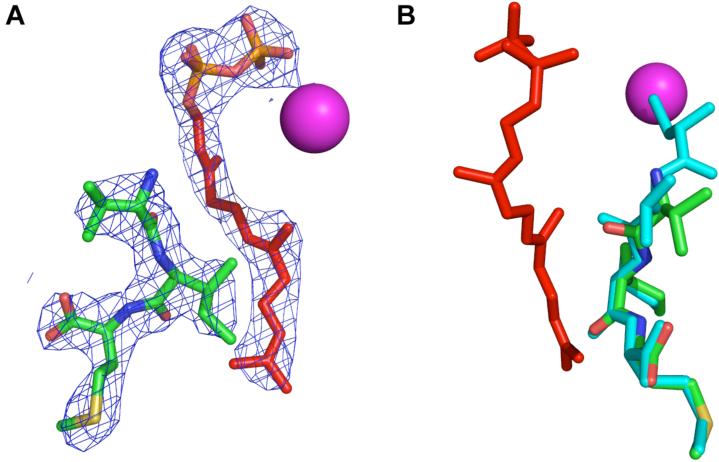
Untagged rPFTase was prepared as described[35] using a baculovirus/SF9 expression system. Crystals of the enzyme containing a farnesylated KKKSKTKCVIM peptide were prepared as described[36]. To form a ternary complex with FPP and 15, the farnesylated product complex was displaced from rPFTase crystals as described by soaking in a stabilizing solution containing 300 μM 15 and 150 μM FPP for one week[2]. Crystals were cryoprotected as described[36] and X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. The crystal diffracted to 2.70 Å resolution, with an overall merging R factor of 7.9% and 99% completeness of data in the highest resolution bin. Data were reduced and scaled with HKL2000[37], and phases were determined using molecular replacement as implemented by PHASER[38] with the PDB file 1D8D as the search model with ligands removed to reduce model bias. Iterative model building and refinement were carried out in COOT[39] and REFMAC5[40], respectively. The final Rcryst/Rfree was 21.6/25.3. PYMOL was used to perform the superpositions, calculate rmsd values, and prepare Figure 7. The structure was deposited to the PDB with accession code 3DPY.
Figure 7.
Crystallographic analysis of analogue 15 and FPP bound to rPFTase. (A, left image): 2Fo-Fe electron density map of 15 and FPP contoured at 1.5σ. (B, right image): Superposition of 15 and VIM peptide. Analogue 15 is shown in green (carbon), red (oxygen), blue (nitrogen), and yellow (sulfur). FPP is shown in red. The CVIM peptide is shown in cyan. The Zn atom in the enzyme active site is in pink.
RESULTS
Synthesis and photolytic properties of caged isoprenoid diphosphates 5 and 9
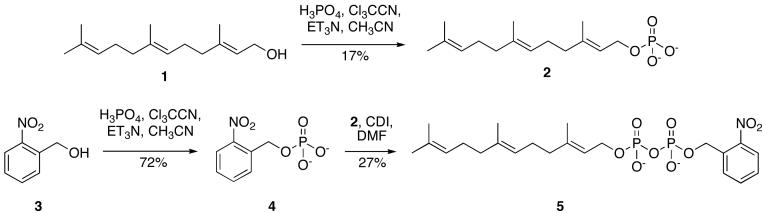
The synthesis of an o-nitrobenzyl caged farnesyl diphosphate (FPP) derivative (5) was accomplished in three steps beginning from commercially available 1 (Figure 2). Farnesol (1) was converted to the farnesyl phosphate (2) by the procedure described by Cramer et al[41]. 2-Nitrobenzyl phosphate (4) was prepared according to a procedure described by Kaplan and coworkers from commercially available 2-nitrobenzyl alcohol[42]. Upon activation with CDI, 4 was coupled to 2 and purified via a two-step purification using ion-exchange chromatography and RP-HPLC to give the desired product 5 in 27 % yield.
Figure 2.
Synthesis of β-(2-nitrobenzyl)farnesyl diphosphate.
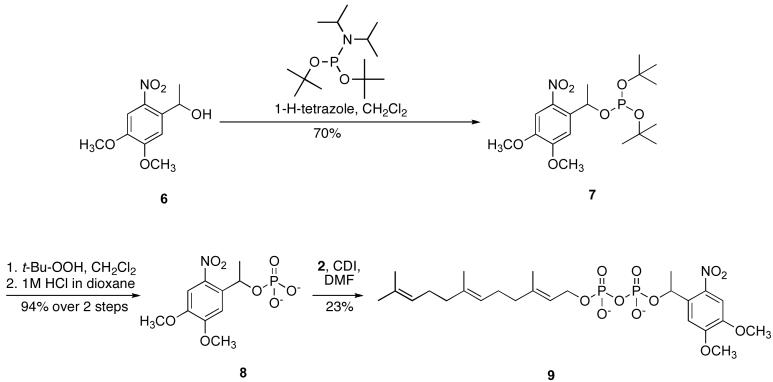
Because the irradiation of the o-nitrobenzyl FPP derivative results in the release of a potentially reactive byproduct, 2-nitrosobenzaldehyde[18], a second photoactivatable FPP analogue was synthesized (Figure 3). To avoid the formation of reactive aldehydes upon photolysis, the 1-(4,5-dimethoxy)phenethyl photolabile protecting group was used, which undergoes photocleavage with kinetics comparable to those of the related o-nitrobenzyl caging group, but releases the less reactive ketone 2-nitrosoacetophenone as a by product[18]. Commercially available 1-(4,5-dimethoxy-2-nitrophenyl)ethanol was phosphorylated with di-t-butyl diisopropylphosphoramidite in CH2Cl2 in the presence of tetrazole to form 7 in 70% yield. Oxidation with t- butylhydroperoxide and deprotection under acidic conditions yielded 8, which was then activated with CDI and coupled to farnesyl phosphate (2). The desired product, 9, was obtained following purification via ion-exchange chromatography and RP-HPLC in 23 % yield. Phosphate analysis of solutions of 5 and 9 in 25 mM NH4HCO3 was performed to obtain accurate concentrations for subsequent experiments.
Figure 3.
Synthesis of β-1-(4,5-dimethoxy-nitrophenyl)ethyl farnesyl diphosphate.
Irradiation of 5 and 9 stock solutions with 300-400 nm (λmax = 350nm) light lead to the release of farnesyl diphosphate. The time course of this reaction was followed by RP-HPLC analysis of aliquots removed at varying intervals (see supporting information). The single-photon uncaging efficiencies, Qu1, were determined from the following relationship: Q1u = (Iσt90%)-1, where I is the irradiation intensity of the apparatus used in ein•cm-2•s-1 (determined by potassium ferrioxalate actinometry), σ is the decadic extinction coefficient (103 ε, molar extinction coefficient) in cm-2•mol-1, and t90% is the irradiation time in seconds for 90% conversion to product. These data are summarized in Table 1. It should be noted that the values for 5 and 9 are comparable to those previously reported for other caged molecules that have been employed in biochemical studies; additionally, our results are also consistent with the previously noted observation that compounds with the nitrobenzyl photolabile group have a greater quantum yield and uncage faster (5, k = 0.43 min-1, Qu1 = 0.70) than identical compounds incorporating a dimethoxy-nitrophenethyl caging group[18] (9, k = 0.12 min-1, Qu1 = 0.08).
Table 1.
Photochemical properties of caged analogues described here and related compounds.
| Caged Compound | λirrad (nm) | Qu1 (mol·ein-1) | Uncaging Rate (sec-1) |
|---|---|---|---|
| NB-FPP (5) | 350 | 0.70 | 25.8 |
| DMNPE-FPP (9) | 350 | 0.08 | 7.20 |
| KKKSKTKC(CNB)VIM (16) | 350 | 0.16 | 8.40 |
| DMNPE-ATP[16] | 350 | 0.07 | 18.0 |
| DMNPE-Doxycycline[51] | 347 | 0.01 | NR |
| DMNB-TRβ agonist[47] | 366 | NR | 0.03 |
λirrad, wavelength data was obtained at; Qu1, quantum efficiency; NR, not reported; NB, nitrobenzyl; DMNPE, dimethoxynitrophenethyl; CNB, α-carboxy-nitrobenzyl; DMNB, dimethoxynitrobenzyl;
Substrate studies of caged isoprenoid diphosphates 5 and 9 with yPFTase
The specific activity of the enzyme in the presence of 5 or 9 and their photolysis products was evaluated using a continuous fluorescence assay developed by Pompliano and coworkers[28] to assess their compatibility. This assay is based on the observation that dansyl-modified CAAX peptides undergo a fluorescence enhancement upon prenylation. Table 2 shows the rate of enzymatic prenylation (μM/min) is approximately 14% of its rate with FPP when only 5 is present as the isoprenoid substrate. Once 5 was irradiated for 12 min (~3 times the half-life for uncaging), saturating amounts of FPP were released, based on the total return in enzymatic activity. This result also demonstrates that the byproducts of the photolysis reaction do not interfere with catalysis in any way. Similar results were observed with derivative 9, although this analogue appears to be a poorer substrate prior to uncaging (7.6% compared with FPP). Substrate reactions were also analyzed by HPLC (see supporting information). The PFTase catalyzed prenylation resulted in the same prenylated product regardless of whether the FPP source was pure FPP or the product of photolysis of 5 or 9.
Table 2.
Activity[a] of yPFTase in the presence of compounds 5 and 9.
| Substrate | Enzyme activity (μM/min) | Normalized activity (%) |
|---|---|---|
| FPP | 0.765 ± 0.007 | 100 |
| Compound 5 | 0.104 ± 0.006 | 14 |
| Irradiated Compound 5 | 0.681 ± 0.012 | 89 |
| Compound 9 | 0.058 ± 0.001 | 7.6 |
| Irradiated Compound 9 | 0.557 ± 0.006 | 73 |
When present, FPP and/or caged FPP were each at 10 μM. Given the rate of photolysis, the irradiated samples would be expected to contain 10 μM free FPP and side products. The CAAX peptide substrate is N-Dansyl-GCVIA
Synthesis of caged CAAX peptides 15 and 16
Alkylation of the cysteine's thiol group within the CAAX peptide with a photolabile group offers an alternative means of creating a catalytically caged prenyltransferase substrate. Peptides 12 and 13 (Figure 4) were synthesized on resin using an ABI Peptide Synthesizer employing DIC, HOBt coupling chemistry. This peptide sequence was chosen because it is the C-terminal sequence of K-Ras4B, a naturally prenylated mammalian protein implicated in oncogenesis[43]. Upon cleavage from the resin, the cysteine thiol was alkylated with 14 under acidic conditions to form the desired caged analogues 15 and 16. Stock solutions were made by dissolving in 20 mM Tris-HCl, 10 mM EDTA and stored in the dark at -20°C to minimize oxidation and photodecomposition.
Figure 4.
Synthesis of caged peptide substrates.
Photolytic properties of caged CAAX peptide 16
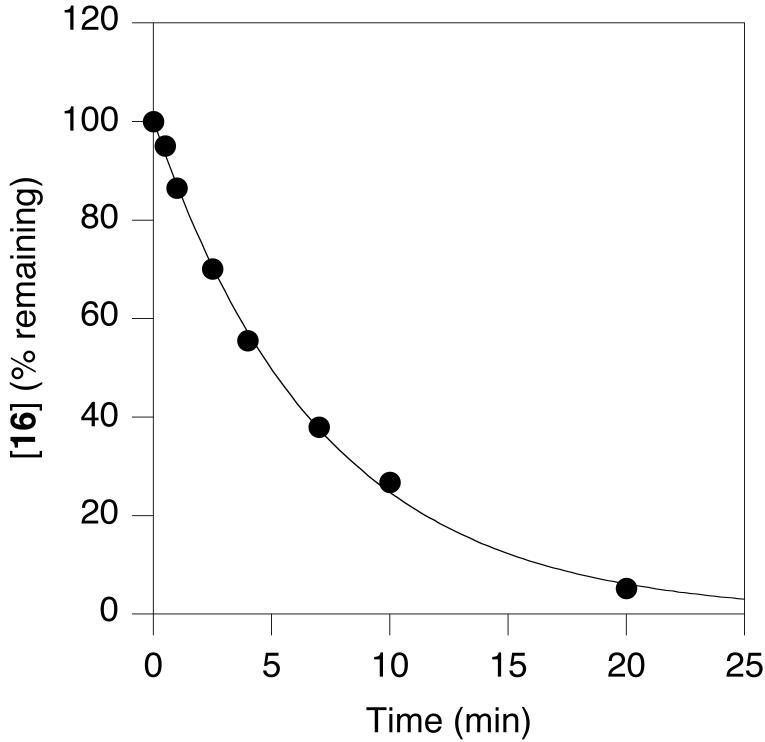
RP-HPLC analysis of an irradiated stock solution of 16 with 300-400 nm light (λmax = 350) at increasing time intervals showed the production of 13, the parent peptide. A plot of the remaining caged peptide (determined by integration of the peak area in the chromatogram) as a function of irradiation time is shown in Figure 5. Non-linear regression analysis using a first-order kinetic model gave a rate constant of 0.14 min-1. Qu1, the corresponding quantum efficiency of uncaging was 0.16 (Table 1).
Figure 5.
Photolysis rate determination for 16.
Substrate studies and binding of caged CAAX peptide with PFTase
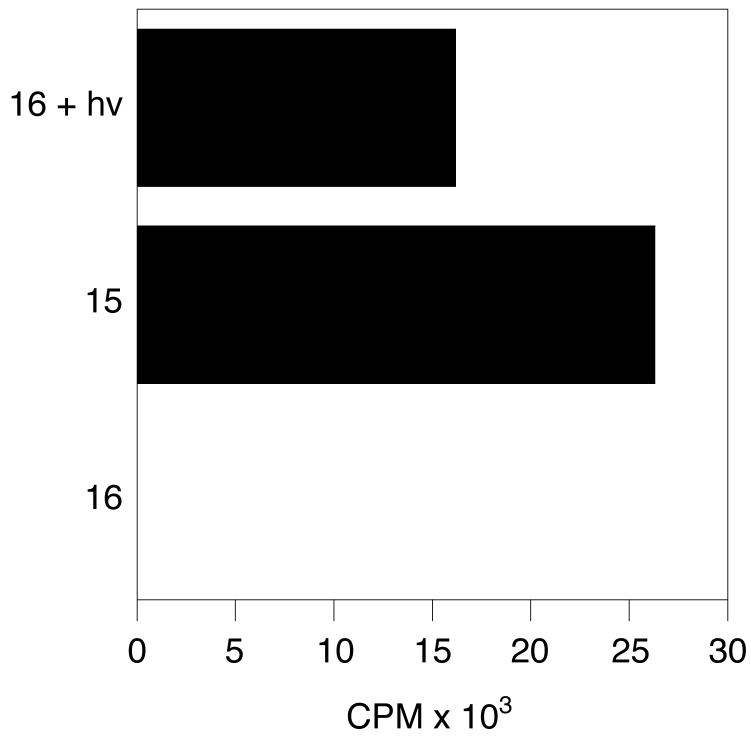
A filter binding assay, where the positively-charged amino acids upstream of the CAAX sequence will bind to p81 phosphocellulose paper (Whatman) and incorporation of a prenyl group onto that peptide can be detected through a radiolabel on the isoprenoid chain[44], was used to evaluate the ability of 16 to act as a PFTase substrate before and after photolysis. Prior to irradiation with 350 nm light, there is no enzyme catalyzed prenylation of 16 with radiolabeled FPP (Figure 6, bottom column). The amount of tritiated isoprenoid incorporated into the photoreleased peptide is comparable to the amount incorporated into the pure CAAX peptide, 13 (Figure 6, top and middle columns).
Figure 6.
PFTase catalyzed prenylation of 16 before and after photolysis with 3H-FPP.
Binding of a caged enzyme substrate prior to photolysis is crucial for time-resolved enzyme experiments. If the caged substrate does not bind, the rate of enzyme reaction initiation is severely limited by diffusion of the released substrate and binding to the active site; in the solid state (i.e. protein crystals) this could be quite slow. There are relatively few examples in the literature where caged substrates bind but do not react prior to photolysis[23, 45]. This in part may be explained by the fact that this is not a requirement for many types of applications of caged substrates such as those referenced in the introduction. Initially, the continuous fluorescence assay using N-Dansyl-GCVLS and FPP as the substrates was used to evaluate the ability of 16 to bind to rPFTase. An IC50 value of 4.7 ± 1.8 μM was calculated (see supporting information), as compared to 165 nM for the tetrapeptide CVIM[46]. While this is a significant decrease in binding ability, it demonstrates that the caged CAAX peptide 16 is able to bind the enzyme active site when present at μM concentrations. Next, X-ray crystallography was employed to determine if the isoprenoid diphosphate substrate was able to bind simultaneously with the caged CAAX peptide. Because the upstream residues of the caged CAAX peptide were not critical for obtaining co-crystals of caged CAAX peptide and PFTase, analogue 15 was utilitized due to availability. The structure of the rPFTase:FPP:15 complex was solved at a resolution of 2.7 Å, and is shown in Figure 7; a superposition with an uncaged CAAX peptide is shown (panel B) for comparison.
The electron density map clearly indicates that both 15 and FPP are bound in the enzyme active site (Figure 7, panel A). Comparison of the protein structures from rPFTase:FPP:15 and rPFTase:FPT-II:CVIM (PDB 1D8D) complexes shows a 0.174 Å rms deviation. Focusing in on an 8 Å sphere surrounding the substrate binding pocket, the rms deviation increases to 0.203 Å. Taken together, these data indicate that no significant movement of the protein side chains, including those needed for catalysis, has occurred. The VIM residues of 15 are essentially superimposable with those in the ternary complex (PDB 1D8D), demonstrating that the alkylation of the cysteine residue with the carboxynitrobenzyl caging group does not affect the position of the C-terminal residues. The N-terminal residues of 15, including the caged cysteine, are disordered and thus not visible in the electron density map. This suggests that 15 can adopt many different binding conformations due to the loss of the strong thiolate-Zn interaction (due to S-alkylation) that normally restricts movement of the cysteine residue within the substrate peptide. The FPP that is bound in the presence of 15 occupies the isoprenoid binding pocket of the active site. The first isoprene unit of FPP is slightly rotated (0.546 Å rms deviation calculated over all FPP atoms, PDB 1KZO), while the diphosphate and second two isoprene units are superimposable. In conclusion, the incorporation of a caging moiety at the cysteine residue of the CAAX peptide does not invoke major changes in the binding capacity or conformation of either enzyme substrate or the protein itself.
DISCUSSION
There are a number of characteristics that make a caged compound suitable for studying a particular enzyme. It is preferable to perform the photocleavage at wavelengths above 300 nm in order to minimize enzyme inactivation and cellular toxicity[15]. Additionally, the photochemical byproducts should not interfere with photolysis nor enzyme catalysis. Finally, the caged substrate must be soluble in aqueous media[16]. Caged isoprenoid diphosphates 5 and 9 meet these criteria. In addition, their uncaging kinetics are comparable to many other commercially available and previously reported caged enzyme substrates/inhibitors and signaling molecules (see Table 1)[47-52]. Caged ATP (Molecular Probes) has been widely used to study the mechanism of ATPases[49, 50] and the molecular basis of skeletal muscle fiber contraction[48, 52]. Cambridge et. al. developed a dimethoxynitrophenethyl-caged analogue of doxycycline, a known analogue of the tetracycline gene activation system, and demonstrated its efficacy in spatially restricted photoactivated gene expression[51].
One requirement for caged compounds is that they are truly inactive as substrates for the target enzyme. While analogues 5 and 9 are considerably slower substrates than FPP, they are still processed by the enzyme at measurable rates (14% and 7.6% respectively relative to FPP) thus limiting their utility. As noted above, analogue 5 is a better substrate than compound 9 indicating that rate decreases with increasing steric bulk on the diphosphate moiety. This suggests that it may be possible to further decrease the rate of processing of isoprenoids caged on the diphosphate by employing larger caging groups including 3',5'-dimethoxybenzoin[53], bromohydroxy quinoline[33, 54], or bromohydroxy coumarin[55]; such experiments are currently underway. Incorporation of a photolabile group into the isoprene portion of farnesyl diphosphate as a different way to minimize background catalysis is also being explored. Nevertheless, these caged FPP analogues may be useful for studies with other FPP-utilizing enzymes or FPP binding proteins. For example, recent work by Das and coworkers has implicated FPP as a transcriptional activator of the thyroid hormone nuclear receptor-β (TRβ)[56]. A photo-caged thyroid hormone receptor analogue, a known agonist of TRβ, has been applied to time-dependent gene expression profiling[47]. Similar experiments with caged-FPP could be used to gain a better understanding of FPP as TRβ agonist. The quantum efficiency and rates of photolysis of caged FPP analogues 5 and 9 are quite comparable to these aforementioned probes, showing their applicability towards a number of biochemical applications including experiments performed in live cells.
Given our limited success in preparing caged isoprenoid substrates suitable for experiments with PFTase, we next examined a caged peptide substrate. Alkylation of the thiol present in peptide 13 with a α-carboxy-nitrobenzyl group yielded a caged molecule (16) that was not processed by the enzyme at any detectable level although it retained the ability to bind to the enzyme. The tripeptide substructure VIM of 15 bound in a manner indistinguishable from the unmodified peptide CVIM. However, based on the IC50 value for 16 compared to that for the unalkylated precursor CVIM, the caged molecule binds approximately 30-fold less avidly. Thus, the caged substrate 16 and related analogues possess all the requisite features necessary for a variety of investigations where temporal control of initiation of the reaction catalyzed by PFTase is required including time-resolved x-ray crystallography and analysis of prenylation reactions in living cells.
CONCLUSIONS AND FUTURE DIRECTIONS
The molecules designed and reported here are the first examples of caged forms of isoprenoid diphosphates as well as the first documented caged substrate for a bisubstrate enzyme that is capable of binding simultaneously with the other substrate. Caged isoprenoid diphosphates 5 and 9 were each prepared in four steps from farnesol. Kinetic studies demonstrate that they are poor substrates for PFTase but efficiently release FPP upon irradiation that can be utilized for catalysis. Given the plethora of enzymes that use FPP as a substrate, including the diverse family of sesquiterpene cyclases, these probes should be useful for a variety of studies. The caged CAAX peptide, 16, photoreleases the parent peptide with similar kinetics to 5 and 9. It does not function as a substrate, but is able to bind with efficient capacity and nearly identical conformation as compared to the photoreleased peptide and does not interfere with the binding of the isoprenoid diphosphate substrate. These results open the door to a wide variety of experiments where temporal control is necessary. A number of such experiments are currently underway in these laboratories.
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health Grants GM58442 (M.D.D.) and GM52382 (L.S.B.) as well as the National Institutes of Health Predoctoral Training Grant T32-GM08700 (A.J.D.). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38
REFERENCES
- 1.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic Analysis of CaaX Prenyltransferases Complexed with Substrates Defines Rules of Protein Substrate Selectivity. J Mol Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 3.Dolence JM, Cassidy PB, Mathis JR, Poulter CD. Yeast protein farnesyltransferase: steady-state kinetic studies of substrate binding. Biochemistry. 1995;34:16687–16694. doi: 10.1021/bi00051a017. [DOI] [PubMed] [Google Scholar]
- 4.Furfine ES, Leban JJ, Landavazo A, Moomaw JF, Casey PJ. Protein farnesyltransferase: kinetics of farnesyl pyrophosphate binding and product release. Biochemistry. 1995;34:6857–6862. doi: 10.1021/bi00020a032. [DOI] [PubMed] [Google Scholar]
- 5.Rozema DB, Poulter CD. Yeast protein farnesyltransferase. pKas of peptide substrates bound as zinc thiolates. Biochemistry. 1999;38:13138–13146. doi: 10.1021/bi990794y. [DOI] [PubMed] [Google Scholar]
- 6.Sousa SF, Fernandes PA, Ramos MJ. Unraveling the mechanism of the farnesyltransferase enzyme. J Biol Inorg Chem. 2005;10:3–10. doi: 10.1007/s00775-004-0612-6. [DOI] [PubMed] [Google Scholar]
- 7.Dolence JM, Poulter CD. A Mechanism for Posttranslational Modifications of Proteins by Yeast Protein Farnesyltransferase. Proc Natl Acad Sci USA. 1995;92:5008–5011. doi: 10.1073/pnas.92.11.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenevich S, Hosokawa A, Cramer CJ, Distefano MD. Transition State Analysis of Model and Enzymatic Prenylation Reactions. J Am Chem Soc. 2007;129:5796–5797. doi: 10.1021/ja069119j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pais JE, Bowers KE, Fierke CA. Measurement of the a-Secondary Kinetic Isotope Effect for the Reaction Catalyzed by Mammalian Protein Farnesyltransferase. J Am Chem Soc. 2006;128:15086–15087. doi: 10.1021/ja065838m. [DOI] [PubMed] [Google Scholar]
- 10.Ohkanda J, Knowles DB, Blaskovich MA, Sebti SM, Hamilton AD. Inhibitors of protein farnesyltransferase as novel anticancer agents. Curr Top Med Chem. 2002;2:303–323. doi: 10.2174/1568026023394281. [DOI] [PubMed] [Google Scholar]
- 11.Crul M, de Klerk GJ, Beijnen JH, Schellens JH. Ras biochemistry and farnesyl transferase inhibitors: a literature survey. Anti-cancer drugs. 2001;12:163–184. doi: 10.1097/00001813-200103000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Bell IM. Inhibitors of Farnesyltransferase: A Rational Approach to Cancer Chemotherapy? J Med Chem. 2004;47:1869–1878. doi: 10.1021/jm0305467. [DOI] [PubMed] [Google Scholar]
- 13.Sousa SF, Fernandes PA, Ramos MJ. Farnesyltransferase inhibitors: a detailed chemical view on an elusive biological problem. Curr Med Chem. 2008;15:1478–1492. doi: 10.2174/092986708784638825. [DOI] [PubMed] [Google Scholar]
- 14.Rowinsky EK. Lately, it occurs to me what a long, strange trip it's been for the farnesyltransferase inhibitors. J Clin Oncol. 2006;24:2981–2984. doi: 10.1200/JCO.2006.05.9808. [DOI] [PubMed] [Google Scholar]
- 15.Young DD, Deiters A. Photochemical control of biological processes. Org Biomol Chem. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 16.Ellis-Davies GC. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew Chem Int Ed. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 18.Pelliccioli AP, Wirz J. Photoremovable protecting groups: reaction mechanisms and applications. Photochem Photobiol Sci. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 19.Curley K, Lawrence DS. Caged regulators of signaling pathways. Pharmacol Ther. 1999;82:347–354. doi: 10.1016/s0163-7258(98)00055-2. [DOI] [PubMed] [Google Scholar]
- 20.Adams SR, Tsien RY. Controlling cell chemistry with caged compounds. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 21.Parker LL, Kurutz JW, Kent SB, Kron SJ. Control of the yeast cell cycle with a photocleavable alpha-factor analogue. Angew Chem Int Ed. 2006;45:6322–6325. doi: 10.1002/anie.200602439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathert P, Rasko T, Roth M, Slaska-Kiss K, Pingoud A, Kiss A, Jeltsch A. Reversible inactivation of the CG specific SssI DNA (cytosine-C5)-methyltransferase with a photocleavable protecting group. Chembiochem. 2007;8:202–207. doi: 10.1002/cbic.200600358. [DOI] [PubMed] [Google Scholar]
- 23.Schlichting I, Rapp G, John J, Wittinghofer A, Pai EF, Goody RS. Biochemical and crystallographic characterization of a complex of c-Ha-ras p21 and caged GTP with flash photolysis. Proc Natl Acad Sci USA. 1989;86:7687–7690. doi: 10.1073/pnas.86.20.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaon I, Turek TC, Weller VA, Edelstein RL, Singh SK, Distefano MD. Photoactive Analogs of Farnesyl Pyrophosphate Containing Benzoylbenzoate Esters: Synthesis and Application to Photoaffinity Labeling of Yeast Protein Farnesyltransferase. J Org Chem. 1996;61:7738–7745. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- 25.Mayer MP, Prestwich GD, Dolence JM, Bond PD, Wu HY, Poulter CD. Protein farnesyltransferase: production in Escherichia coli and immunoaffinity purification of the heterodimer from Saccharomyces cerevisiae. Gene. 1993;132:41–47. doi: 10.1016/0378-1119(93)90512-2. [DOI] [PubMed] [Google Scholar]
- 26.Fu HW, Moomaw JF, Moomaw CR, Casey PJ. Identification of a cysteine residue essential for activity of protein farnesyltransferase. Cys299 is exposed only upon removal of zinc from the enzyme. J Biol Chem. 1996;271:28541–28548. doi: 10.1074/jbc.271.45.28541. [DOI] [PubMed] [Google Scholar]
- 27.Cassidy PB, Dolence JM, Poulter CD. Continuous fluorescence assay for protein prenyltransferases. Methods Enzymol. 1995;250:30–43. doi: 10.1016/0076-6879(95)50060-x. [DOI] [PubMed] [Google Scholar]
- 28.Pompliano DL, Gomez RP, Anthony NJ. Intramolecular fluorescence enhancement: a continuous assay of Ras farnesyl:protein transferase. J Am Chem Soc. 1992;114:7945–7946. [Google Scholar]
- 29.Han Y, Albericio F, Barany G. Occurrence and Minimization of Cysteine Racemization during Stepwise Solid-Phase Peptide Synthesis(1)(,)(2) J Org Chem. 1997;62:4307–4312. doi: 10.1021/jo9622744. [DOI] [PubMed] [Google Scholar]
- 30.King DS, Fields CG, Fields GB. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Pept Protein Res. 1990;36:255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 31.Grewer C, Jager J, Carpenter BK, Hess GP. A new photolabile precursor of glycine with improved properties: A tool for chemical kinetic investigations of the glycine receptor. Biochemistry. 2000;39:2063–2070. doi: 10.1021/bi9919652. [DOI] [PubMed] [Google Scholar]
- 32.Adams SR, Kao JPY, Grynkiewicz G, Minta A, Tsien RY. Biologically useful chelators that release Ca2+ upon illumination. J Am Chem Soc. 1988;110:3212–3220. [Google Scholar]
- 33.Fedoryak OD, Dore TM. Brominated hydroxyquinoline as a photolabile protecting group with sensitivity to multiphoton excitation. Org Lett. 2002;4:3419–3422. doi: 10.1021/ol026524g. [DOI] [PubMed] [Google Scholar]
- 34.Hatchard CG, Parker CA. A new sensitive chemical actinometer. II. Potassium ferrioxalate as a standard chemical actinometer. Proc Roy Soc. 1956;A235:518–536. [Google Scholar]
- 35.Chen WJ, Moomaw JF, Overton L, Kost TA, Casey PJ. High level expression of mammalian protein farnesyltransferase in a baculovirus system. The purified protein contains zinc. J Biol Chem. 1993;268:9675–9680. [PubMed] [Google Scholar]
- 36.Long SB, Casey PJ, Beese LS. The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 .ANG. resolution ternary complex structures. Structure. 2000;8:209–222. doi: 10.1016/s0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J App Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Cramer F, Rittersdorf W, Boehm W. Chemistry of high-energy phosphates. XVIII. Synthesis of phosphoric acid esters and pyrophosphoric acid esters of the terpene alcohols. Annu Rev. 1962;654:180–188. [Google Scholar]
- 42.Kaplan JH, Forbush B, 3rd, Hoffman JF. Rapid photolytic release of adenosine 5'-triphosphate from a protected analogue: utilization by the Na:K pump of human red blood cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 43.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 44.Khan SG, Mukhtar H, Agarwal R. A rapid and convenient filter-binding assay for ras p21 processing enzyme farnesyltransferase. J Biochem Biophys Methods. 1995;30:133–144. doi: 10.1016/0165-022x(94)00072-l. [DOI] [PubMed] [Google Scholar]
- 45.Cohen BE, Stoddard BL, Koshland DE., Jr. Caged NADP and NAD. Synthesis and characterization of functionally distinct caged compounds. Biochemistry. 1997;36:9035–9044. doi: 10.1021/bi970263e. [DOI] [PubMed] [Google Scholar]
- 46.Graham SL, deSolms SJ, Giuliani EA, Kohl NE, Mosser SD, Oliff AI, Pompliano DL, Rands E, Breslin MJ, Deana AA, et al. Pseudopeptide inhibitors of Ras farnesyl-protein transferase. J Med Chem. 1994;37:725–732. doi: 10.1021/jm00032a004. [DOI] [PubMed] [Google Scholar]
- 47.Link KH, Cruz FG, Ye HF, O'Reilly KE, Dowdell S, Koh JT. Photo-caged agonists of the nuclear receptors RARgamma and TRbeta provide unique time-dependent gene expression profiles for light-activated gene patterning. Bioorg Med Chem. 2004;12:5949–5959. doi: 10.1016/j.bmc.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 48.He Z, Stienen GJ, Barends JP, Ferenczi MA. Rate of phosphate release after photoliberation of adenosine 5'-triphosphate in slow and fast skeletal muscle fibers. Biophys J. 1998;75:2389–2401. doi: 10.1016/s0006-3495(98)77683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartung K, Froehlich JP, Fendler K. Time-resolved charge translocation by the Ca-ATPase from sarcoplasmic reticulum after an ATP concentration jump. Biophys J. 1997;72:2503–2514. doi: 10.1016/S0006-3495(97)78895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gropp T, Cornelius F, Fendler K. K+-dependence of electrogenic transport by the NaK-ATPase. Biochim Biophys Acta. 1998;1368:184–200. doi: 10.1016/s0005-2736(97)00162-4. [DOI] [PubMed] [Google Scholar]
- 51.Cambridge SB, Geissler D, Keller S, Curten B. A caged doxycycline analogue for photoactivated gene expression. Angew Chem Int Ed. 2006;45:2229–2231. doi: 10.1002/anie.200503339. [DOI] [PubMed] [Google Scholar]
- 52.Allen DG, Lannergren J, Westerblad H. The use of caged adenine nucleotides and caged phosphate in intact skeletal muscle fibres of the mouse. Acta Physiol Scand. 1999;166:341–347. doi: 10.1111/j.1365-201x.1999.00571.x. [DOI] [PubMed] [Google Scholar]
- 53.Corrie JET, Trentham DR. Synthetic, Mechanistic and Photochemical Studies of Phosphate Esters of Substituted Benzoins. J Chem Soc Perkin Trans. 1992:2409–2417. [Google Scholar]
- 54.Zhu Y, Pavlos CM, Toscano JP, Dore TM. 8-Bromo-7-hydroxyquinoline as a photoremovable protecting group for physiological use: mechanism and scope. J Am Chem Soc. 2006;128:4267–4276. doi: 10.1021/ja0555320. [DOI] [PubMed] [Google Scholar]
- 55.Furuta T, Takeuchi H, Isozaki M, Takahashi Y, Kanehara M, Sugimoto M, Watanabe T, Noguchi K, Dore TM, Kurahashi T, Iwamura M, Tsien RY. Bhc-cNMPs as either water-soluble or membrane-permeant photoreleasable cyclic nucleotides for both one- and two-photon excitation. Chembiochem. 2004;5:1119–1128. doi: 10.1002/cbic.200300814. [DOI] [PubMed] [Google Scholar]
- 56.Das S, Schapira M, Tomic-Canic M, Goyanka R, Cardozo T, Samuels HH. Farnesyl Pyrophosphate Is a Novel Transcriptional Activator for a Subset of Nuclear Hormone Receptors. Mol Endocrin. 2007;21:2676–2686. doi: 10.1210/me.2007-0080. [DOI] [PubMed] [Google Scholar]