SUMMARY
The smallest known DNA transposases are those from the IS200/IS605 family. Here we show how the interplay of protein and DNA activates TnpA, the Helicobacter pylori IS608 transposase, for catalysis. First, transposon end binding causes a conformational change that aligns catalytically important protein residues within the active site. Subsequent precise cleavage at the left and right ends, the steps that liberate the transposon from its donor site, does not involve a site-specific DNA binding domain. Rather, cleavage site recognition occurs by complementary base pairing with a TnpA-bound subterminal transposon DNA segment. Thus, the enzyme active site is constructed from elements of both protein and DNA, reminiscent of the interdependence of protein and RNA in the ribosome. Our structural results explain why the transposon ends are asymmetric and how the transposon selects a target site for integration, and allow us to propose a molecular model for the entire transposition reaction.
INTRODUCTION
DNA transposition is a ubiquitous phenomenon occurring in all kingdoms of life during which discrete segments of DNA called transposons move from one genomic location to another. In both eukaryotes and prokaryotes, DNA transposition has been a significant part of evolution. Many eukaryotic genomes are littered with transposons or their inactive remnants (Lander et al., 2001), primarily scattered between genes. In bacteria, transposable elements can carry antibiotic resistance genes and, when combined with conjugation, are major drivers of broad genome remodeling and the emergence of antibiotic resistant strains. The discovery and engineering of DNA transposons active in vertebrate cells (Miskey et al., 2005) has led to their use in identifying oncogenes and tumor suppressors and characterizing genes of unknown function, through their ability to disrupt genes or regulatory regions. They also have exciting potential as gene delivery systems for gene therapy applications.
A variety of structurally and mechanistically distinct transposase enzymes have evolved to carry out transposition by several different pathways (Curcio & Derbyshire, 2003). In all cases, these enzymes possess a nuclease activity that allows them to cleave DNA in order to excise transposon DNA and subsequently splice it into a new location. Depending on the system (Dyda et al., 1994; Grindley et al., 2006), different types of nucleophiles can be used by transposases to cleave DNA by attacking a phosphorus atom of a backbone phosphate group: water, generally activated by enzyme-bound metal ions; a hydroxyl group at the 5′ or 3′ end of a DNA strand; or a hydroxyl-group bearing amino acid in the active site of the transposase itself, such as serine or tyrosine. When a catalytic serine or tyrosine is used, the enzyme becomes attached to DNA through a covalent phosphotyrosine or phosphoserine bond.
One group of transposases that use catalytic tyrosines are the Y1 transposases (Ronning et al., 2005). These are members of a vast superfamily of nucleases characterized by a conserved His-hydrophobic-His (HUH) motif (Koonin & Ilyina, 1993) that provides two ligands to a divalent metal ion cofactor. HUH nucleases always cut a DNA strand with a polarity resulting in a 5′ phosphotyrosine linkage and a free 3′ OH group. In contrast to many HUH superfamily members which are monomeric and have two catalytic tyrosines, Y1 transposases have only one catalytic tyrosine and form obligatory dimers.
We have recently determined the structure of a Y1 transposase from the insertion sequence IS608 (Ronning et al., 2005), originally identified in Helicobacter pylori (Kersulyte et al., 2002), a bacterium that causes gastric inflammation leading to ulcers and occasionally to stomach cancer. Insertion sequences (IS) are the simplest autonomous transposable elements. While they tend to be short (< 2.5 kb) and carry only those genes needed for transposition, if placed flanking a DNA segment, many are able to mobilize the intervening genes (Mahillon & Chandler, 1998). In addition to dispersing antibiotic resistance genes, IS transposition can indirectly lead to antibiotic-resistant bacterial strains. For example, a metronidazole-resistant strain of H. pylori has arisen because the nitroreductase gene needed for pro-drug activation has been disrupted by an IS605-related transposition event (Debets-Ossenkopp et al., 1999).
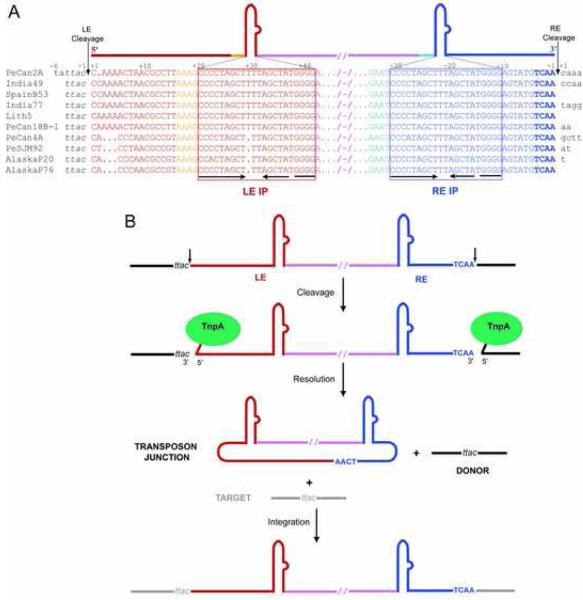
ISs can be classified into groups or families based on the general features of their DNA sequences and associated transposases (www-is.biotoul.fr). A particularly interesting family consists of the IS200/IS605 elements (Kersulyte et al., 2002) which do not have inverted sequences at their ends characteristic of many prokaryotic and eukaryotic transposons. Rather, imperfect palindromic (IP) sequences are located close to the transposon ends (Figure 1A). In the case of one family member IS608, the left end (LE) and right end (RE) have almost identical IP sequences that form DNA stem loop structures (Ronning et al., 2005). Intriguingly, these IPs are asymmetrically located: the IP at RE (IPRE) is closer to the RE cleavage site at the transposon end than the IP at LE (IPLE) is to the LE cleavage site, and the nucleotide sequences between the IPs and the cleavage sites at the two ends are unrelated (Figure 1A). Among IS608 copies from different H. pylori strains, the sequence between IPRE and the RE cleavage site is completely conserved whereas it is variable between IPLE and the LE cleavage site.
Figure 1. IS608 Transposition.
(A) Alignment (adapted from Kersulyte et al. [2002]) of the termini and flanking sequences of IS608 elements from Helicobacter pylori. The strain is indicated on the left; all the work reported here was done with IS608 from PeCan2A (top line). Flanking DNA is in black, bases of the left end (LE) are shown in red and orange, and bases of the right end (RE) are shown in shades of blue. Boxed sequences underlined with inverted arrows delineate the Imperfect Palindromes (IP) at each end. The bases are numbered such that, at each end, the cleavage site is between base−1 and base+1 where bases 5′ of the cleavage site are negative, and those 3′ are positive. (B) Model of single-stranded transposition from Guynet et al. (in press). Intermediates in the reaction are a circular transposon junction and a precisely sealed donor backbone.
Another curious feature of IS200/IS605 family members is that, unlike many DNA transposons which integrate essentially randomly, they always insert just 3′ of a specific four or five nucleotide (nt) sequence. The form of IS608 that is inserted is an excised circular intermediate (also known as a transposon junction; Figure 1B) in which the RE is linked directly to the LE (Guynet et al., in press). Upon excision, IS608 precisely reseals the gap left behind in the donor DNA, and insertion occurs without target site duplications (Ton-Hoang et al., 2005). Thus, these seamless reactions proceed without loss or gain of nucleotides or the need for host cell DNA repair factors.
All IS608 cleavage and rejoining steps are carried out by a single 155-amino acid transposase, TnpA (Guynet et al., in press). In the TnpA dimer, there are two active sites, assembled in trans, in which the tyrosine nucleophile of one monomer is near the HUH motif of the other. TnpA recognizes and cleaves only the top strand of each transposon end (Ton-Hoang et al., 2005), resulting in yet another level of asymmetry: due to the conserved polarity of cleavage, after nucleophilic attack by the active site tyrosine on the phosphodiester backbone of DNA at LE, TnpA is covalently linked to the transposon 5′ end whereas cleavage at RE results in the attachment of TnpA to flanking donor DNA (Figure 1B).
We have previously shown how TnpA recognizes and pairs its transposon ends (Ronning et al., 2005), a necessary prelude to the cycle of coordinated DNA strand cleavage and rejoining steps that constitute transposition. Here, based on five different crystal structures of complexes between TnpA and various IS608 LE and RE sequences, and associated biochemical experiments, we describe the entire transposition cycle - activation of TnpA upon end binding, recognition of the cleavage sites at the very edges of the transposon, and target site selection – and the absolute dependence of these steps on the inherent asymmetry of the transposon ends.
RESULTS
TnpA complexes with the LE26 DNA hairpin
We have previously described the crystal structures of TnpA alone and complexed with a 22-mer hairpin (RE22) representing IPRE (Figure 2A; Ronning et al., 2005). While these were informative, in both structures, the catalytic tyrosine, Y127, was tucked away from the HUH motif and hydrogen-bonded to S110 where it could not act as a nucleophile. As there are several crystal structures of HUH nucleases with fully assembled active sites (Guasch et al., 2003; Larkin et al., 2005), it was clear that a conformational change was required. The other puzzling aspect of active site assembly was how the catalytically required metal ion would bind. In other HUH nuclease crystal structures, the metal cofactor (Mg2+ or Mn2+) is coordinated by three protein ligands: the two HUH histidines and either a third histidine or a glutamate, depending on the nuclease. Neither of our structures suggested an obvious candidate for the third metal ion ligand.
Figure 2. Conformational Change Induced in TnpA by a Four Nucleotide Extension of the Imperfect Palindrome.
(A) Active site of TnpA bound to the RE22 hairpin (shown in blue). The two monomers of the TnpA dimer are colored green and orange. In this inactive conformation, the nucleophilic Y127 of one monomer is hydrogen-bonded to Ser110 of the other. (B) The same region of TnpA when bound to LE26 (shown in red and yellow) in which the IP is extended by 4 nt in the 5′ direction. Base A+18 has displaced Y127. Note that in the hairpin regions, LE and RE differ only by one T at the tip. The conformational change also necessarily shifts the residues that follow helix αD; we are only able to trace these in one of the monomers in the TnpA/LE26 structure.
To shed light on these issues, we crystallized TnpA with a series of longer DNA substrates that contained the IPs and extended towards the transposon ends. (In our base numbering convention [Figure 1], the cleavage site at each transposon end is between base−1 and base+1. Base numbers increase with distance from the cleavage site with negative numbers on the 5′ side of each cleavage site. Nucleotides (nt) flanking LE are italicized and the last four nt on RE are in bold.) We first co-crystallized and solved the 2.1 Å resolution structure of TnpA complexed with a 26-mer hairpin DNA (LE26) representing IPLE with a 4 nt 5′ extension (Table S1, Figure 2B). In the TnpA/LE26 complex, IPLE binds precisely as seen for IPRE. The only difference between IPLE and IPRE, an extra T in the LE hairpin loop, is turned toward the solvent and does not contact TnpA. However, as a consequence of adding four nt in the direction of the transposon end, there has been a major conformational change affecting both TnpA monomers in which helix αD carrying Y127 has swung across the face of the β-sheet to adopt a completely new location (Movie S1, Figure 2). The pivot point for the movement is around two residues, G117 and G118, in the loop immediately preceding helix αD (Figure S1). As a result of the helix movement, the nucleophilic -OH group of Y127 shifts 12 Å to a position now consistent with active site arrangements of other HUH nucleases.
The crucial element of LE26 that appears to drive the conformational change in helix αD is base A+18 in the 4 nt 5′ extension. In the TnpA/LE26 complex, the previously observed hydrogen bond between Y127 and S110 has been disrupted, and the space that had been occupied by Y127 in the TnpA/RE22 structure is now occupied by A+18 (Figure 2). The new hydrogen bond between A+18 and S110 is the only additional base-specific interaction that TnpA makes with LE26 when compared with RE22. The ability of A+18 to evict helix αD from its previous location is due in part to the dearth of interactions between helix αD and the rest of TnpA. In the TnpA/RE22 complex, helix αD is held in place by only one hydrogen bond and some scattered van der Waals contacts between hydrophobic groups. As a result of the intrusion of base A+18 in the TnpA/LE26 complex, the hydroxyl group of Y127 is no longer hydrogen-bonded to anything and thus is ready to attack the scissile phosphate.
Another residue of interest located on helix αD is Q131 which is highly conserved among IS200 transposases and whose mutation to alanine dramatically reduces transposition in vivo (Figure S2). As a consequence of the conformational change in helix αD, Q131 is now close to the HUH histidines where it appears appropriately placed to complete the divalent metal ion binding site. To test this, we soaked preformed TnpA/LE26 crystals overnight in buffer containing 5 mM MnCl2 (Table S1). The anomalous difference Fourier electron density calculated from data measured on these crystals in the neighborhood of the active site indeed shows Mn2+ coordinated by H64, H66, and Q131 (Figure S2).
We also compared the metal ion binding affinity of TnpA/RE22 and TnpA/LE26 complexes using isothermal titration calorimetry. The experiments were performed with Mn2+ as our previous experience with an HUH nuclease suggested that Mn2+ might bind with higher affinity than Mg2+(Hickman et al., 2002). As shown in Figure S2, TnpA/RE22 has no detectable affinity for Mn2+ whereas the divalent metal ion is clearly bound by TnpA/LE26. Consistent with the role of Q131 as the crucial third metal ion ligand, we cannot detect Mn2+ binding by the TnpA(Q131A)/LE26 complex. Taken together, these data indicate that the 4 nt 5′ extension of LE26 relative to RE22 causes a rearrangement in TnpA leading to active site assembly in which Y127 and the third metal ion ligand are simultaneously transported to where they are needed for catalysis.
Cleavage activity of TnpA is dependent on the 4 nt 5′ extension of the IPs
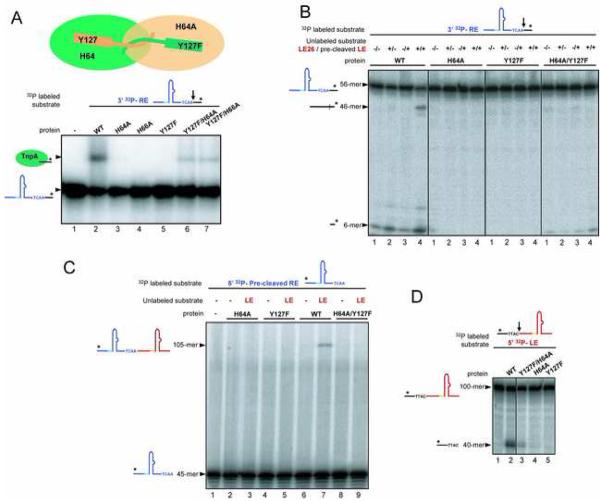
Our observation that a short 5′ extension of IPLE causes a conformational change in TnpA that appears to assemble the active site led us to ask if, in the LE26-bound form, TnpA was competent for catalysis. We have previously shown that TnpA cleaves 70- and 80-mer ssDNA RE and LE substrates which span the IPs and the cleavage sites (Ronning et al., 2005; Ton-Hoang et al., 2005). To investigate the effect of adding the four nt to IP sequences, we modified the cleavage reaction by providing the IPs and the cleavage sites on separate oligonucleotides. TnpA was first bound to oligonucleotides containing the IP sequences, with or without the 4 nt 5′ extensions, and substrates that spanned the cleavage sites from bases −4 to +4 were then added. Upon cleavage, TnpA forms a phosphotyrosine link to the four nt on the 5′ end of the break (Ronning et al., 2005; Ton-Hoang et al., 2005), and the resulting covalent complexes can be detected by SDS-PAGE on the basis of the increase in the molecular weight of TnpA.
As shown in Figure 5A, on both LE (lanes 5 and 6) and RE (lanes 3 and 4), formation of covalent complexes between TnpA and the cleavage substrates is dependent on the 4 nt 5′ extension. This is consistent with the crucial role of the additional nucleotides in inducing active site assembly. Furthermore, cleavage is dependent on the “correct” combination of LE and RE oligonucleotides: when IPLE with the 4 nt 5′ extension was mixed with the RE cleavage substrate (lane 9), or IPRE with the 4 nt 5′ extension was mixed with the LE cleavage substrate (lane 11), covalent complexes were not detected. This indicates that TnpA is able to differentiate between the LE and RE cleavage sites in a manner that depends on the particular IP bound.
Figure 5. Transposon End Cleavage Can Be Re-Directed.
(A) SDS-PAGE gel showing the results of cleavage assays. Upon cleavage of an 8-mer that spans the LE cleavage site, TnpA (green oval) becomes covalently attached to four nt and can be resolved from unmodified TnpA. TnpA cleaves LE and RE cleavage substrates (cs) only when the appropriate 5′ extension is added to IP sequences. No covalent complex is formed with the Y127F active site mutant. (B) Modification of cleavage specificity. Shown are the results of LE cleavage assays using LE26 and LEcs, and two variants.
RE31 and RE35 complexes
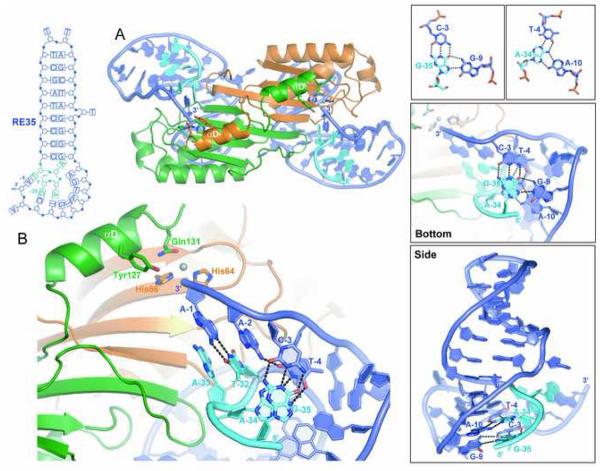
To address how the transposon ends reach the TnpA active sites, we determined the 2.9 Å resolution structure of TnpA co-crystallized with a RE 31-mer (RE31) that contains IPRE and the 9 nt in the 3′ direction reaching the end of the transposon (Table S1). In marked contrast to the effect seen on LE where extending the IP sequence towards the LE cleavage site leads to TnpA activation, in the TnpA/RE31 complex, each TnpA monomer is in the inactive conformation. Although IPRE is bound as seen in the TnpA/RE22 complex, the nucleotides 3′ of IPRE protrude out into solvent and become completely disordered after a few residues (Figure S4).
We therefore extended RE31 by adding 4 nt in the 5′ direction. Co-crystals of TnpA bound to RE35 diffracted to 2.4 Å resolution (Table S1), and the structure revealed two important differences when compared to that of TnpA/RE31 (Figure 3A, B; Figure S4). First, the active site has been assembled: helix αD has undergone the conformational change seen in the TnpA/LE26 complex and Mn2+ that was included in the crystallization buffer is bound. Secondly, all RE35 nucleotides are ordered and the conformation is such that its 3′ end, the last nt of RE, is now located in the active site.
Figure 3. The IS608 Right End is Directed into the TnpA Active Site by Internal DNA Interactions, not by Protein-DNA Recognition.
(A) Overall view of theTnpA/RE35 structure. (B) Close-up of the active site showing the base pairs between the four bases at the RE of IS608 (TCAA) and bases 5′ of IPRE (GAAT) from base G−35 to T−32, in light blue). The grey sphere is bound Mn2+. (C) Two base triplets (top) are central to the RE35 conformation (shown in two orthogonal views in the middle and bottom panels).
In the TnpA/RE35 complex, RE35 resembles a distorted letter “C” in which IPRE is bound on one side of TnpA, and the 4 nt 5′ extension and the 9 nt that lead to the cleavage site both curl around the protein surface into the active site of the same monomer on the opposite face (Figure 3C). Furthermore, there are specific base pairing interactions between the 4 nt 5′ extension of the IPRE and 3 nt at the 3′ end of the transposon that direct the terminal 3′ OH group, whose oxygen would be part of the scissile phosphate, into the active site (Figure 3B). The resulting arrangement of active site elements resembles the co-crystal structure of TraI, an HUH protein, bound to ssDNA that spans the F plasmid nic site in which the scissile phosphate was captured in the active site (Larkin et al., 2005).
A second important role of the 4 nt 5′ extension of IPRE is now clear: the last four nt of the transposon RE, TCAA, that immediately precede the cleavage site are held in place by a set of Watson-Crick base pairs with the 5′ extension of IPRE (Figure 3C). The base pairs (A−1:T−32, C−3:G−35, and T−4:A−34) do not match up linearly in the transposon sequence and the resulting twisted structure was unexpected. One base of the 5′ extension, A−33, recapitulates the role of A+18 in the TnpA/LE26 complex in that it has displaced Y127 during the transition from the inactive to the active conformation, and is now tucked into the pocket previously occupied by Y127.
Another surprising aspect of the way in which the 3′ end of the transposon is brought into the active site is that two base pairs, C−3:G−35 and T−4:A−34, are, in fact, part of base triplets (Figures 3C, S4). G−9, G−35, and C−3 form a G−G·C triplet which is stacked on the A−A·T triplet formed by A−10, A−34, and T−4. Although base triples have been frequently observed in RNA structures, to our knowledge, this is the first time DNA base triplets have been observed bound to a protein in a biologically relevant context. An interesting question is whether RE folds by itself to achieve the observed configuration or it is induced by TnpA.
The LE ternary complex
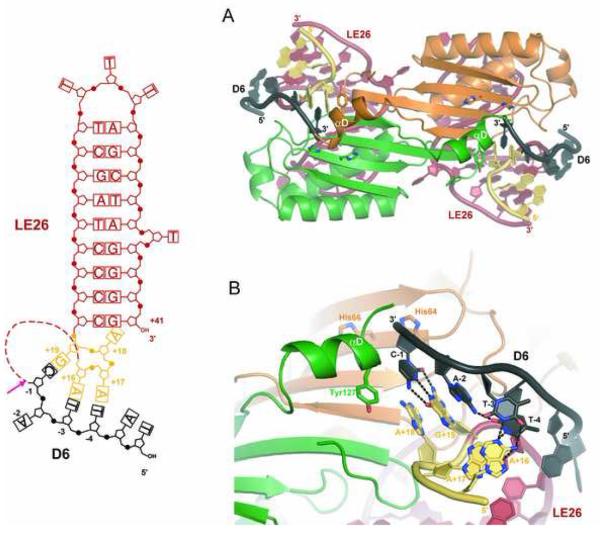
Attempts to co-crystallize TnpA with longer LE sequences that extend 5′ from IPLE towards the LE cleavage site have thus far been unsuccessful, due to the poor biophysical properties of these complexes. However, the observation that IS608 always integrates precisely 3′ of TTAC led us to explore the possibility that this sequence might somehow be recognized by TnpA. Although short TTAC-containing oligonucleotides do not bind to TnpA on their own, they do so in the presence of LE26 (data not shown), and we obtained crystals of TnpA bound to LE26 and the 6-mer TATTAC (denoted D6) that diffracted to 1.9 Å resolution (Table S1).
In the ternary LE complex, we observe the TTAC bases of D6 in the same position and orientation as the TCAA bases at the very end of RE35 in the TnpA/RE35 complex. The TTAC tetranucleotide is located in the active site where it is held in place by the 4 nt 5′ extension of IPLE (Figure 4). In this case, base pairs occur between A+16:T−3, A+17:T−4, and G+19:C−1 (Figure 4B). These orient the scissile phosphate (whose position can be inferred from the terminal 3′ OH group of D6) for nucleophilic attack by Y127. The overall DNA conformation is very similar to that seen in TnpA/RE35 except that the nucleotides between the cleavage site and LE26 are missing; the analogous regions of the TnpA/LE26/D6 and TnpA/RE35 complexes are virtually superimposable.
Figure 4. Ternary Complex of TnpA Bound to LE26 and the LE Donor Flank Echoes Right End Recognition.
(A) Overall view of the TnpA/LE26/D6 structure. On the left is a schematic of the oligonucleotides used where the dotted line indicates the break between LE26 and the donor flank, D6 (in black). The red arrow indicates the LE cleavage site. (B) Close-up of the active site showing that although the mode of interaction resembles the TnpA/RE35 structure, the specific bases differ.
The observation that the LE ternary complex structurally echoes the TnpA/RE35 complex is explained by the conserved polarity of the cleavage reactions. Although the interactions are similar on both ends, the flanking TTAC sequence recognized by the 4 nt 5′ extension of IPLE differs from the transposon 3′ end TCAA sequence that is recognized by the IPRE 5′ extension. The key is that the compensating changes to retain Watson-Crick base pairing are present in the 5′ extensions of the IPs.
In the TnpA/LE26/D6 complex, helix αD appears in yet another inactive position away from the active site. We suspect this is a crystallization artifact as the activated position of helix αD is blocked by a crystal lattice contact and we know that, in solution, helix αD is able to adopt the activated conformation in LE ternary complexes as TnpA pre-bound to LE26 can cleave an 8-mer that spans the cleavage site (Figure 5A). The previously seen inactive conformation (Ronning et al., 2005) is also unavailable as base A+18 of LE26 occupies the pocket where Y127 sits in uncomplexed TnpA and in the TnpA/RE22 and TnpA/RE31 complexes. Finally, the relevance of the observed position of helix αD in the TnpA/LE26/D6 complex is suspect as any extension of D6 in the 3′ direction (i.e., uncleaved LE) would immediately be in steric conflict with helix αD. This variability in the position of helix αD adds to the body of data that suggests it is a mobile structural element that tends to wander (Figure S5).
We note that the TnpA/LE26/D6 structure represents not only how TnpA recognizes its LE cleavage site but also how a TTAC-containing target site is captured. Since the circular transposon intermediate that can be integrated into target DNA, a transposon junction (Guynet et al., in press), contains the LE26 sequence but not the LE TTAC flank (Figure 1B), TnpA bound to the transposon junction is capable of binding (and subsequently cleaving) a TTAC-containing target. Therefore, recognition of TTAC of the target and of the LE flank is identical.
Changing cleavage specificity
The recognition of the integration target DNA sequence by protein-bound transposon DNA, rather than by TnpA directly, raises the obvious possibility that TnpA-catalyzed insertions might readily be re-targeted to other tetranucleotide sequences. This seems possible in principle, as three of the four nt in the 5′ extensions of the IPs do not appear to play any role beyond recognizing TTAC on LE or TCAA on RE. Thus, their replacement should not impede transposition.
To test the potential for re-directing, we changed either two or three of the nucleotides in the 5′ extension of IPLE, and introduced the compensating base-pairing partners in 8-mer cleavage substrates (which represent both the LE cleavage site and the target site). In the variant 5′ extensions, A+18 was retained as its role is to displace Y127 from its inactive location and it is not involved in DNA recognition. As shown in Figure 5B for two variant target sites, the cleavage specificity of TnpA can indeed be effectively changed, and we are in the process of examining transposon re-targeting in vivo.
Substrate cleavage and junction formation by active site mutants of TnpA
All our crystal structures of TnpA show the dimer in a trans active site configuration in which Y127 of one monomer is close to the HUH motif of the other. To confirm that this shared active site arrangement is used during catalysis, we performed several types of in vitro assays which recapitulate biochemical steps of IS608 transposition (Guynet et al., in press) using mixtures of single active site mutant versions of TnpA. We reasoned that, if the shared active arrangement is indeed used, mixing an inactive Y127F mutant with an inactive H64A mutant should result in measurable activity as any heterodimers would have one active site which combines the H64A and Y127F mutations yet the other would have the HUH motif and Y127 intact (Figure 6A).
Figure 6. Mixed Mutant Dimers Suggest Transposon End Cleavage is in Trans and Resolution is in Cis.
(A) Results of RE cleavage assays using mixed active site point mutants. The radiolabeled oligonucleotide is detected. (B) RE cleavage and donor backbone formation by mixed active site mutants was assessed by mixing a RE cleavage substrate (56-mer) with a LE flank (40-mer). The product of RE cleavage is a 6-mer, and the 46-mer is the sealed donor backbone that forms between the LE flank and the RE cleavage product. (C) Transposon junction formation by mixed active site mutants was assessed by mixing a LE cleavage substrate (100-mer which is cleaved to yield a 60-mer transposon LE) with a precleaved transposon RE (45-mer). The transposon junction is formed by the WT protein (lane 7) but not by mixed active site mutants (lane 9). (D) LE cleavage is catalyzed by mixed active site mutants (lane 3), but not by the single point mutants (lanes 4 and 5).
Figure 6A shows the results of RE cleavage experiments in which a 56-mer RE substrate that includes six flanking nt is incubated with wild-type TnpA, single active site point mutants, or mixed active site mutants. As expected (Ronning et al., 2005), no covalent products are observed with the H64A, H66A, or Y127F mutants (lanes 3-5). However, when the Y127A and H64A mutants are mixed, a covalent complex between TnpA and the 6-mer cleavage product can be detected (lanes 6 and 7), suggesting that a catalytically competent trans active site has assembled. The 6-mer product can also be directly detected after digestion with proteinase K. As shown in Figure 6B, in control experiments with the 56-mer RE cleavage substrate, wild-type TnpA generates the 6-mer product whereas the Y127F and H64A mutants do not (lanes 1 in first three panels). However, mixing these mutants results in the cleavage of the RE substrate to generate the 6-mer (fourth panel, lane 1), consistent with a trans active site configuration.
One of the hallmarks of IS608 transposition is that, upon transposon excision, the donor backbone is precisely sealed (Figure 1B). To determine whether mixed mutant dimers can generate sealed donor backbones, the 56-mer RE cleavage substrate with six flanking nt was mixed with a 40-mer LE backbone (i.e., 40 nt of flanking sequence ending with TTAC-3′) in the presence of LE26. The LE donor substrate is necessarily pre-cleaved since, in the mixed mutant dimers, only one active site is functional; LE26 is also required because the 5′ extension of its IP is needed to localize the TTAC of the LE donor substrate in the active site. As shown in Figure 6B, wild-type TnpA covalently joins the 40-mer LE donor substrate to the 6-mer product of RE cleavage to produce a 46-mer sealed donor backbone (panel 1, lane 4), but no such product is observed with the mixed mutant dimers (panel 4, lane 4).
Another reaction that occurs during IS608 transposition is the formation of a circular intermediate (Figure 1B) in which RE is linked directly to LE (Ton-Hoang et al., 2005). To test whether mixed mutant dimers can form such transposon junctions, we mixed a pre-cleaved 45-mer RE substrate that ends in TCAA-3′, representing the product of RE cleavage on the transposon side, with a 100-mer LE cleavage substrate that contains 60 nt of LE flanked by 40 nt (Figure 6C). Wild-type TnpA (lane 7) produces 105-mer junctions by catalyzing the attack of the RE 3′ OH of TCAA on the LE 5′ phosphotyrosine linkage that is formed upon LE cleavage. While mixed mutant dimers can cleave LE (Figure 6D, lane 3), junctions cannot be detected (Figure 6C, lane 9).
These observations show that, while mixed mutant dimers can cleave both LE and RE, a result consistent with a trans active site arrangement for cleavage, they cannot form sealed donor backbones or transposon junctions. This suggests that the mixed mutant dimers are deficient in the resolution steps of transposition, as they are unable to resolve phosphotyrosine intermediates to form the appropriately joined products.
DISCUSSION
The overwhelming body of evidence indicates that IS608 transposes using ssDNA (Kersulyte et al., 2002; Ton-Hoang et al., 2005; Ronning et al. 2005), and we have recently shown that ssDNA LE and RE oligonucleotides are readily cleaved, form transposon junctions and sealed donor backbones, and integrate pre-formed transposon junctions into target DNA in the presence of TnpA (Guynet et al., in press). With our structural results demonstrating how TnpA is activated and how it recognizes the transposon ends and its cleavage sites, together with the observation that helix αD is a mobile structural unit, we propose the following model for IS608 transposition (Figure 7, Movie S2):
Figure 7. Model for IS608 Transposition.
Upon LE and RE binding (A), Y127 of each monomer cleaves the transposon ends and becomes covalently attached to the 5′ side of the gap (B). Movement of αD helices from trans to cis and resolution of the phosphotyrosine intermediates results in a transposon junction and a sealed donor backbone (C). For integration (D), the donor backbone is replaced by target DNA containing TTAC which is recognized by an element of LE DNA. Cleavage (E), movement of the αD helices, and resolution of the phosphotyrosine intermediates (F) results in transposon insertion into target DNA immediately 3′ of TTAC.
Transposition starts with TnpA locating and pairing the transposon ends by binding to the hairpins formed by the top strand LE and RE IPs (Figure 7A). IPLE and IPRE differ by only one base in the hairpin loop, and our structures of the TnpA dimer bound to two identical DNA molecules most likely reliably reflect features of TnpA bound to one IPLE and one IPRE. Binding the transposon IPs induces the conformational change seen in the TnpA/LE26 complex during which helix αD moves into the activated position thereby assembling the active site.
Divalent metal ion binding to the assembled active site localizes and polarizes the scissile phosphate, preparing it for nucleophilic attack by Y127 (Hickman et al., 2002; Larkin et al., 2005). On LE, upon cleavage, Y127 becomes covalently linked to the 5′ end of the transposon while the 3′ end of flanking donor remains bound through base-base interactions between its TTAC and the four nt just 5′ of IPLE (as in the TnpA/LE26/D6 complex). We have no evidence that the 15 transposon nt between this 5′ extension and the 5′ end of the transposon interact with TnpA, consistent with their variability among different IS608 isolates (Berg et al., 2002).
On RE, cleavage results in a 5′ phosphotyrosine linkage between Y127 and the flanking donor DNA while the 3′ end of the transposon stays bound through base-pairing interactions between the terminal TCAA tetranucleotide and the four nt 5′ extension of IPRE (as in the TnpA/RE35 complex). The entire RE (represented by RE35) is ordered and its DNA conformation is stabilized by internal hydrogen bonds, consistent with its complete sequence conservation.
The subsequent formation of the transposition intermediates, a transposon junction and the sealed donor backbone, would arise straightforwardly if the two αD helices now trade places, pivoting on G117 and G118 and bringing the covalently linked DNA strands with them (Figure 7B, Movie S2). This model requires the 3′ OH groups to remain in their “active sites of origin” to act as the nucleophiles to resolve the swapped phosphotyrosine linkages. This immobility seems likely as our structures of both LE and the RE complexes suggest that 3′ ends would remain bound as long as the IPs with their four nt 5′ extensions are present.
By this mechanism, in one active site of the dimer, a sealed donor backbone would be generated by nucleophilic attack by the stationary LE donor flanking 3′ OH (TTAC-OH) on the swapped RE donor flank 5′ phosphotyrosine linkage. In the other active site, attack by the resident RE 3′ OH (TCAA-OH) on the 5′ phosphotyrosine linkage of the swapped transposon LE would result in a transposon junction. This proposed reaction scheme is supported by the three previously unexplainable aspects of asymmetry seen at the transposon ends: the tolerance of sequence variability at LE but not at RE; the cleavage polarities that dictate that one phosphotyrosine linkage joins TnpA to the transposon end whereas the other covalently attaches TnpA to flanking DNA; and, finally, the spacing differences between the cleavage sites and the IPs. On RE, upon cleavage TnpA becomes attached to the RE flank which is part of the donor plasmid and presumably can move freely. It is therefore easy to imagine that the traveling helix αD can bring covalently attached flanking DNA to the adjacent active site. However, on LE, it is the transposon 5′ end that is attached to helix αD. Thus, LE must have a sufficiently long and flexible spacer between the 4 nt IPLE extension and the 5′ end of the transposon that can act as a tether with some slack so helix αD can move from one active site to the other. The slack is necessary because LE cannot move as a unit as it is anchored to the TnpA dimer by the IPLE hairpin.
The notion of a switch from the trans to a cis active site arrangement is supported by the crystal structure (PDB ID 2fyx) of a closely related transposase from Deinococcus radiodurans, which was captured in the inactive configuration but with a cis helix αD arrangement. Although we have no structural evidence that the cis configuration occurs during IS608 transposition, our biochemical data showing that mixed mutant dimers are defective in the resolution steps provides strong support that both transposon junction and sealed backbone formation occurs with helix αD in cis: in this configuration, mixed mutant dimers have no functional active sites. As shown in Movie S2, the switch of helices from one active site to the other can be easily modeled as long as the RE flank and the nucleotides between IPLE and the LE cleavage site are free to move.
For the next step, integration, we do not know if the excised transposon junction intermediate remains bound to TnpA or is released and rebound. If, during recombination, the junction is released upon formation, it seems likely that TnpA resets to the trans configuration before transposon junction recapture, as we have only observed uncomplexed TnpA in that state. If the transposon junction stays bound, the cleavage steps required for integration may start with TnpA in the cis configuration and switch to trans for resolution. From the point of view of the overall mechanism, the outcomes of these two possibilities are equivalent.
We propose that integration of a transposon junction intermediate into a TTAC-containing target is a mechanistic replay of excision. After transposon junction formation, the sealed donor backbone dissociates from the complex and is replaced by a TTAC-containing target sequence (Figure 7C). At one active site, the target DNA would bind through interactions between its TTAC tetranucleotide and the four nt 5′ extension of IPLE. Upon cleavage, the TTAC target flank would stay bound to the complex and the 5′ end would be covalently attached to Y127. At the other active site, the transposon junction would be cleaved, leaving TCAA (the RE 3′ end of IS608) bound to the active site and the LE (5′ end of IS608) linked through a 5′ phosphotyrosine linkage to Y127. A switch in the active site arrangements leads directly to the resolution steps in which the top strand of IS608 is inserted into a new target site: the stationary 3′ OH of the TTAC target flank attacks the swapped 5′ phosphotyrosine link at LE while the RE TCAA 3′ OH attacks the swapped 5′ phosphotyrosine linkage of the target flank that was created during target cleavage (Figure 7D).
One of the most baffling questions about IS608 transposition was how its transposase, a 155 residue, single-domain protein, could carry out all of the required steps. Part of the answer is that TnpA is sneakier than we thought: it uses bound transposon DNA to recognize its cleavage sites both on donor and target DNA obviating the need for an additional DNA binding domain. Furthermore, TnpA works only on one DNA strand and does not have to deal with the complementary strand (Kersulyte et al., 2002; Ton-Hoang et al., 2005; Ronning et al. 2005; Guynet et al., in press).
If TnpA acts solely on ssDNA, how do its substrates arise? ssDNA formation might be promoted by plasmid supercoiling combined with the propensity of the IPs to form stem loops; if this occurs, the cleavage sites might, with some frequency, dissociate from their complementary strands. On the other hand, IS608 might excise in vivo only when DNA becomes single stranded during a normal cellular process such as lagging strand synthesis. If transposition occurs only during DNA replication, excised TnpA-bound sealed transposon circles might readily find a suitable TTAC-containing ssDNA target. Restricting transposition to one stage in the cell cycle would also ensure that the reaction is substrate-limited, thus preventing wanton and possibly destructive movement. Since ssDNA is also generated during conjugative DNA transfer, conjugating plasmids might be preferred targets for IS608 with the added advantage of immediate horizontal transfer.
The importance of ssDNA substrates for IS200 transposition is illustrated by recent work on Deinococcus radiodurans which can survive severe ionizing radiation damage using a mechanism called “extended synthesis-dependent strand annealing” to reconstitute its shattered chromosomes (Zahradka et al., 2006). During recovery, as much as 15% of newly synthesized DNA is single-stranded and, remarkably, the transposition frequency of ISDra2, an IS200/IS605 family member, increases 500-fold (Mennecier et al., 2006), suggesting that ssDNA is normally rate-limiting.
One of our most surprising results is that IS608 uses DNA sequences in the transposon itself to recognize its cleavage sites. This mode of recognition suggests that TnpA may be re-directed to new target sites, opening up an unanticipated avenue of transposon targeting with an array of genomic and biotechnological applications.
The interdependence of protein and DNA in creating suitable substrates is reminiscent of the intertwining of protein and RNA in ribosomes (Noller, 2005). This "self-recognition" has also been reported in other RNA-dependent systems. For example, the structural basis of the matchmaking functions of mitochondrial RNA binding proteins (MRPs) has recently been reported (Schumacher et al., 2006).
There is a striking parallel between the IS608 transposition pathway and the mobility of group II introns, particularly with that of the bacterial L1.LtrB system (Belfort et al., 2005; Lambowitz & Zimmerly, 2004). For example, the mobility of RNA introns is dependent on an intron-encoded multifunctional protein which is needed to bind the RNA. Upon splicing, exons bordering the intron are joined to each other, reminiscent of the resealing of donor DNA ends by TnpA. The lariat intermediate formed during intron splicing is similar to the circular IS608 transposon junction intermediate, both of which are covalently sealed. Perhaps the most significant parallel involves targeting, as these introns select their target, or “retrohoming” site, by base-pairing interactions between segments of the lariat intermediate and the target DNA strand. This is the basis for the development of re-targeted introns (Karlberg et al., 2001) that can be used to disrupt chromosomal genes. In intron mobility, the key catalytic component is the RNA itself, which catalyzes both the splicing and reverse splicing events. Because DNA has no known self-cleavage and strand transfer activity, IS608 has to rely on the assistance of TnpA to carry out the necessary cleavage and joining reactions. Our work demonstrates that a targeting mechanism previously thought to be the property of RNA-based mobile systems also occurs in the context of a mobile DNA element.
From a structural and functional perspective, the closest characterized relatives of TnpA are IS91-related transposases (Garcillán-Barcia et al., 2001). These are widespread in bacteria, have eukaryotic homologs (Kapitonov & Jerka, 2001), and have been implicated in the movement of so-called Common Regions (Stokes et al., 1997; Toleman et al., 2006) which may be responsible for a large fraction of the spread of antibiotic resistance genes. While IS91-like elements transpose by a mechanism that is likely different from IS608, they also contain subterminal IPs and insert just 3′ of short conserved sequences. It will be interesting to determine if their mode of target recognition resembles that shown here for IS608. If so, the potential for their re-targeting could provide new possibilities toward the site-specific modification of eukaryotic genomes.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
TnpA was purified as previously described (Ronning et al., 2005). DNA was synthesized on an Applied Biosciences 394 DNA/RNA synthesizer or purchased from IDT (Coralville, IA). Sequences used for crystallization are shown in Figures 2-4. Oligonucleotides containing inverted repeat sequences were resuspended in TE, heated to 95°C for 15 min, and then rapidly cooled on ice and placed at −20°C until use. All crystals were obtained by vapor diffusion in hanging drops by mixing complexes 1:0.8-1.0 (v/v) with well solutions as described below. TnpA/LE26 crystals were frozen in paratone, and the rest were flash frozen in liquid propane.
TnpA/LE26
TnpA at 8 mg/ml was mixed with LE26 at a final protein:DNA molar ratio of 1:1.1, and dialyzed against 20 mM Tris pH 7.5, 0.3 M sodium malonate, 0.2 mM TCEP, and 2 mM EDTA. Well solutions consisted of 0.2 M sodium formate and 15-20% PEG 3350.
TnpA/RE31
TnpA at 5 mg/ml was mixed with RE31 at a molar ratio of 1:1.1 and dialyzed against 20 mM Tris pH 7.5, 0.2 M sodium malonate, 10 mM MnCl2, and 0.2 mM TCEP. Well solutions consisted of 15-20% PEG 3350 and 0.1 M MES pH 6.5. Crystals were cryoprotected using well solution supplemented with 10% glycerol.
TnpA/RE35
The complex was prepared as for TnpA/RE31, and the well solution consisted of 20-25% PEG 3350, 0.1 M MES pH 5.5, and 0.1 M ammonium acetate. Crystals were frozen after stepwise replacement of water for 10% glycerol in the mother liquor.
TnpA/LE26/D6
TnpA (5 mg/ml) was mixed with LE26 and D6 at a 1:1.1:1.3 molar ratio, dialyzed against 20 mM Tris pH 7.5, 0.2 M MgCl2, 0.2 mM TCEP, and mixed with well solutions consisting of 6-10% PEG 600 and 50 mM sodium citrate pH 5.0. Crystals were cryoprotected by transfer into 10% PEG 600, 0.1 M MgCl2, 25 mM sodium citrate,10 mM Tris pH 7.5, and 15% glycerol.
Data Collection and Structure Determination
Data on the TnpA/LE26 and TnpA/RE31 crystals were collected on beamline 22-ID at APS on a MAR300 CCD detector. All other diffraction data were collected at 95 K on a rotating anode source equipped with multilayer focusing optics using Cu Kα radiation and an Raxis IV image plate detector. Data were integrated and scaled with Denzo and Scalepack (Otwinowski and Minor, 1997; Table S1). The structures were solved with molecular replacement by AMoRe (Navaza 2001), providing clear solutions in all cases. Search models and solution statistics are in Table S1, as are refinement statistics. For refinement, manual model building with program O (Jones, et al., 1991) was alternated with cartesian simulated annealing, positional and restrained B-factor refinement cycles using CNS 1.1 (Brünger et al. 1998). TnpA residues 133-155 could not be traced in the RE31, RE35, and LE26/D6 complexes. In the LE26 and LE26/Mn2+ complex structures, all protein residues could be included in one monomer. Both LE26 complexes showed weak electron densities and high temperature factors for residues 132-141 between helices αD and αE, suggesting high mobility. All molecular figures and animations were made with Pymol (DeLano, 2002). Coordinates have been deposited in the Protein Data Bank with the accession codes xxxx (TnpA/LE26), xxxx (TnpA/LE26+Mn2+), xxxx (TnpA/RE31), xxxx (TnpA/RE35), and xxxx (TnpA/LE26/D6).
Isothermal Titration Calorimetry (ITC)
Complexes of TnpA and TnpA/Q131A prebound to RE22 or LE26 (TnpA concentration of 0.035 - 0.038 mM) were prepared by dialysis against 10 mM HEPES pH 7.5, 0.2M NaCl, 5 mM DTT, 5 mM MgCl2, and 10% glycerol. Mn2+ binding was measured by ITC as described (Hickman et al., 2002) except that aliquots of 2 μl MnCl2 (5 mM in dialysis buffer) were used.
Covalent Complex Formation (Figure 5)
Wild type or the Y127F mutant TnpA at 1 mg/ml was mixed with hairpin oligos and cleavage substrates at a final protein:DNA molar ratio of 1:1.1:1.1. Protein/hairpin mixtures were dialyzed overnight against 20 mM Tris pH 7.5, 0.2 M sodium malonate, 20 mM MgCl2, prior to addition of the substrate and incubation for 1 h at 20 °C. Samples were heat-denatured in SDS-containing sample buffers and analyzed on 4-12% SDS-PAGE gels. TnpA and TnpA attached to the 4-mer product of cleavage were detected by Coomassie staining.
Purification of Mixed TnpA Mutants and Activity Assays
Each point mutant (Y127F, H64A, H66A) was introduced into pBS107, expressed in E. coli Rosetta (DE3) cells (Novagen), and separately purified as previously described (Ton-Hoang et al., 2005). To form mixed mutant dimers, equimolar amounts of either Y127F and H64A or Y127F and H66A were incubated together on ice for 20 min. RE cleavage reactions (Figure 6A) were performed by 30 min incubation at 37°C of 28 nM of a 3′-32P-labelled RE 70-mer cleavage substrate (see Supplemental Material for sequence) with 10μM of total protein in a final volume of 10 μl in 20 mM HEPES pH 7.5, 160 mM NaCl, 5 mM MgCl2, 10 mM DTT, 20 μg/ml BSA, 0.5 μg of poly-dIdC, and 20% glycerol. Reaction products were separated on a 16% SDS-PAGE gel and subsequently analysed by phosphorimaging. Similar reaction conditions (Guynet et al., in press) were used for RE cleavage and donor backbone formation (Figure 6B), transposon junction formation (Figure 6C), and LE cleavage (Figure 6D). All DNA sequences are listed in Supplemental Material. Products were separated on an 8% sequencing gel and analyzed by phosphorimaging.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Program of the National Institute of Diabetes, Digestive, and Kidney Diseases of the NIH. The Chandler lab was supported by CNRS (France) and EU contract LSHM-CT-2005-019023. C.G. received a training grant from the French MENRT. Data were also collected at the SER-CAT 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. We thank Dr. Wei Yang for insightful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM. In: Mobile Introns: Pathways and Proteins in Mobile DNA II. Craig NL, et al., editors. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grossekunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and Nmr System - a New Software Suite For Macromolecular Structure Determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: From Mu to kangaroo. Nature Rev. Mol. Cell Biol. 2003;4:1–13. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Debets-Ossenkopp YJ, Pot RGJ, van Westerloo DJ, Goodwin A, Vandenbroucke-Grauls CMJE, Berg DE, Hoffman PS, Kusters JG. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylor NCTC11637. Antimicrob. Agents Chemother. 1999;43:2657–2662. doi: 10.1128/aac.43.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMol Molecular Graphics System. 2002 at www.pymol.org.
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Garcillán-Barcia M, Bernales I, Mendiola MV, de la Cruz F. Single-stranded DNA intermediates in IS91 rolling-circle transposition. Mol. Microbiol. 2001;39:494–501. doi: 10.1046/j.1365-2958.2001.02261.x. [DOI] [PubMed] [Google Scholar]
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. Single-stranded transposition of IS608: In vitro reconstitution of a new transposition mechanism. Mol. Cell. doi: 10.1016/j.molcel.2007.12.008. in press. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ronning DR, Kotin RM, Dyda F. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus. Rep. Mol. Cell. 2002;10:327–337. doi: 10.1016/s1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nature Biotech. 2001;19:1162–1167. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 2002;184:992–1002. doi: 10.1128/jb.184.4.992-1002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Ilyina TV. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile Group II Introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Struct. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Lee HH, Yoon JY, Kim HS, Kang JY, Kim KH, Kim DJ, Ha JY, Mikami B, Yoon HJ, Suh SW. Crystal structure of a metal ion-bound IS200 transposase. J. Biol. Chem. 2006;281:4261–4266. doi: 10.1074/jbc.M511567200. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennecier S, Servant P, Coste G, Bailone A, Sommer S. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 2006;59:317–325. doi: 10.1111/j.1365-2958.2005.04936.x. [DOI] [PubMed] [Google Scholar]
- Miskey C, Izsvák Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell. Mol. Life Sci. 2005;62:629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. 2001;D57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Noller HF. Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. part A. Vol. 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Karamooz E, Zíková A, Trantírek L, Lukes J. Crystal structures of T. bruncei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Stokes HW, O'Gorman DB, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- Toleman MA, Bennett PM, Walsh TR. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature. 2006;443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.