Abstract
Macrophage migration inhibitory factor (MIF) is a cytokine that is involved in the regulation of inflammation as well as cell proliferation and differentiation. Deactivation of MIF by antibodies or inhibition of MIF binding to its receptor, CD74, attenuates tumor growth and angiogenesis. To discover small-molecule inhibitors of MIF’s biological activity, virtual screening was performed by docking 2.1 million compounds into the MIF tautomerase active site. After visual inspection of 1200 top-ranked MIF-ligand complexes, 26 possible inhibitors were selected and purchased, and 23 of them were assayed. The in vitro binding assay for MIF with CD74 revealed that 11 of the compounds have inhibitory activity in the µM regime including four compounds with IC50 values below 5 µM. Inhibition of MIF tautomerase activity was also established for many of the compounds with IC50 values as low as 0.5 µM; Michaelis-Menten analysis was performed for two cases and confirmed the competitive inhibition.
Introduction
Macrophage migration inhibitory factor (MIF) is an immunoregulatory and proinflammatory cytokine that is released by many cell types including macrophages and T-cells. Cytokines have been shown to be involved in the pathology of many human inflammatory diseases. As a cytokine that is detectable in circulation as well as in inflamed sites, MIF is implicated in several inflammatory and autoimmune diseases including rheumatoid arthritis, atherosclerosis, asthma, and lupus.1–3 MIF also is involved in multiple aspects of tumor growth including control of cell proliferation and promotion of angiogenesis.4,5 The central role of MIF in tumorigenesis has been further supported by genetic data showing that individuals with high expression alleles of the MIF gene are at greater risk for the development of invasive prostate cancer.6
The mechanism by which MIF acts as a proinflammatory mediator and thereby controls local and systemic immune responses is still unknown. An increasing body of evidence suggests that: (a) MIF is indirectly promoting angiogenesis by stimulating tumor cells to produce angiogenic factors, such as IL-8 and VEGF,5 (b) MIF directly downregulates the expression and function of the tumor-suppressor protein p53,7 (c) MIF is activating MAPKs,8,9 thereby enhancing cellular responses,10 and (d) MIF counter-regulates the expression of glucocorticoids,11,12 which suppress the expression and release of many proinflammatory molecules. Recent studies have shown that MIF signal transduction is initiated by binding to a transmembrane protein, CD74.13,14 Inhibition of MIF-CD74 binding has been shown to attenuate tumor growth and angiogenesis.4
Apart from its immunoregulatory role, MIF is also a phenylpyruvate tautomerase. Possible relationships between the tautomerase and immunological/inflammatory activities of MIF remain under investigation.15,16 Though MIF may exert some biological function via an enzyme mechanism,17 the catalytic activity of mammalian MIF is likely vestigial.18 However, there is evidence that the interaction of MIF with its receptor, CD74, occurs in the vicinity of the active site and that MIF tautomerase inhibition is correlated with inhibition of MIF-CD74 binding.16
The emerging role of MIF in angiogenesis and tumorigenesis as well as in inflammatory diseases indicates that modulating the cytokine’s activity can result in new therapies.2,19,20 Specifically, inhibition of the biological activities of MIF by antibodies or genetic deletion leads to reduced cellular proliferation and inhibition of tumor growth and angiogenesis.4,21,22 Moreover, as reviewed by Orita et al.,2 immunoneutralization of MIF and deletion of the MIF gene have been shown to have therapeutic benefits towards inflammatory diseases and also to suppress tumor growth.
Although injectable biological agents such as anti-cytokine antibodies or soluble cytokine receptors have effectively inhibited MIF activities, these strategies have significant associated risks and limitations in addition to high cost and inconvenience of application.19 Alternatively, MIF could be effectively targeted by oral formulation of small-molecule inhibitors. Biochemical and structure-function analysis of MIF has laid the basis for structure-guided drug design. The crystal structure for MIF revealed a new structural superfamily;23,24 the 114-residue MIF monomer has a β/α/β motif and three monomers associate to form a symmetrical trimer. The trimer is toroidal with a solvent-filled central channel. MIF also was found to show structural homology with two prokaryotic tautomerases, and phenylpyruvate and D-dopachrome were discovered to be MIF tautomerase substrates.17,25 Site-directed mutagenesis and crystallography have defined the MIF catalytic site.24 Each MIF trimer has three tautomerase active sites, which are well-defined cavities located at the interfaces of the monomer subunits. The N-terminal proline of MIF resides in the tautomerase binding pocket and has an unusually low pKa of 5.6–6;26 this renders the proline nucleophilic and allows it to effect the tautomerization of substrates.17
Several small-molecule tautomerase inhibitors of MIF have been reported,2,16,27–31 but none effectively inhibit MIF’s cytokine activity. The first small-molecule inhibitor of MIF’s biological activity was N-acetyl-p-benzoquinone (NAPQI), a metabolite of acetaminophen, which binds covalently to MIF’s N-terminal proline.16 NAPQI is a 40-µM tautomerase inhibitor, and it also decreases macrophage TNFα production as well as other biological actions of MIF.16 Recently, another covalent inhibitor, 4-iodo-6-phenylpyrimidine (4-IPP), was reported including a crystal structure for its covalent complex with MIF.30 Since covalent inhibitors are often not optimal as lead candidates, drug-like molecules that bind reversibly to the MIF N-terminal region have also been sought. A tryptophan Schiff base that is a 1.65-µM MIF tautomerase inhibitor was found;31 it decreases the number of THP-1 (human acute monocytic leukemia) cells that bind fluorescent MIF by 45% at 10 µM. Subsequently, a dihydroisoxazole derivative showed some inhibitory activity in assays of counter-regulation of glucocorticoid inhibition of TNFα, PGE2 and COX-2 production,29 but it shows low activity in a MIF-CD74 binding assay (vide infra). In view of this limited success with the discovery of small molecules that disrupt the biological activities MIF, we initiated an extensive virtual screening and assaying campaign to discover more potent and diverse lead compounds. As reported here, this effort resulted in the discovery of 11 novel compounds with IC50 values in the µM-range including 4 with IC50s below 5 µM. Such potent MIF-CD74 inhibitors not only provide leads for development of preclinical candidates for the treatment of cancer and inflammatory diseases, but also can be used as validation tools to further investigate the biology of MIF.
Methods
Virtual Screening
The crystal structure of MIF complexed with the substrate p-hydroxyphenyl pyruvate (HPP) (PDB ID: 1CA7)24 was employed in the docking calculations performed with Glide 4.0 (Schrödinger, LLC). Hydrogen atoms were added to the crystal structure and the complex was submitted to a series of restrained, partial minimizations using the OPLS-AA force field32 within the “Protein and Ligand Preparation” module of Glide. The key residue Pro1, which is postulated to act as a catalytic base to effect substrate tautomerization, was kept unprotonated, consistent with the observation that in the presence of no ligand or of an inhibitor, Pro1 has a pKa of 5.6–6.26 To compensate for the fixed protein structure, which is not expected to be optimal for a particular ligand, the van der Waals radii for non-polar ligand atoms were scaled by a factor of 0.8, thereby decreasing penalties for close contacts. Receptor atoms were not scaled. For the protein preparation, grid generation, and ligand docking procedures, the default Glide settings were used.
Drug-like subsets of the HitFinder collection from the Maybridge database [www.maybridge.com] and the ZINC database33 were used for the virtual screening. The HitFinder collection was pre-processed with the Glide module “LigPrep”, which prepared the ligands in multiple protonation and tautomerization states. This procedure resulted in ca. 24,000 structures as compared to the initial 14,400 of the HitFinder collection. The ZINC “drug-like” set had already been pre-processed and includes multiple protonation and tautomerization states.33 The complete virtual library, pre-filtered for properties based on Lipinski’s rules,34 totaled about 2.1 million compounds. All structures were docked and scored using the Glide standard-precision (SP) mode.35 The 40,000 top-ranked structures from ZINC and the 1,000 top-ranked ones from the Maybridge database resulting from the SP filter were re-docked and re-scored using the Glide extra-precision (XP) mode.36
In essence, Glide performs a thorough conformational search for a ligand; then it determines all reasonable orientations (“poses”) for each low-energy conformer in the designated binding site. In the process, torsional degrees of the ligand are relaxed, though the protein conformation is fixed. The SP “scoring function” is applied to judge the poses by considering, for example, hydrophobic and electrostatic interactions, hydrogen bonding, steric clashes, desolvation and internal energy of the ligand, and possible trapped or bridging water molecules in the binding site. In XP mode, the poses are further relaxed by complete energy minimizations. The resultant more accurate structures provide a basis for more detailed evaluation of contributions from explicit water molecules in the binding site and hydrophobic interactions.
Assayed Compounds
Compounds were purchased from commercial vendors. Their structures were confirmed by 1H NMR and mass spectroscopy. NMR spectra were recorded on a Bruker Avance DRX-500 (500 MHz) and ESI mass spectra were obtained on a Waters Micromass-ZQ at the Yale Chemical Instrumentation Center. Spectra are provided in the Supporting Information.
MIF-CD74 Binding Assay
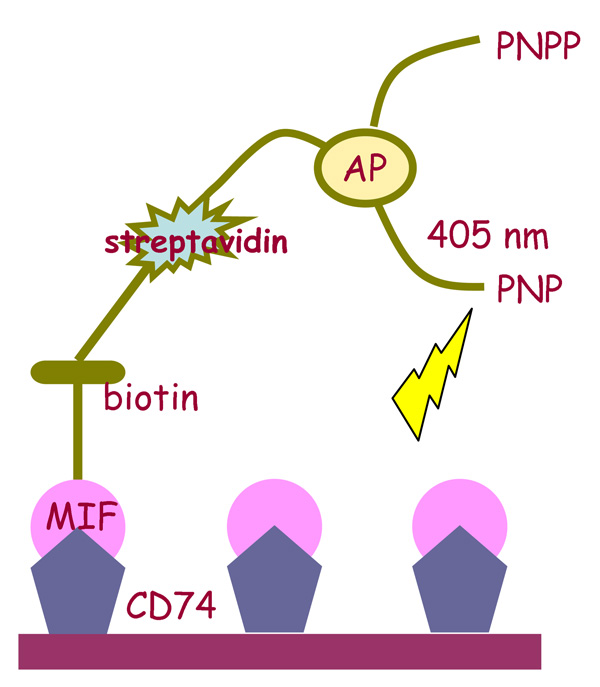
The interaction between MIF and its receptor, CD74, was analyzed following methodologies previously described (Figure 1).13 96-well plates were coated with recombinant MIF receptor ectodomain (sCD74 = CD7473–232), washed 4 times, and blocked with Superblock (Pierce Biotechnology) at 4 °C overnight. Test compounds were pre-incubated with biotinylated human MIF (2 ng/µl) (Roche Applied Sciences) prepared recombinantly37 for 30 minutes at room temperature. The mixtures then were added to wells for overnight incubation at 4 °C. The wells were washed 4 times and strepavidin-conjugated alkaline phosphatase (R&D Systems) was added for one hour incubation at room temperature. After additional washes, 60µl/well of p-nitrophenyl phosphate (Sigma) substrate was added. Absorbance at 405 nm was plotted as percent A405 relative to wells containing biotinylated human MIF alone. Each plot represented at least three independently performed assays, and each data point depicts a SEM ≤ 10%. Each test compound was analyzed over 10–12 concentrations, and over a ~500-fold concentration range.
Figure 1.
Schematic of the MIF-CD74 binding assay using biotinylated MIF and immoboilized CD74 ectodomain. For the bound complexes, alkaline phosphatase (AP) converts the substrate p-nitrophenylphosphate (PNPP) to p-nitrophenol (PNP), which absorbs light at 405 nm. In the presence of an inhibitor, MIF-CD74 binding and the product signal are reduced.
In Vitro Characterization of MIF Tautomerase Activity
Tautomerase activity was assessed using the substrate 4-hydroxyphenylpyruvate (4-HPP) following a previously reported procedure.38 Briefly, 4-HPP was dissolved in 50 mM ammonium acetate at pH 6.0, allowed to equilibrate to room temperature, and stored at 4 °C. The substrate and buffer were mixed in 96-well plates, followed by addition of purified rhMIF. Tautomerase activity was determined at 25 °C by adding 4-HPP to a quartz cuvette containing 0.435 M boric acid at pH 6.2 and the pre-incubated mixture of rhMIF) and test compound, in triplicate, and measuring the increase in absorbance at 305 nm over 30–90 s. Compounds were tested over a 5000-fold concentration range. In the same manner, Michaelis-Menten analysis was carried out for two of the inhibitors at concentrations of 0, 3 and 5 µM with HPP as substrate; Lineweaver-Burk plots demonstrated the competitive inhibition.
Results and Discussion
Validation of the Docking Method
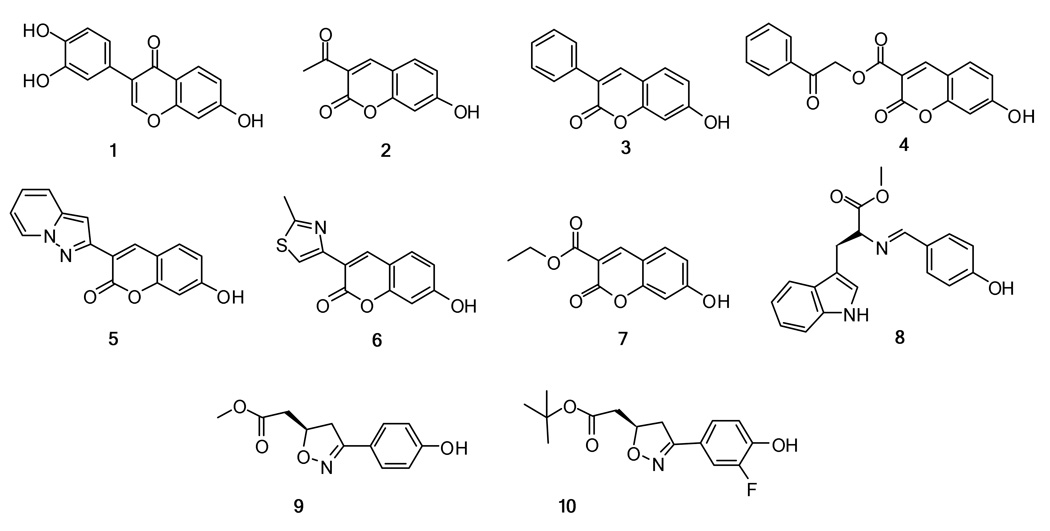
To test the performance of the Glide docking and scoring, 10 known MIF tautomerase inhibitors were processed with Glide as described above (Figure 2). The selected inhibitors were a chromene-4-one derivative (1),2 coumarin derivatives (2–7),2,27 an L-tryphophan Schiff base (8),31 9 (ISO-1),39 and 10 (ISO-17).40 The Glide XP scores showed significant correlation with the assay data for these known inhibitors (see Figure 3 and Table 1). Compounds 1–7 were assayed by Orita et al.,2 who used D-dopachrome as the substrate of MIF and Ki values were determined. Compounds 8–10 were assayed with the same protocol, but L-dopachrome methyl ester was the substrate and IC50 values were determined. To accommodate the data from the two different assays, the regression analysis included an indicator variable (0 or 1) for the log IC50s of compounds 8–10; this yielded an offset of 0.936 log unit for the three compounds.
Figure 2.
Structures of 10 reported MIF tautomerase inhibitors used to validate the docking procedure.
Figure 3.
Glide XP Score versus log Ki or log IC50 for 10 known tautomerase inhibitors.
Table 1.
Inhibition Constants, Ki, Half-maximal Inhibitory Concentrations, IC50, and Glide XP Gscores for Known MIF Tautomerase Inhibitors.
It also was possible to make comparisons of the structures for three of the MIF-inhibitor complexes obtained from Glide XP with prior work. Specifically, Orita et al. reported a crystal structure for the complex with 7 as well as structures for the complexes of 1 and 3 from docking with the program DOCK 4.0.1.2,27
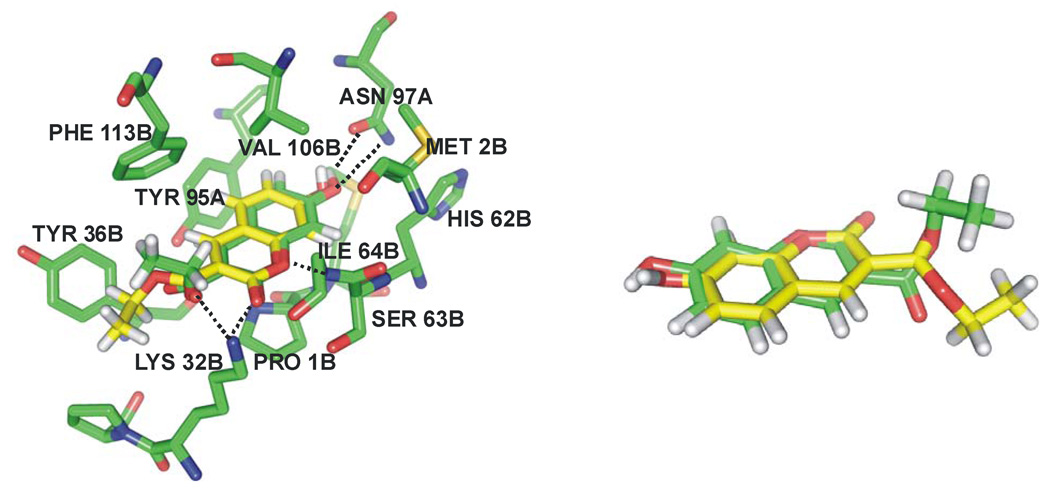
The crystal structure for the complex with 7 agrees very well with the conformation obtained by docking in the MIF binding site for the coumarin ring system (Figure 4). However, Glide predicts a syn geometry for the two carbonyl groups of compound 7, likely due to the two hydrogen bonds that are formed with Lys32 of MIF in this configuration. The apparent discrepancy with the anti geometry in the crystal structure may be attributable to the fact that the ethyl ester moiety is disordered and showed poor electron density.27 It also should be noted that the twisted ester moiety with a nearly 90° OCOC dihedral angle in both structures is highly unlikely; acyclic esters are predominantly Z with a dihedral angle near 0°. The RMSD between the Glide-calculated pose and the crystal structure of compound 7 is 0.59 Å when taking into account only the heavy atoms of the coumarin ring system, and 1.43 Å for all heavy atoms in the molecule. In general, the key features in the crystal structure are represented well by the Glide pose. The hydroxyl group at C7 of 7 forms a hydrogen bond with the side chain of Asn97. In fact, as drawn in Figure 2, the 10 known inhibitors all have a phenolic terminus, which is expected to participate in hydrogen bonds with the side-chain amido group of Asn97. Furthermore, the backbone NH of Ile64 makes hydrogen bonds with both oxygens of the lactone fragment in the coumarin ring. The coumarin ring also appears to have favorable T-shaped aryl-aryl interactions with Tyr95 and Phe113. And, again, the Glide pose features hydrogen bonds between the ammonium group of Lys32 and both carbonyl oxygens of 7.
Figure 4.
Comparison of the docked (yellow) and the observed crystal structure27 (green) for the complex of MIF with 7. The RMSD for heavy atoms in the coumarin ring is 0.56 Å, while for all heavy atoms it is 1.43 Å. The exocyclic ester group is disordered in the crystal structure.
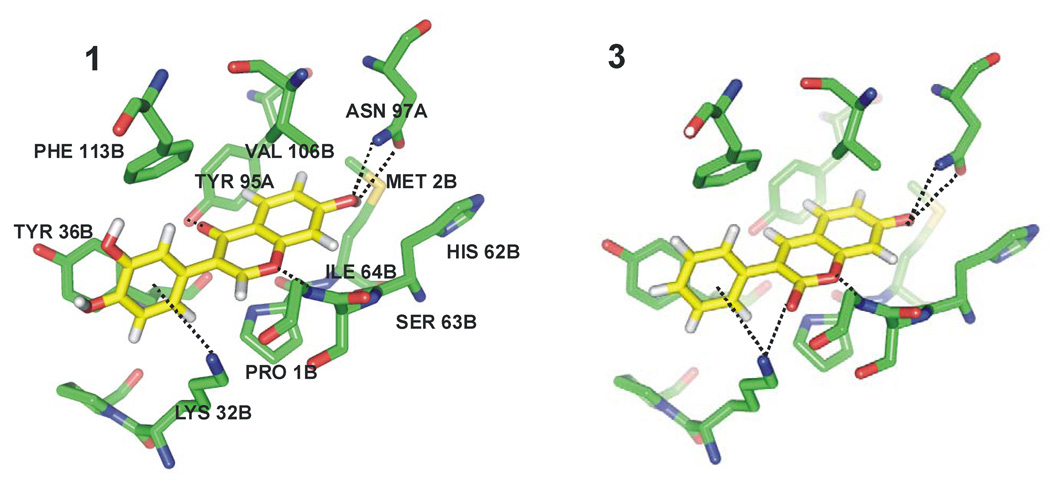
Significant similarities exist for the structures of the complexes with 7 and for those predicted by Glide for 1 and 3 (Figure 5). As in the previous models from DOCK,2,27 the bicyclic ring systems are positioned in the same manner, and both inhibitors form hydrogen bonds between a hydroxyl group and the side chain of Asn97 and between the ether oxygen of the chromen-4-one or of the coumarin ring and the backbone NH of Ile64. Aryl-aryl interactions in the binding pocket are notable between Tyr95 with the chromen-4-one or coumarin ring in a T-shaped geometry, while a parallel, stacked arrangement is predicted between the phenyl appendage and Tyr36. Also in accord with the prior docking results, 1 forms a hydrogen bond between its C4 carbonyl oxygen and the hydroxyl group of Tyr95; the O-O distance is 2.5 Å in the previous theoretical model2 and 2.6 Å in the Glide pose. A cation-π interaction is indicated between the phenyl ring of both compounds and the ammonium group of Lys32, and compound 3 also features a hydrogen bond between its carbonyl oxygen and the ammonium group of Lys32. In all, the structural results and the activity correlation in Figure 3 provided optimism for the virtual screening exercise.
Figure 5.
Docking poses from Glide XP for tautomerase inhibitors 1 and 3. The side chain of Ile64B has been removed for clarity.
Virtual Screening
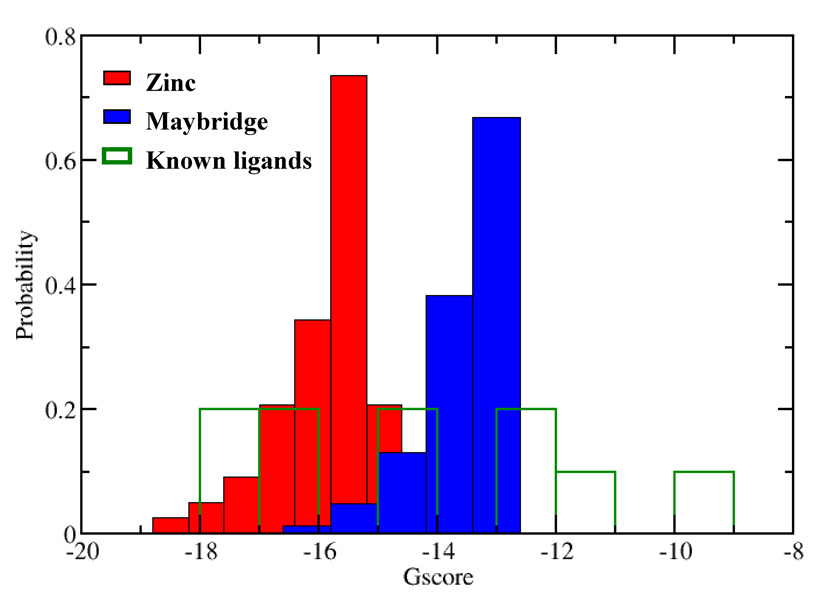
The docking was then extended to the 2.1 million structures obtained from the ZINC and Maybridge databases. The distribution of the Glide XP scores for the top-ranked 1,000 compounds from the ZINC database, the top-ranked 1,000 compounds from the Maybridge set, and the 10 known tautomerase inhibitors is shown in Figure 6. It is evident that the ZINC collection delivers many significantly lower XP Gscores that are likely to result in more potent inhibitors. This fact undoubtedly arises from the far larger size and diversity of the ZINC library (ca. 2.1 M compounds from 10 vendors) in comparison to the Maybridge Hitfinder collection (14,400 compounds). The average molecular weight for the top-scoring 1,000 compounds from the ZINC database is 321.7, while it is 306.1 for the top 1,000 Maybridge compounds. Thus, although the top ZINC compounds have on average one additional non-hydrogen atom, the greater structural variety of the larger database is undoubtedly an important asset.
Figure 6.
Distributions of the Glide XP scores for the top-ranked 1,000 ZINC compounds, the top-ranked 1,000 Maybridge compounds, and the ten known tautomerase inhibitors.
The complexes for the top-ranked 1,000 compounds from the ZINC database resulting from the XP processing, the top-ranked 100 compounds from ZINC with just the SP filter, and the top-ranked 100 XP hits from the Maybridge database were submitted to display and visual inspection. Many well-ranked structures were ruled unlikely owing to poor conformations including twisted amide and ester groups or overly short non-bonded contacts. Final selection of compounds for assaying also took into account chemical diversity, properties predicted by the QikProp program,41 and QikProp alerts concerning potentially undesirable chemical substructures. Structural diversity in compound selection is desired to provide alternative lead series for optimization. Previously reported cores, substituted phenols and especially coumarin derivatives,2 were avoided in the selection with some exceptions for interesting substituents.
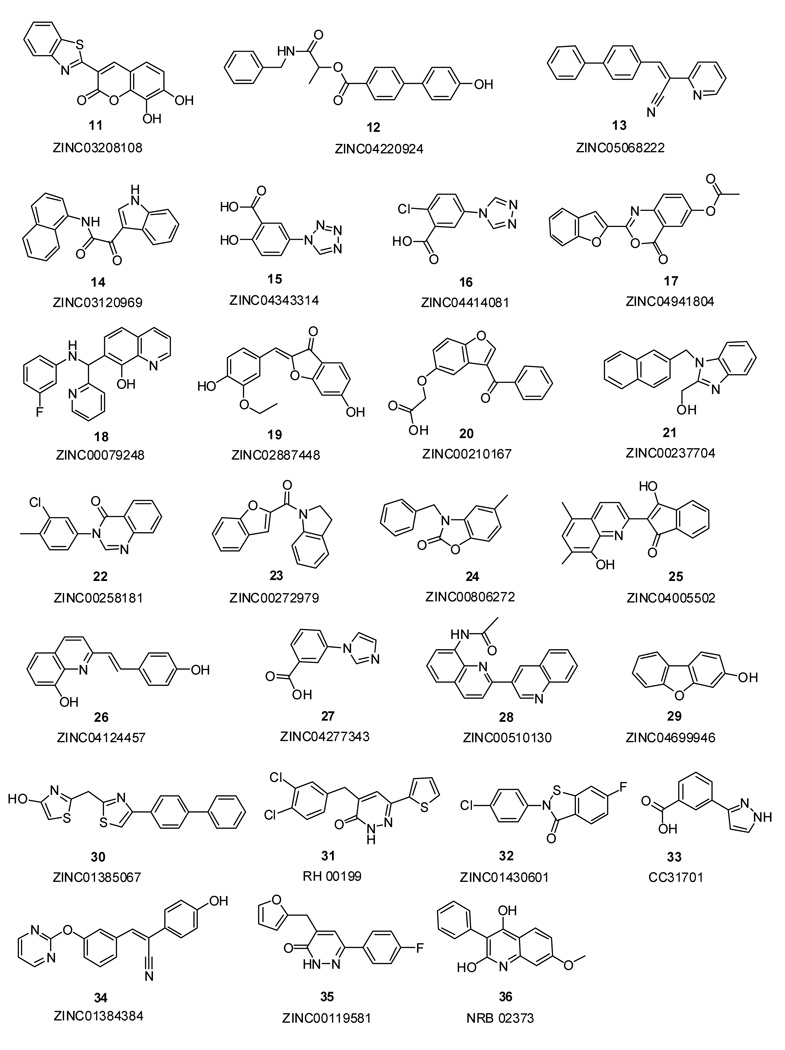
Finally, 34 potential inhibitors were designated for purchase; however, only 24 turned out to be commercially available. Compounds 11–30, 32, 34, 35 were chosen from the ZINC database and compound 31 from the Maybridge collection (Figure 7). For the unavailable compounds, a similarity search was performed using SciFinder Scholar [www.cas.org]. Commercially available compounds that showed 97% or greater similarity to the desired ones were re-docked using Glide, as described above. If the Gscore of a compound was within the range of previously selected XP or SP compounds, it was purchased. Compounds 33 and 36 were chosen in this manner. Thus, 26 compounds were finally acquired including five (14, 15, 16, 27, 33) which were selected based on their SP performance and visualization, though they were not in the top 1000 from the XP scoring. Ultimately 23 compounds were assayed. The chemical structures and purity of the compounds were verified with 1H NMR and mass spectroscopy at the Yale Chemical Instrumentation Center. Compound 34 was impure and gave incorrect NMR and mass spectra, while 35 and 36 were delivered in insufficient quantities for spectral analysis and were thus not assayed.
Figure 7.
Chemical structures of the purchased compounds and their database codes.
In vitro Assays
The 23 compounds were first assayed in vitro for disruption of human MIF binding to its receptor, CD74. Eleven of the 23 compounds were discovered to be active with half maximal inhibitory concentrations (IC50) in the µM range including four below 5 µM (Table 2). The IC50 of the most potent compound, 24, was 1.5 µM. Thirteen of the compounds were also assayed for MIF tautomerase inhibition, and 23, 24, and 32 were found to have IC50 values of 3, 0.5 and 4 µM, respectively. It should be noted that 9, a previously reported tautomerase inhibitor in the dopachrome-based assay,39 was inactive in the 4-HPP tautomerase assay and marginally active in the MIF-CD74 binding assay. It shows maximal inhibition of 40% at 10 µM, so an IC50 could not be obtained (Table 2). As other references, a biologically neutralizing anti-MIF IgG1 monoclonal antibody (clone12302, R&D Systems) was found to be a 0.4 µM inhibitor in the binding assay, while 4-IPP is inactive in the binding assay, but it is a 4.5 µM inhibitor in the 4-HPP tautomerization assay.30 Notably, the dihydroindole and benzooxazolone derivatives 23 and 24 are potent in both assays.
Table 2.
Experimental Activities from the Tautomerase and MIF-CD74 Inhibition Assays, and Glide Scores and Rankings.
| Compound | IC50 (µM) MIF-CD74 binding assaya |
IC50 (µM) 4-HPP tautomerase assaya |
XP Gscore |
SP Gscore |
XP Rank | SP Rank |
|---|---|---|---|---|---|---|
| 11 | NA | −17.89 | 32 | |||
| 12 | NA | −15.77 | 443 | |||
| 13 | NA | −15.90 | 389 | |||
| 14 | NA | −9.71 | 21 | |||
| 15 | 550 | NA | −9.50 | 52 | ||
| 16 | 250 | NA | −9.86 | 8 | ||
| 17 | 2.5 | 730 | −16.21 | 285 | ||
| 18 | 2.5 | 530 | −16.57 | 182 | ||
| 19 | NA | −15.74 | 458 | |||
| 20 | 1500 | 980 | −15.06 | 977 | ||
| 21 | NA | −16.41 | 223 | |||
| 22 | NA | −15.92 | 379 | |||
| 23 | 4.0 | 3.0 | −15.39 | 696 | ||
| 24 | 1.5 | 0.5 | −15.89 | 394 | ||
| 25 | NA | −16.24 | 278 | |||
| 26 | 8 | max 40% | −15.13 | 938 | ||
| 27 | NA | −9.63 | 31 | |||
| 28 | NA | −17.99 | 26 | |||
| 29 | 900 | max 38% | −15.8 | 430 | ||
| 30 | 65 | 300 | −16.73 | 150 | ||
| 31 | 250 | 430 | −18.15 | 1 | ||
| 32 | NA | 4.2 | −15.89 | 393 | ||
| 33 | 15 | max 30% | −9.67 | 27 | ||
| 9 | max 40% | NA | ||||
| 4-IPP | NA | 4.5 | ||||
| anti-MIF antibody | 0.4 |
NA for measured, but not active. Blank for not measured.
In order to check the expected competitive inhibition of the tautomerase activity, Michaelis-Menten analysis was performed for inhibitors 23 and 24 with 4-HPP as the substrate. Linear Lineweaver-Burk plots were generated using seven substrate concentrations and the inhibitors at concentrations of 0, 3, and 5 µM. For 23 the corresponding three Km values are 222 ± 20, 322 ± 35, and 533 ± 50 µM, and for 24 they are 235 ± 20, 322 ± 40, and 444 ± 50 µM. The values in the absence of the inhibitors are consistent with a previously reported Km of 170 ± 40 µM.42 In all cases, the y-intercept was the same indicating no change in the maximal rate for the catalyzed reaction, Vmax. In conjunction with the increasing Km values with increasing inhibitor concentrations, competitive inhibition is indicated.
There is not a straightforward correspondence between activity in the MIF-CD74 and tautomerase assays, and it would not be expected. In the simplest model for MIF-CD74 binding occurring near the tautomerase active site, the structure of the inhibitors affects how much they protrude from the active site. The leftmost residues in Figure 5 are on the surface of MIF. The extent, if any, to which the inhibitors protrude and the concomitant variability in how surface residues such as Lys32 rearrange would be expected to modulate the MIF-CD74 interaction. Small, potent tautomerase inhibitors like 4-IPP might have little effect on CD74 binding, while larger, weaker tautomerase inhibitors like 17 and 18 could be more disruptive of the interaction with CD74, as observed. Conversely, molecules that show no or little tautomerase inhibitory activity could still interfere with the binding of MIF to CD74, e.g., by binding in the same vicinity but having little effect on the binding of tautomerase substrates. In general, the MIF-CD74 antagonists do provide some tautomerase inhibition; the exceptions are 15, 16, and 9 which are very weak MIF-CD74 antagonists. This can be interpreted as additional support for the notion that the interaction of MIF with its receptor, CD74, occurs in the vicinity of the active site.16 However, there is no question that detailed understanding of the inhibitory effects of specific compounds awaits experimental structures for MIF-CD74-inhibitor complexes. The fact that MIF is trimeric with three catalytic sites also adds potential complexity for interpretation of the tautomerase results, which may depend on the effects on activity at one site upon binding at another site.
What is absolutely clear from the results in Table 2 is that through virtual screening, which targeted the tautomerase active site of MIF, there has been substantial success in finding both promising inhibitors of MIF’s tautomerase activity and of its binding to CD74. The reported leads also provide opportunities for pursuing analog series that can be expected to yield valuable insights on the relationship of activity and structure.
As can be seen in Table 2, most compounds that were chosen for purchase and assaying did not receive the highest XP rankings for the reasons noted above. However, the #1 XP-ranked compound from the Maybridge database (31) was purchased and found to be a 250-µM inhibitor in the receptor assay, while 28 and 11, which were ranked 26th and 32nd from the ZINC database were inactive. The particularly promising compounds 23 and 24 were #696 and #394 in the ZINC XP evaluation. In addition, three of the five compounds selected based on the SP scoring, which were not in the top 1000 for XP, did show some activity. Overall, the docking results were provocative and essential, but skilled human filters are still necessary in our experience.
Calculated Properties and Structural Basis for Inhibition
Some predicted properties from QikProp for the most active MIF-CD74 inhibitors are summarized in Table 3.41 The rms errors for QikProp predictions are 0.5–0.6 log unit. When QikProp is run on 1700 known oral drugs, 90% have MW < 470, QP log P < 5.0, QP log S > −5.7, and QP PCaco > 22 nm/s.43,44 The results in Table 3 compare favorably with these limits, so optimism can be expressed for the utility of the compounds as drug leads. Development starting from 30 and 31 would need to take into account their low predicted solubilities.
Table 3.
Predicted Properties from QikProp for the Most Active Compounds.
| Compound | MWa | QP log Pb | QP log Sc | QP PCacod |
|---|---|---|---|---|
| 16 | 223.618 | 1.55 | −2.50 | 61.3 |
| 17 | 321.289 | 2.09 | −3.74 | 584.3 |
| 18 | 345.375 | 4.61 | −5.46 | 2362.6 |
| 23 | 263.295 | 3.33 | −3.76 | 3998.2 |
| 24 | 239.273 | 2.97 | −3.17 | 2660.7 |
| 26 | 263.295 | 3.22 | −4.01 | 840.4 |
| 30 | 350.452 | 5.07 | −6.46 | 1605.7 |
| 31 | 337.223 | 4.75 | −6.10 | 1594.1 |
| 33 | 188.185 | 1.31 | −2.18 | 69.0 |
Molecular weight.
Predicted octanol/water log P.
Predicted aqueous solubility; S in mol/L.
Predicted Caco-2 cell permeability in nm/s.
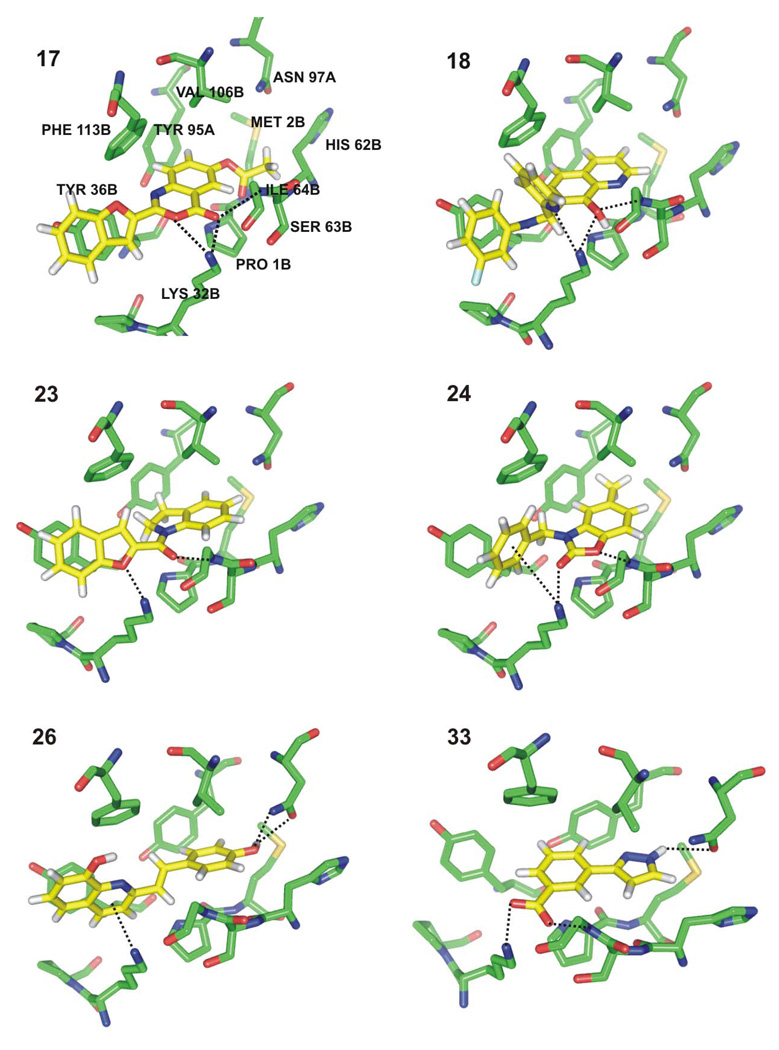
Figure 8 illustrates the computed poses for complexes between MIF and the six most potent MIF-CD74 inhibitors, 17, 18, 23, 24, 26, and 33. The compounds represent six unique chemotypes for potential lead optimization. Aryl-aryl interactions, hydrogen bonds, and seemingly favorable van der Waals contacts provide the binding energy. The compounds are predicted to bind in the MIF catalytic pocket in a similar manner to the known tautomerase inhibitors in Figure 4 and Figure 5. 17, 18, 23, 24, and 33 form a hydrogen bond with the backbone nitrogen of Ile64 as well as with the ammonium group of Lys32. Of the nine compounds with a MIF-CD74 inhibitory activity at or below 250 µM, only 26 is predicted to form the two hydrogen bonds with Asn97 that are expected for the known inhibitors 1–10. However, the pyrazolyl NH of 33 participates in a hydrogen bond with the carbonyl oxygen of the Asn97 side chain. The imidazole analogue 27 was purchased thinking that it was likely zwitterionic and that the protonated nitrogen might participate in an analogous hydrogen bond with Asn97; 15 and 16 provided additional variations of the theme and show some activity. Though the remaining most-active compounds lack a hydrogen-bonding group in the proximity of Asn97, introduction of such a group could be explored during lead optimization. Incidentally, it seems possible that alternative poses are viable in some cases by flipping the ligand end-to-end in the binding site, e.g., the benzofuran unit could be proximal to Asn97 in the complexes for 17 and 23.
Figure 8.
Docking poses for six of the most potent MIF-CD74 inhibitors: 17, 18, 23, 24, 26, 33. The side chain of Ile64 has been removed for clarity.
Notably, all of the purchased compounds have conjugated ring systems that were expected to be well buried in the active site. As shown in the examples in Figure 8, Tyr95 is involved in an edge-to-face aryl-aryl interaction in all cases, while Tyr36 and Phe113 are also providing stacking and edge-to-face interactions with all inhibitors except 33. The latter compound is the only charged inhibitor depicted in Figure 8; its carboxylate group is predicted to participate in a salt bridge with Lys32 and to accept an additional hydrogen bond from the backbone NH of Ile64. Though 33 has a molecular weight of only 188, it appears to have excellent hydrogen-bonding and aryl-aryl compatibility with the MIF active site. Another useful design element is potential cation-π interactions with Lys32, as with 24 and 26.
Conclusions
Virtual screening of drug-like subsets of the publicly available ZINC and Maybridge databases was performed to seek inhibitors of the biological activity of the cytokine MIF. The docking procedure was validated on three known MIF tautomerase inhibitors by comparing the Glide binding poses with previously calculated models for two of these compounds as well as a crystal structure. The structures from Glide reproduced well the expected geometries of the complexes. Furthermore, the Glide XP scores showed significant correlation with tautomerase assay data for ten known inhibitors. Following the virtual screening, the complexes for the top-ranked 1,200 compounds were visually inspected and 26 compounds were purchased. One compound was impure and gave incorrect spectra and two others were delivered in inadequate quantities. Finally, 23 compounds were assayed in vitro for disruption of MIF-CD74 binding and 13 for MIF tautomerase inhibition. Notably, 11 of the 23 compounds were found to inhibit the binding of MIF to its receptor in the µM range including four with IC50 values below 5 µM.
Visualization of the computed structures shows potentially favorable aryl-aryl interactions, hydrogen bonding, and van der Waals contacts between the inhibitors and the active site of MIF. However, a tautomerase inhibitor is not necessarily a MIF-CD74 antagonist. While compounds 32 and 4-IPP inhibit the enzymatic activity, they were inactive in the MIF-CD74 binding assay. On the other hand, compounds 17, 18, and 26, while being MIF-CD74 inhibitors in the low µM realm, are weak tautomerase inhibitors. Deeper understanding of the structure-activity relationships would greatly benefit from crystallographic investigations of MIF-CD74-inhibitor complexes. Nevertheless, judging by the success of the present research, it appears likely that the MIF-CD74 inhibitors do bind in the MIF tautomerase active site.
The present work has led to discovery of small-molecule inhibitors of MIF-CD74 binding, which are the most potent that have been reported to date. The structural diversity of the inhibitors has provided valuable alternative series for on-going lead optimization. Potent, selective MIF-CD74 inhibitors are anticipated to provide promising candidates for the treatment of cancer and inflammatory diseases. Moreover, they can be used as validation tools to further explore the biology of MIF.
Supplementary Material
NMR and mass spectral data for the assayed compounds. These materials are available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
Gratitude is expressed to the National Institutes of Health (AI043210, AR049610, AR050498, GM032136), the National Foundation for Cancer Research, and the Alliance for Lupus Research for support, and to Dr. Julian Tirado-Rives for computational assistance. ZC is also grateful for support from an AACR Judah Folkman Fellowship for Cancer Research in Angiogenesis.
Abbreviations
- MIF
Macrophage Migration Inhibitory Factor
- 4-HPP
4-hydroxyphenylpyruvate
- 4-IPP
4-iodo,6-phenylpyrimidine
- NAPQI
N-acetyl-p-benzoquinone
- ISO-1
(S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester
- ISO-17
(S,R)-[3-(3-Fluoro-4-hydroxy-phenyl)-4,5-dihydro-isoxazol-5-yl]-acetic acid tert-butyl ester
- MAPK
mitogen-activated protein kinase
- IL-8
interleukin-8
- PGE2
prostaglandin E2
- VEGF
vascular epithelial growth factor
- rhMIF
recombinant human MIF
- TNF
tumor necrosis factor
- PNPP
p-nitrophenyl phosphate
- PNP
p-nitrophenol
- AP
alkaline phosphatase
- RMSD
root mean-squared deviation
References
- 1.Morand EF, Leech M, Bernhagen J. MIF: a new cytokine link between rheumatoid arthritis and atherosclerosis. Nat. Rev. Drug Discov. 2006;5:399–411. doi: 10.1038/nrd2029. [DOI] [PubMed] [Google Scholar]
- 2.Orita M, Yamamoto S, Katayama N, Fujita S. Macrophage migration inhibitory factor and the discovery of tautomerase inhibitors. Curr. Pharm. Des. 2002;8:1297–1317. doi: 10.2174/1381612023394674. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez E, Gomez LM, Lopez-Nevot MA, Gonzalez-Gay MA, Sabio JM, Ortego-Centeno N, de Ramon E, Anaya JM, Gonzalez-Escribano MF, Koeleman BP, Martin J. Evidence of association of macrophage migration inhibitory factor gene polymorphisms with systemic lupus erythematosus. Genes Immun. 2006;7:433–436. doi: 10.1038/sj.gene.6364310. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 5.Hagemann T, Robinson SC, Thompson RG, Charles K, Kulbe H, Balkwill FR. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol. Cancer Ther. 2007;6:1993–2002. doi: 10.1158/1535-7163.MCT-07-0118. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Siegler KL, Vera PL, Iczkowski KA, Bifulco C, Lee A, Gregersen PK, Leng L, Bucala R. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 2007;8:646–652. doi: 10.1038/sj.gene.6364427. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, Bucala R. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: Regulatory role in the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 2002;99:345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos L, Lacey DC, Yang Y, Leech M, Morand EF. Activation of synovial cell p38 MAP kinase by macrophage migration inhibitory factor (MIF) J. Rheumatol. 2004;31:1038–1043. [PubMed] [Google Scholar]
- 9.Mitchell RA, Metz CN, Peng T, Bucala R. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J. Biol. Chem. 1999;274:18100–18106. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 10.Kusuhara M, Chait A, Cader A, Berk BC. Oxidized LDL stimulates mitogen-activated protein kinases in smooth muscle cells and macrophages. Arterioscler. Thromb. Vasc. Biol. 1997;17:141–148. doi: 10.1161/01.atv.17.1.141. [DOI] [PubMed] [Google Scholar]
- 11.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 12.Leech M, Metz C, Bucala R, Morand EF. Regulation of macrophage migration inhibitory factor by endogenous glucocorticoids in rat adjuvant-induced arthritis. Arthritis Rheum. 2000;43:827–833. doi: 10.1002/1529-0131(200004)43:4<827::AID-ANR13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendrat K, Al-Abed Y, Callaway DJE, Peng T, Calandra T, Metz CN, Bucala R. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry. 1997;36:15356–15362. doi: 10.1021/bi971153a. [DOI] [PubMed] [Google Scholar]
- 16.Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. U.S.A. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosengren E, Bucala R, Aman P, Jacobsson L, Odh G, Metz CN, Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol. Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- 18.Swope MD, Lolis E. Macrophage migration inhibitory factor: Cytokine, hormone, or enzyme? Rev. Physiol. Biochem. Pharmacol. 1999;139:1–32. doi: 10.1007/BFb0033647. [DOI] [PubMed] [Google Scholar]
- 19.Bucala R, Lolis E. Migration inhibitory factor: A critical component of autoimmune inflammatory diseases. Drug News Perspect. 2005;18:417–426. doi: 10.1358/dnp.2005.18.7.939345. [DOI] [PubMed] [Google Scholar]
- 20.Lolis E, Bucala R. Macrophage migration inhibitory factor. Expert Opin. Ther. Targets. 2003;7:153–164. doi: 10.1517/14728222.7.2.153. [DOI] [PubMed] [Google Scholar]
- 21.Chesney J, Metz CN, Bacher M, Peng T, Meinhardt A, Bucala R. An essential role for macrophage migration inhibitory factor (MIF) in angiogenesis and the growth of a murine lymphoma. Mol. Med. 1999;5:181–191. [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa H, Nishihira J, Sato Y, Kondo M, Takahashi N, Oshima T, Todo S. An antibody for macrophage migration inhibitory factor suppresses tumour growth and inhibits tumour-associated angiogenesis. Cytokine. 2000;12:309–314. doi: 10.1006/cyto.1999.0562. [DOI] [PubMed] [Google Scholar]
- 23.Sun HW, Bernhagen J, Bucala R, Lolis E. Crystal structure at 2.6-Å resolution of human macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubetsky JB, Swope M, Dealwis C, Blake P, Lolis E. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 25.Rosengren E, Åman P, Thelin S, Hansson C, Ahlfors S, Björk P, Jacobsson L, Rorsman H. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 1997;417:85–88. doi: 10.1016/s0014-5793(97)01261-1. [DOI] [PubMed] [Google Scholar]
- 26.(a) Stamps SL, Fitzgerald MC, Whitman CP. Characterization of the role of the amino-terminal proline in the enzymatic activity catalyzed by macrophage migration inhibitory factor. Biochemistry. 1998;37:10195–10202. doi: 10.1021/bi9806955. [DOI] [PubMed] [Google Scholar]; (b) Soares TA, Goodsell DS, Ferreira R, Olson AJ, Briggs JM. Ionization state and molecular docking studies for the macrophage migration inhibitory factor: the role of lysine 32 in the catalytic mechanism. J. Mol. Recognit. 2000;13:146–156. doi: 10.1002/1099-1352(200005/06)13:3<146::AID-JMR497>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Orita M, Yamamoto S, Katayama N, Aoki M, Takayama K, Yamagiwa Y, Seki N, Suzuki H, Kurihara H, Sakashita H, Takeuchi M, Fujita S, Yamada T, Tanaka A. Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: Discovery and X-ray crystallography. J. Med. Chem. 2001;44:540–547. doi: 10.1021/jm000386o. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase activity by dopachrome analogs. Bioorg. Med. Chem. Lett. 1999;9:3193–3198. doi: 10.1016/s0960-894x(99)00561-2. [DOI] [PubMed] [Google Scholar]
- 29.Lubetsky JB, Dios A, Han J, Aljabari B, Ruzsicska B, Mitchell R, Lolis E, Al-Abed Y. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J. Biol. Chem. 2002;277:24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 30.Winner M, Meier J, Zierow S, Rendon BE, Cricklow GV, Riggs R, Bucala R, Leng L, Smith N, Lolis E, Trent JO, Mitchell RA. A Novel, Macrophage Migration Inhibitory Factor Suicide Substrate Inhibits Motility and Growth of Lung Cancer Cells. Cancer Res. 2008;68:7253–7257. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dios A, Mitchell RA, Aljabari B, Lubetsky J, O'Connor K, Liao H, Senter PD, Manogue KR, Lolis E, Metz C, Bucala R, Callaway DJE, Al-Abed Y. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J. Med. Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 33.Irwin JJ, Shoichet BK. ZINC - A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 35.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 36.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 37.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 38.Stamps SL, Taylor AB, Wang SC, Hackert ML, Whitman CP. Mechanism of the phenylpyruvate tautomerase activity of macrophage migration inhibitory factor: Properties of the P1G, P1A, Y95F, and N97A mutants. Biochemistry. 2000;39:9671–9678. doi: 10.1021/bi000373c. [DOI] [PubMed] [Google Scholar]
- 39.Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz C, Pavlov VA, Miller EJ, Tracey KJ. ISO-1 binding to the tautomerase active site of MIF inhibits its proinflammatory activity and increases survival in severe sepsis. J. Biol. Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 40.Cheng KF, Al-Abed Y. Critical modifications of the ISO-1 scaffold improve its potent inhibition of macrophage migration inhibitory factor (MIF) tautomerase activity. Bioorg. Med. Chem. Lett. 2006;16:3376–3379. doi: 10.1016/j.bmcl.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen WL. QikProp, 3.0. New York: Schrodinger, LLC; 2006. [Google Scholar]
- 42.Taylor AB, Johnson WH, Jr, Czerwinski RM, Li H-S, Hackert ML, Whitman CP. Crystal Structure of Macrophage Migration Inhibitory Factor Complexed with (E)-2-Fluoro-p-hydroxycinnamate at 1.8 Å Resolution: Implications for Enzymatic Catalysis and Inhibition. Biochemistry. 1999;38:7444–7452. doi: 10.1021/bi9904048. [DOI] [PubMed] [Google Scholar]
- 43.Proudfoot JR. The evolution of synthetic oral drug properties. Bioorg. Med. Chem. Lett. 2005;15:1087–1090. doi: 10.1016/j.bmcl.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 44.Thakur VV, Kim JT, Hamilton AD, Bailey CM, Domaoal RA, Wang L, Anderson KS, Jorgensen WL. Optimization of pyrimidinyl- and triazinyl-amines as non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg. Med. Chem. Lett. 2006;16:5664–5667. doi: 10.1016/j.bmcl.2006.08.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR and mass spectral data for the assayed compounds. These materials are available free of charge via the Internet at http://pubs.acs.org.