Abstract
The Y-STR DYS19 is firmly established in the repertoire of Y-chromosomal markers used in forensic analysis yet is poorly understood at the molecular level, lying in a complex genomic environment and exhibiting null alleles, as well as duplications and occasional triplications in population samples. Here, we analyse three null alleles and 51 duplications and show that DYS19 can also be involved in inversion events, so that even its location within the short arm of the Y chromosome is uncertain. Deletion mapping in the three chromosomes carrying null alleles shows that their deletions are less than ~300 kb in size. Haplotypic analysis with binary markers shows that they belong to three different haplogroups and so represent independent events. In contrast, a collection of 51 DYS19 duplication chromosomes belong to only four haplogroups: two are singletons and may represent somatic mutation in lymphoblastoid cell lines, but two, in haplogroups G and C3c, represent founder lineages that have spread widely in Central Europe/West Asia and East Asia, respectively. Consideration of candidate mechanisms underlying both deletions and duplications provides no evidence for the involvement of non-allelic homologous recombination, and they are likely to represent sporadic events with low mutation rates. Understanding the basis and population distribution of these DYS19 alleles will aid in the utilisation and interpretation of profiles that contain them.
Keywords: Y chromosome, Y-STR, DYS19, Duplication, Deletion, Inversion
Introduction
Over the last decade [1], the Y chromosome has become firmly established as a powerful system in forensic analysis, showing particular utility in male–female DNA mixtures. Y-chromosomal short tandem repeats (Y-STRs) have proven to be informative DNA markers, featuring in a large database (the Y Reference Haplotype Database; http://www.yhrd.org [2]) that allows rapid interrogation of population-specific frequencies of haplotypes and provides some information about the likely geographical origins of individuals.
The first forensically useful Y-STR and one that has since become incorporated in every commercial forensic Y-chromosomal haplotyping kit and in the minimal haplotype listed in the Y Reference Haplotype Database (YHRD) is DYS19 [3]. This tetranucleotide repeat marker was pioneered as an exclusion tool in a rape case [4] and as a marker in deficiency paternity testing [5], and was also the first Y-STR to be used in dating of anthropologically important events [6].
Despite its ubiquity in the fields of Y-chromosomal forensic analysis and evolutionary studies, DYS19 is surprisingly poorly understood at the molecular level compared to other Y-STRs. It has a number of paralogues elsewhere on the Y chromosome [7] and is deleted (‘null’ alleles [8]), duplicated [9, 10], or even triplicated [8, 9] on some chromosomes. The YHRD (release 22) contains 104 duplications (~0.2%), one triplication, and four null alleles [two Bhutan, one Hrodna (Belarus), one Tver (Russia)] among its ~53,000 haplotypes.
Deletions and duplications of Y-STRs are of forensic relevance because they can be interpreted as allele drop-outs and DNA contamination, respectively, which may affect the evidential value of a DNA profile. It is therefore important to understand the molecular mechanisms and rates of the processes underlying these deletions and duplications and how they are distributed in human populations.
Variability involving DYS19 can be seen against a background of the generally high degree of structural variability of the Y chromosome. Cytogenetic and molecular studies have demonstrated that many large-scale structural variants exist, including deletions [11-13], duplications [11, 12, 14, 15], and inversions [12]. Underlying this structural polymorphism is a high rate of mutation through non-allelic homologous recombination (NAHR) between very similar paralogous sequences, which are particularly frequent on the Y [16]. These paralogues also act as substrates for the frequent non-reciprocal transfer of sequence information via gene conversion events [17, 18].
The availability of the near-complete sequence of the euchromatic region of a single Y chromosome [16] offers an opportunity to analyse the genomic context of DYS19 in more detail, to investigate the homologies involving sequences around the marker and to address candidate mechanisms for duplications and deletions. The availability of a robust and well-resolved phylogeny based on slow-mutating binary markers such as single-nucleotide polymorphisms (SNPs) [19, 20], allows us to interpret these molecular events in a phylogeographic context.
Here, we analyse Y chromosomes carrying null and duplicated alleles of DYS19 and use deletion mapping to show that its position on the chromosome itself is polymorphic. We use haplotyping with binary markers and multiple Y-STRs, together with deletion mapping and bioinformatic prediction, to address the molecular basis of the underlying rearrangements and to identify examples that descend from a common ancestor. This analysis shows that DYS19 duplications are apparently not mediated by repeat-mediated recombination events and identifies two founder lineages carrying DYS19 duplications that have reached high frequencies in particular haplogroups and populations.
Materials and methods
DNA samples
DNA samples from a total of 55 unrelated men were from collections of the authors and were obtained with appropriate informed consent. Some samples form part of sets described previously [21-24], and four were from the Centre d'Etude du Polymorphisme Humain-Human Genome Diversity Project (CEPH-HGDP) panel [25]. With the exception of these four lymphoblastoid DNAs, all samples were derived from either blood or buccal scrapes.
Deletion mapping
Y-specific sequence-tagged sites (STSs-primer sequences available from the literature [16, 26]) were amplified by polymerase chain reaction (PCR) and analysed by agarose gel electrophoresis. An STS was considered to be deleted when absent in the presence of a larger Y-specific control amplicon coamplified in the same PCR reaction. The PCR system was as described [13], and cycling conditions were: 94°C 30 s, 60°C 30 s and 70°C 30 s for 33 cycles.
Y chromosome haplotyping and identification of DYS19 deletions and duplications
Twenty-six Y-specific STRs (DYS19, DYS385a/b, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS425, DYS426, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, YCAII-a/b and Y-GATA-H4.1) were typed in a 20-plex [27] and an additional 14-plex [22]. PCR products were resolved on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems) and analysed using GeneMapper software (Applied Biosystems). Allele nomenclature was as described [22] and in accordance with International Society of Forensic Genetics recommendations [28]. To allow us to combine our data with published datasets, we consider here only 15 of the 26 loci (DYS19, DYS388, DYS389I, DYS389II-I, DYS390, DYS391, DYS392, DYS393, DYS426, DYS437, DYS439, DYS434, DYS435, DYS436 and DYS438).
DYS19 deletions and duplications were initially ascertained using a number of published multiplexes [22, 27, 29] and commercial kits, i.e. AmpFlSTR®YFiler® PCR Amplification kit (Applied Biosystems), PowerPlex® Y System (Promega) and Mentype® Argus Y-MH (Biotype) and were confirmed in repeated amplifications. Deletions (as opposed to small-scale primer site mutations) were verified by use of the two non-overlapping primer pairs 3F/3R and 2F/2R [7]. DYS19 was considered to be duplicated when its two peaks in an electropherogram were of approximately equal height and area; see “Results” for details of peak area and height ratios.
Binary markers were typed in a hierarchical fashion, using either the SNaPshot minisequencing protocol (Applied Biosystems) on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems) or primer extension on the Sequenom mass spectrometry system (Sequenom, San Diego, CA, USA). Amplification and extension primers were based on ones published previously [30, 31], with additional primers based on published sequences [19].
Identifying and testing candidate rearrangement-sponsoring repeats
Perfect direct repeats as candidates for sponsoring deletions and duplications were identified using the REPuter program [32] at http://bibiserv.techfak.uni-bielefeld.de/reputer/.
Having identified a candidate pair of partial L1 long interspersed nuclear element (LINE) sequences (see “Results”, final section), we sought junction products by PCR in deletion and duplication chromosomes. Deletion sponsored by the repeats would allow the generation of a junction PCR product with the primer pair pL1f (5′-aac tga aag aga gag gaa ctt tgg-3′) and dL1r (5′-cta gtg tcg gaa tta ttt caa tg-3′) (Fig. 1d), while duplication would allow us to detect a junction product with the primer pair pL1r (5′-tga act ccc att cac aat tgc-3′) and dL1f (5′-ggt act atc aat aac act ggc-3′).
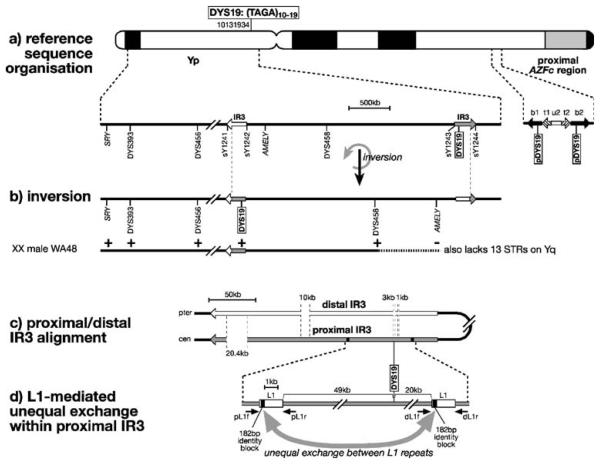
Fig. 1.
Complex genomic environment of DYS19 and putative mechanisms for inversions, deletions and duplications. a Reference sequence organisation around DYS19, showing position of the Y-STR on an idiogram of the Y chromosome (with genome position of start of the marker given according to build 36.1 of the reference assembly), and below, a schematic view of the region around DYS19 and its paralogues (pDYS19) on Yq. Arrows indicate inverted repeats (IR3 elements) or large repeat units in the region proximal to AZFc [42]. STSs and other markers used in mapping are shown below. b Structure of Yp following IR3-mediated inversion, with breakpoints indicated by dotted lines and mapping of presence (+) or absence (−) of markers in XX male WA48. The horizontal dotted line indicates the region of uncertainty of the breakpoint, resulting from the wide marker spacing. c Alignment of the IR3 inverted repeats; in the reference sequence organisation, DYS19 lies within the proximal IR3, corresponding to a 3-kb gap in the distal IR3. d Putative mechanism of DYS19 deletion or duplication mediated by unequal exchange (curved grey arrow) between flanking direct partial L1 repeats within the proximal IR3. Small arrows indicate PCR primers used to seek junction PCR products in deletion and duplication chromosomes
Y-STR network construction and dating
Weighted median-joining networks [33] were constructed from 14-locus (DYS388, DYS389I, DYS389II-I, DYS390, DYS391, DYS392, DYS393, DYS426, DYS437, DYS439, DYS434, DYS435, DYS436 and DYS438) or 6-locus (DYS389I, DYS389II-I, DYS390, DYS391, DYS392 and DYS393) Y-STR haplotypes using Network 4.0 (http://www.fluxus-engineering.com/sharenet.htm). Weighting [34] was used to remove some reticulations (closed structures) within the network by taking into account the range of different mutation rates of the markers, reflected indirectly by their allele length variances among all chromosomes included in each network. Note that our conclusions are not affected by the choice of weighting scheme.
Time-to-most-recent-common-ancestor (TMRCA) of the clusters within the network of Fig. 2 was estimated using the rho statistic within Network, taking the per-STR per-generation mutation rate to be 2.0×10−3 [35] and the generation time 31 years [36].
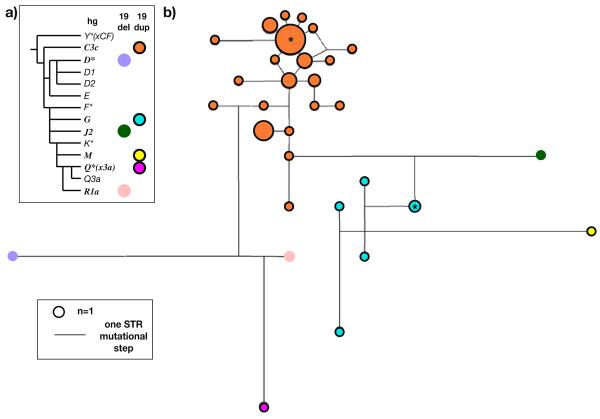
Fig. 2.
Haplogroups and Y-STR haplotypes of DYS19 deletion and duplication chromosomes. a Binary marker phylogeny of the Y chromosome, with haplogroups (hg) [19] containing DYS19 deletion and duplication chromosomes indicated by coloured circles. b) Weighted median joining network [33] containing the 14-locus (DYS388, DYS389I, DYS389II-I, DYS390, DYS391, DYS392, DYS393, DYS426, DYS437, DYS439, DYS434, DYS435, DYS436 and DYS438) Y-STR haplotypes of 3 deletion and 51 duplication chromosomes. Circles represent haplotypes, with area proportional to frequency and coloured according to haplogroup as in a. Nodes used as roots in TMRCA estimations are indicated by asterisks
Results
Inversion encompassing DYS19
According to the reference Y chromosome sequence [16], DYS19 lies on Yp, within the proximal member of a pair of ~300-kb inverted repeat sequences (IR3; Fig. 1a), separated by ~3.6 Mb. However, these IR3 elements are known to sponsor recurrent paracentric inversions [12, 16], which could in principle transpose DYS19 into the distal IR3 region.
During analysis of the partial Y chromosome carried by a translocation XX man, we found evidence to demonstrate that this does indeed occur. Male WA48 was identified in a screen for men lacking the Amelogenin Y sex-test locus on Yp. As well as AMELY, he lacks a Y-chromosomal long arm, shown by the absence of 13 Y-STRs mapping to Yq. He carries distal Yp markers (translocated onto the short arm of one of his X chromosomes), including the sex-determining SRY gene and the distal Y-STRs DYS393 and DYS456. Based on the reference sequence organisation, we would therefore expect him to lack the Yp STRs DYS458 and DYS19 (Fig. 1a), since they are proximal to the absent AMELY. However, both are actually present (Fig. 1b). While we cannot rule out a more complex rearrangement, the most parsimonious explanation for this discrepant pattern of markers is a paracentric inversion, transferring DYS19 to the distal IR3 element, followed by translocation of a terminal segment of Y-chromosomal material (between ~6.5 and ~8.6 Mb in size) onto the X chromosome.
A maximum-likelihood estimate of the per-generation rate of IR3-sponsored inversion is 9.2×10−4 [12], with chromosomes carrying the two different orientations scattered among different branches of the Y phylogeny and among different populations. This suggests that, in any given Y haplogroup or population, both the position and orientation of DYS19 on Yp are in fact uncertain.
DYS19 deletions
To investigate deletion of DYS19, we identified three Y chromosomes each carrying a DYS19 null allele on an otherwise complete Y-STR haplotype (Table S1 of the Electronic Supplementary Material). These null alleles were ascertained using the DYS19 primers employed in the 20-plex PCR [27]. To exclude the possibility of small-scale mutations affecting only the primer binding sites, we confirmed all three deletions using an independent non-overlapping primer pair [7].
Analysis of deletions is, in principle, more straightforward than that of duplications, since the absence of a portion of the chromosome is easier to score than its presence in two copies, which normally requires quantitative analysis [e.g. quantitative PCR (Q-PCR)] or direct physical mapping (e.g. high-resolution fluorescent in situ hybridization). However, in this case, the fact that DYS19 lies within one of a pair of closely related (99.75% similar [16]) IR3 elements makes deletion mapping around this STR particularly difficult. We analysed the three deletion chromosomes using the four unique STSs (sY1241–1244) that mark the boundaries of the two IR3 elements (Fig. 1a); in each case, all four boundaries are present, showing that the deletion breakpoints must be contained within an IR3 element (either proximal or distal). The deletions therefore cover an extent of less than ~300 kb, but further definition of their extents through STS analysis is not straightforward because the presence of one intact IR3 masks any internal deletion within the other. Not only are the two IR3 elements almost identical in sequence, but their roles in mediating inversions suggest that designing internal proximal- or distal-specific STSs would not be profitable.
DYS19 duplications
To investigate duplication of DYS19, we identified 51 chromosomes each showing two alleles of different lengths but approximately equal peak heights and areas in capillary electrophoresis (Table S1 of the Electronic Supplementary Material). Note that duplicated alleles are under-ascertained, since cases where both copies carry identical repeat copy numbers cannot be identified without quantitative analysis.
The amount and quality of available DNA precluded Q-PCR-based approaches to understanding the molecular basis of the DYS19 duplications. If a duplicated region contains more than one Y-STR, then duplications of multiple STRs within a physical interval can help to delimit its length; this was the case for the AZFa interval [14], which contains nine Y-STRs including DYS388, DYS389I, DYS389II, DYS438 and DYS439. However, the only other STR lying on Yp included in our Y-STR multiplexes is DYS393, some 7 Mb distal to DYS19 in the reference sequence. Unsurprisingly, no examples of co-duplication of this locus are observed in our set. DYS458, present in the Y-filer (Applied Biosystems) kit, lies ~2.4-Mb distal to DYS19 in the reference sequence. DYS458 data available for four chromosomes (19dup23, 24, 50 and 51) show no evidence of duplication (not shown), so the extents of the duplicated regions therefore remain undetermined.
Haplotyping of deletions and duplications
Typing of binary markers shows that the three deletion chromosomes belong to three different haplogroups (D*, J2 and R1a; Fig. 2a), and therefore must represent independent events.
A similar analysis places the 51 duplication chromosomes in four different haplogroups (Fig. 2a), indicating that there are at least four independent duplications of DYS19. Within a 14-locus Y-STR network (Fig. 2b), the six hgG and 43 hgC3c duplication chromosomes each form clusters suggesting single duplication origins and identity-by-descent within each of these haplogroups; time to most recent common ancestor estimates are 2,330±840 years and 1,780±630 years, respectively, for these clusters. The issue arises of whether or not all chromosomes within these two haplogroups might carry DYS19 duplications—chromosomes presenting only a single peak (allele) in an electropherogram can nonetheless be duplicated for the STR, so ascertainment is not straightforward. While it is neither more or less likely that a duplication occurred before or after the haplogroup-defining SNP, the knowledge would be interesting because then typing the SNP would, in effect, be ascertaining the duplication. For haplogroup G, it is clearly not the case that all chromosomes carry the duplication: for example, a published dataset of 56 hgG haplotypes [37] contains no DYS19 two-allele cases, despite displaying DYS19 repeat numbers between 14 and 17 and therefore providing ample opportunity for ascertainment of duplications. A recent study [10] has shown that ten Italian chromosomes carrying DYS19 duplications all belong to hgG2*(xG2a,G2b); it may be that duplications are confined to this sublineage and indeed that all hgG2* (xG2a,G2b) chromosomes carry duplications. However, the published evidence [10] does not make clear how many hgG chromosomes not belonging to this sublineage were tested for DYS19 duplication, so it is difficult to assess this possibility. For haplogroup C3c, duplication chromosomes are more predominant, with 46 of 126 chromosomes (37%) showing two distinct alleles (unpublished; [38, 39]); however, the fact that no examples of chromosomes belonging to one particular haplotype cluster within hgC3c (the ‘Manchu haplotype’ [40]) show two DYS19 alleles, again despite high diversity of DYS19 (alleles 14 to 18), suggests that DYS19 duplication chromosomes actually form a subset of hgC3c.
Singletons carrying duplications are found in hgs M (19dup50) and Q(xQ3a) (19dup51), and interpretation of these cases is more problematic. While they may represent genuine germ-line mutations, in the absence of other examples within these haplogroups (indicating identity-by-descent), it remains possible that these chromosomes bear somatic DYS19 mutations with approximately balanced cell populations carrying the two different alleles. Both cases are from the CEPH-HGDP panel and therefore derived from lymphoblastoid cell lines, in which somatic STR mutations have been noted in the past [41]. Comparisons of peak area ratios for the two DYS19 alleles support this idea. For all other chromosomes, the average value for the shorter allele divided by the longer is 1.12 (range 1.04 to 1.24); however, for 19dup50 and 19dup51, the ratios are, respectively, 1.39 and 1.93.
Possible mechanisms of deletions and duplications
Are these deletions and duplications caused by NAHR, mediated by directly repeated sequences? It is difficult to address this question for the duplications because we know little about their extents. However, for the deletions, we know that any sponsoring repeats must lie within an IR3 element. We sought candidate repeats by carrying out a sliding self-alignment of the ~300-kb proximal IR3 copy using the program REPuter [32]. This revealed, as the only plausible candidates for sponsoring such local rearrangements, duplicated direct partial L1 LINE repeats ~69 kb apart, flanking DYS19 (Fig. 1d). These sequences show 96% sequence similarity over 1.75 kb and contain the largest block of sequence identity of 182 bp. NAHR between these sequences could lead to deletion or duplication of DYS19. To test this idea, we designed primers flanking each L1 segment and sought junction products by PCR in both deletion and duplication chromosomes. No junction products were obtained (data not shown), so the L1-mediated mechanism is not supported.
An alternative mechanism for duplication/deletion would be non-reciprocal transfer through gene conversion mediated by homology—the genomic context of DYS19 provides potential opportunities for this. As described above, it lies within a repeated IR3 element; furthermore (and as noted by others [7]), two paralogues, lacking variable tetranucleotide repeats, lie on the long arm within the b1 and b2 repeats [42] (part of palindrome 3 [16]) proximal to AZFc (Fig. 1a). In principle, gene conversion between IR3 elements could be responsible for DYS19 deletion or duplication. A sequence alignment of the two IR3 repeats (Fig. 1c) suggests that this possible mechanism is unlikely: DYS19 lies in a region of the proximal element corresponding to a ~3-kb gap in the distal element, and gene conversion is very unlikely to operate over such a large region of heterology. Gene conversion might also operate between a DYS19-containing IR3 element and a b1 or b2 repeat on Yq, each of which contains a DYS19 paralogue (Fig. 1a). However, the paralogues lack the DYS19 (TAGA)n repeat array, and show a mean of only 92% sequence similarity to the DYS19 region itself in regions 200 bp either side of the array; the largest block of sequence identity is only 53 bp in length. Again, this degree of heterology makes gene conversion seem an improbable mechanism [43].
In summary, our investigations do not provide any evidence of an NAHR-based mechanism for rearrangements involving DYS19, whether by unequal exchange or gene conversion. It seems likely that the rearrangements are sporadic events occurring through diverse processes that are probably not mediated by homologous recombination, as has been shown for some Amelogenin Y deletions [13].
Discussion
Recently, with the convenience of the availability of the human genome sequence, the systematic identification of new Y-STRs has become relatively straightforward [44]. However, most of the markers so well established in forensic practice today were developed more laboriously, before this resource was available, and represent a heterogeneous set of loci. With our current knowledge of the genomic sequence context and behaviour of DYS19, it would not have been a strong candidate for selection as a useful new marker from genomic sequence information: it is observed to be deleted, duplicated or triplicated and lies in one of a pair of dynamic short-arm inverted repeats with >99% sequence identity, with very similar long-arm paralogues, making the design of truly specific primers difficult [7]. Despite these apparent disadvantages, DYS19 has become one of the most widely typed markers on the Y chromosome.
Statements about the locations of Y-STRs need to be made carefully, given the fluidity of the organisation of the Y chromosome, and it is clear that the position of DYS19 is uncertain in any given lineage (although this finding does not materially affect its forensic utility). Any study of Y-chromosomal structural variation needs to bear in mind that, although it is an invaluable resource, the reference sequence [16] is not necessarily a relevant starting point for considering mechanisms of rearrangement in other lineages.
The Y chromosome bears a rich complement of ampliconic repeats [16], and these are known to sponsor many recurrent rearrangements, including examples at AZFa [14, 45], AZFb [46], AZFc [26, 42, 47-49] and Amelogenin Y [13, 15]. However, this does not mean that every rearrangement is driven by these NAHR processes. In the case of DYS19 duplications and deletions, we have found no evidence to suggest that they are caused by recurrent NAHR or conversion between paralogous repeats. It seems more likely that they result from sporadic, and perhaps non-homology-mediated, processes with very low mutation rates.
The very large number of chromosomes surveyed for DYS19 copy number variation means that these rare events are nonetheless detected, and this is facilitated by the expansion of particular lineages through the strong genetic drift and possible social selection that operates on the Y. At least two duplication lineages have propagated in East Asia (within hgC3c) and West Asia/Central Europe (within hgG), so that duplications in populations from these regions may be relatively frequent and of importance to forensic practice. In terms of absolute numbers of chromosomes, duplications are more common than deletions, but considering the number of independent events, the different rearrangements exist at similar frequencies. There is no evidence for a deletion lineage that has spread in the manner of the duplication lineages. Note, however, that DYS19 deletions might be under-reported to databases such as the YHRD if they are interpreted by contributors as ‘incomplete’ profiles, signifying technical failure; this effect may be stronger for deletions than for duplications, which are more widely known and better understood.
The YHRD (release 22) contains 96 DYS19 duplications in addition to the examples analysed by us; ten are known to belong to hgG [10]. We used Network analysis (Figure 1 of the Electronic Supplementary Material) to ask if there is evidence that any of the remaining 86 chromosomes belong to either of the predominant clusters in hgG and C3c, combining the YHRD cases with ours. Our hgM and hgQ* (xQ3a) chromosomes are peripheral in the network, with no closely related haplotypes, suggesting that the YHRD contains no further cases belonging to these two lineages and supporting the idea that the hgM and Q*(xQ3a) cases represent somatic mutations. Many of the known hgG and hgC3c chromosomes, however, share haplotypes with YHRD examples or are their single-step mutational neighbours, strongly suggesting that they also share haplogroups. On this conservative basis, at least 18 of the 86 YHRD chromosomes of unknown haplogroup probably belong to the hgG lineage (a total proportion, including chromosomes identified by us, of 23% of duplications), and 43 of 86 chromosomes (total sample proportion 59%) belong to the hgC3c lineage.
The existence of DYS19 deletions and duplications can have a number of practical consequences. Mutation at a single-copy Y-STR is easily recognised and interpreted in a deficiency paternity test [50], but if a duplication is present, results could be confusing: for example, a patrilineal relative apparently carrying allele 16 (but in reality the duplication 16–16) might be compared to a son carrying 16–17. Consideration of peak–height ratios and of the population of origin and background haplotypes of the tested individuals would aid interpretation. In forensic casework, deletions and duplications might be interpreted as allele drop-outs or evidence of DNA contamination [9], but provided that they can be reliably recognised, they could elevate the significance of a match between a suspect and a stain, rather in the way that heteroplasmy of mitochondrial DNA can increase the strength of evidence [51]. Finally, given the high frequencies of duplications in two haplogroups and their associated populations, DYS19 duplications may be useful in the deduction of population of origin of a DNA sample.
Supplementary Material
Acknowledgements
We thank all DNA donors, Aleksander Bagdonavicius (Path Centre Forensic Biology, WA, Australia), Evelyne Heyer and Lluis Quintana-Murci for provision of DNA samples, Matt Hurles for providing PCR primers for SNaPshot analysis and Yali Xue for discussions. M.A.J. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (grant number 057559); P. B. and C.T.S. were supported by the Wellcome Trust. E.J.P. and D.R. C.S. were supported by the Arts and Humanities Research Council and the EC Sixth Framework Programme under contract number ERAS-CT-2003-980409, within the framework of the European Science Foundation EUROCORES programme “The Origin of Man, Language and Languages”. P.dK. was supported by the NWO (Netherlands Organisation for Scientific Research) project 231-70-001 within the same EUROCORES programme.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00414-008-0253-3) contains supplementary material, which is available to authorized users.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Jobling MA, Pandya A, Tyler-Smith C. The Y chromosome in forensic analysis and paternity testing. Int J Legal Med. 1997;110:118–124. doi: 10.1007/s004140050050. [DOI] [PubMed] [Google Scholar]
- 2.Willuweit S, Roewer L, on behalf of the International Forensic Y Chromosome User Group Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Roewer L, Arnemann J, Spurr NK, Grzeschik KH, Epplen JT. Simple repeat sequences on the human Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet. 1992;89:389–394. doi: 10.1007/BF00194309. [DOI] [PubMed] [Google Scholar]
- 4.Roewer L, Epplen JT. Rapid and sensitive typing of forensic stains by PCR amplification of polymorphic simple repeat sequences in case work. Forensic Sci Int. 1992;53:163–171. doi: 10.1016/0379-0738(92)90193-z. [DOI] [PubMed] [Google Scholar]
- 5.Santos FR, Epplen JT, Pena SDJ. Testing deficiency paternity cases with a Y-linked tetranucleotide repeat polymorphism. In: Pena SDJ, Chakraborty R, Epplen JT, Jeffreys AJ, editors. DNA fingerprinting: state of the science. Birkhäuser Verlag; Basel: 1993. pp. 261–265. [DOI] [PubMed] [Google Scholar]
- 6.Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci U S A. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler JM, Schoske R. Duplication of DYS19 flanking regions in other parts of the Y chromosome. Int J Legal Med. 2004;118:178–183. doi: 10.1007/s00414-004-0436-5. [DOI] [PubMed] [Google Scholar]
- 8.Santos FR, Gerelsaikhan T, Munkhtuja B, Oyunsuren T, Epplen JT, Pena SDJ. Geographic differences in the allele frequencies of the human Y-linked tetranucleotide polymorphism DYS19. Hum Genet. 1996;97:309–313. doi: 10.1007/BF02185760. [DOI] [PubMed] [Google Scholar]
- 9.Butler JM, Decker AE, Kline MC, Vallone PM. Chromosomal duplications along the Y-chromosome and their potential impact on Y-STR interpretation. J Forensic Sci. 2005;50:853–859. [PubMed] [Google Scholar]
- 10.Capelli C, Brisighelli F, Scarnicci F, Blanco-Verea A, Brion M, Pascali V. Phylogenetic evidence for multiple independent duplication events at the DYS19 locus. Forensic Sci Int Genet. 2007;1:287–290. doi: 10.1016/j.fsigen.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C. Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet. 1996;5:1767–1775. doi: 10.1093/hmg/5.11.1767. [DOI] [PubMed] [Google Scholar]
- 12.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 13.Jobling MA, Lo IC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, Chang YM, Kraaijenbrink T, Henke J, Guanti G, McKeown B, van Oorschot RA, Mitchell RJ, de Knijff P, Tyler-Smith C, Parkin EJ. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet. 2007;16:307–316. doi: 10.1093/hmg/ddl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch E, Jobling MA. Duplications of the AZFa region of the human Y chromosome are mediated by homologous recombination between HERVs and are compatible with male fertility. Hum Mol Genet. 2003;12:341–347. doi: 10.1093/hmg/ddg031. [DOI] [PubMed] [Google Scholar]
- 15.Murphy KM, Cohen JS, Goodrich A, Long PP, Griffin CA. Constitutional duplication of a region of chromosome Yp encoding AMELY, PRKY, and TBL1Y: implications for sex chromosome analysis and bone marrow engraftment analysis. J Mol Diagn. 2007;9:408–413. doi: 10.2353/jmoldx.2007.060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova R, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome: a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of massive palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 18.Bosch E, Hurles ME, Navarro A, Jobling MA. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 21.Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkin EJ, Kraayenbrink T, van Driem GL, Tshering K, de Knijff P, Jobling MA. 26-locus Y-STR typing in a Bhutanese population sample. Forensic Sci Int. 2006;161:1–7. doi: 10.1016/j.forsciint.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Parkin EJ, Kraayenbrink T, Opgenort JR, van Driem GL, Tuladhar NM, de Knijff P, Jobling MA. Diversity of 26-locus Y-STR haplotypes in a Nepalese population sample: isolation and drift in the Himalayas. Forensic Sci Int. 2007;166:176–181. doi: 10.1016/j.forsciint.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roewer L, Krüger C, Willuweit S, Nagy M, Rodig H, Kokshunova L, Rothamel T, Kravchenko S, Jobling MA, Stoneking M, Nasidze I. Y-chromosomal STR haplotypes in Kalmyk population samples. Forensic Sci Int. 2007;173:204–209. doi: 10.1016/j.forsciint.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu JY, Carcassi C, Contu L, Du RF, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang XY, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian YP, Shu QF, Xu JJ, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 26.Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 27.Butler JM, Schoske R, Vallone PM, Kline MC, Redd AJ, Hammer MF. A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci Int. 2002;129:10–24. doi: 10.1016/s0379-0738(02)00195-0. [DOI] [PubMed] [Google Scholar]
- 28.Gusmão L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM. DNA COMMISSION OF THE International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int. 2006;157:187–197. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, de Knijff P, Jobling MA. High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci Int. 2002;125:42–51. doi: 10.1016/s0379-0738(01)00627-2. [DOI] [PubMed] [Google Scholar]
- 30.Bosch E, Calafell F, González-Neira A, Flaiz C, Mateu E, Scheil H-G, Huckenbeck W, Efremovska L, Mikerezi I, Xirotiris N, Grasa C, Schmidt H, Comas D. Male and female lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 31.Hurles ME, Sykes BC, Jobling MA, Forster P. The dual origin of the Malagasy in island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucl Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 34.Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gusmão L, Sanchez-Diz P, Calafell F, Martin P, Alonso CA, Alvarez-Fernandez F, Alves C, Borjas-Fajardo L, Bozzo WR, Bravo ML, Builes JJ, Capilla J, Carvalho M, Castillo C, Catanesi CI, Corach D, Di Lonardo AM, Espinheira R, Fagundes de Carvalho E, Farfan MJ, Figueiredo HP, Gomes I, Lojo MM, Marino M, Pinheiro MF, Pontes ML, Prieto V, Ramos-Luis E, Riancho JA, Souza Goes AC, Santapa OA, Sumita DR, Vallejo G, Vidal Rioja L, Vide MC, Vieira da Silva CI, Whittle MR, Zabala W, Zarrabeitia MT, Alonso A, Carracedo A, Amorim A. Mutation rates at Y chromosome specific microsatellites. Hum Mutat. 2005;26:520–528. doi: 10.1002/humu.20254. [DOI] [PubMed] [Google Scholar]
- 36.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 37.Cinnioglu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 38.Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C. A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet. 2002;71:466–482. doi: 10.1086/342096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C. Male demography in East Asia: a north–south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, Xu J, Du R, Fu S, Li P, Yang H, Tyler-Smith C. Recent spread of a Ychromosomal lineage in northern China and Mongolia. Am J Hum Genet. 2005;77:1112–1116. doi: 10.1086/498583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banchs I, Bosch A, Guimera J, Lazaro C, Puig A, Estivill X. New alleles at microsatellite loci in CEPH families mainly arise from somatic mutations in the lymphoblastoid cell-lines. Hum Mut. 1994;3:365–372. doi: 10.1002/humu.1380030407. [DOI] [PubMed] [Google Scholar]
- 42.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 43.Lukacsovich T, Waldman AS. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics. 1999;151:1559–1568. doi: 10.1093/genetics/151.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayser M, Kittler R, Erler A, Hedman M, Lee AC, Mohyuddin A, Mehdi SQ, Rosser Z, Stoneking M, Jobling MA, Sajantila A, Tyler-Smith C. A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet. 2004;74:1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intra-chromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J Med Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am J Hum Genet. 2004;74:180–187. doi: 10.1086/381132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balaresque P, Bowden GR, Parkin EJ, Omran GA, Heyer E, Quintana-Murci L, Roewer L, Stoneking M, Nasidze I, Carvalho-Silva DR, Tyler-Smith C, de Knijff P, Jobling MA. Dynamic nature of the proximal AZFc region of the human Y chromosome: multiple independent deletion and duplication events revealed by microsatellite analysis. Hum Mut. 2008 doi: 10.1002/humu.20757. in press. DOI 10.1002/humu.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolf B, Keil W, Brinkmann B, Roewer L, Fimmers R. Paternity testing using Y-STR haplotypes: assigning a probability for paternity in cases of mutations. Int J Legal Med. 2001;115:12–15. doi: 10.1007/s004140000201. [DOI] [PubMed] [Google Scholar]
- 51.Ivanov PL, Wadhams MJ, Roby RK, Holland MM, Weedn VW, Parsons TJ. Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nat Genet. 1996;12:417–420. doi: 10.1038/ng0496-417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.