Abstract
Background
An inverted region on chromosome 17 has been previously linked to many Pick complex diseases. Due to the inversion, an exact causal locus has been difficult to identify, but the microtubule-associated protein tau gene is a likely candidate gene for its involvement in these diseases with tau inclusion.
Objective
To search for variants that confer susceptibility to 4 tauopathies and clinically related disorders.
Design
Genomewide association study.
Setting
University research laboratory.
Participants
A total of 231 samples were genotyped from an unrelated white population of patients with progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), frontotemporal dementia, and frontotemporal dementia with amyotrophy. Unaffected individuals from the same population were used as controls.
Main Outcome Measures
The results from an inverted region of chromosome 17 that contains the MAPT gene. Genotypes of cases and controls were compared using a Fisher exact test on a marker-by-marker basis. Haplotypes were determined by visually inspecting genotypes.
Results
Comparing any particular disease and controls, the association was constant across the inverted chromosome segment. Significant associations were seen for PSP and PSP combined with CBD. Of the 2 haplotypes seen in the region, H1 was overrepresented in PSP and CBD cases compared with controls.
Conclusions
As expected, the markers are highly correlated and the association is seen across the entire region, which makes it difficult to narrow down a disease-causing variant or even a possible candidate gene. However, considering the pathologic abnormalities of these diseases and the involvement of tau mutations seen in familial forms, the MAPT gene represents the most likely cause driving the association.
Pick complex refers to a spectrum of diseases with a variety of overlapping clinical and pathologic features due to a related genetic etiology. A common, although not ubiquitous, overlapping feature of these diseases is the presence of tau inclusions. Thus, the genetic and brain histochemical features of the microtubule-associated protein tau gene (MAPT) (OMIM 157140) provide a compelling reason for thinking that patients with these clinically and pathologically diverse findings should be thought of as a contiguous group. These diseases are characterized clinically by cognitive, behavioral, and movement defects. This study focused on 4 diseases in the spectrum in which tau histochemical and genetic features are believed to be critical: progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and frontotemporal dementia (FTD) with or without amyotrophy.
The clinical signs and symptoms observed in patients with these diseases are correlated with the anatomical distribution of neuronal loss, which can be variable. There are several patterns of inclusions of insoluble proteins in affected individuals, but there is limited correlation between inclusion type and clinical symptoms. Pick complex diseases can be accompanied by tau inclusions, ubiquitin inclusions, or no inclusions at all.1 Pick complex diseases that contain tau inclusions are collectively referred to as tauopathies.
Many families with inherited tauopathies have been linked to the same genomic region and, thus, collectively, these families are said to be affected with FTD and parkinsonism linked to chromosome 17 (FTDP-17).2 MAPT was considered to be a likely candidate gene in this region for its involvement in FTD with tau inclusions and, subsequently, many MAPT mutations have been identified in affected individuals. Tau mutations have been found to affect the level of translated protein or alternative RNA splicing, which may upset the interaction between tau and microtubules, allowing unbound and abnormally phosphorylated tau to polymerize into inclusions.3 Different tau mutations alter biochemical properties of the gene product, but these mutations do not necessarily predict the exact clinical nature of the disease. The same mutation in affected individuals, even in the same family, may result in a different age at onset, combination of symptoms, and clinical diagnosis.4 The variable morphologic features of accumulated tau proteins could be explained by the wide range of mutations that have been found in these diseases. In various tauopathies, the inclusions may differ based on the ratios of particular isoforms and the physical location of accumulation. At least 40 MAPT mutations have been identified in patients with FTD and related diseases.5 Tau inclusions, usually without MAPT mutations, are part of the pathologic definition of CBD and PSP, whereas cases of FTD are often seen without tau mutations or tau inclusions. Another set of cases with FTDP-17 that contain ubiquitin inclusions, but no tau inclusions, was linked to the same region on chromosome 17.6 Further gene resequencing of this set of cases led to the discovery of mutations in the progranulin gene from this region; these mutations are responsible for many cases of FTDP-17.6
In its natural state, MAPT works to stabilize microtubule formation and regulate transport along microtu-bules.4 Dysfunctional tau proteins can interrupt axonal transport by reducing the cell’s ability to control microtubule formation, ultimately leading to neuron dysfunction and death.2 Normally, the tau protein is located in axons but, in diseased cells, it relocates to the cell body and forms insoluble hyperphosphorylized fibrillary inclusions.1 This hyperphosphorylation of tau may lead to a loss of microtubule affinity and a resistance to proteases, leading to aggregation.7 Six major isoforms are produced in the adult human brain through the alternative splicing of exons 2, 3, and 10.4 The 6 isoforms can be divided into 2 groups, depending on the number of microtubule-binding domains. Alternative splicing of exon 10 leads to 4-repeat (4R) or 3-repeat (3R) binding domains.4 The number of binding domains affects the binding of tau to tubulin; 4R tau binds stronger and assembles more efficiently than 3R tau.4 A reduction in binding efficiency may increase the amount of unbound tau in the neuron, leading to aggregation, although increased binding may have an equally damaging effect.4 An accumulation of unbound tau may result if any isoform fails to function, creating insoluble inclusions.2 Inclusions found in affected individuals may contain all 6 isoforms in equal amounts or different ratios of selected isoforms. Many mutations disrupt the splicing of exon 10, leading to unequal 3R and 4R ratios. Tau deposits in PSP and CBD are predominantly 4R, whereas deposits in FTD contain equal levels of 4R and 3R.1
The region containing the MAPT gene has been shown to be genetically complex owing to an inversion commonly found in white populations. Three highly homologous low-copy repeats (LCRs) flank the region.8 The 2 LCRs telomeric of MAPT, LCRs B and C, are inverted relative to the centromeric LCR A.8 Low-copy repeats A and B flank the MAPT haplotype, suggesting that the inversion was caused by nonallelic homologous recombination.8 Figure 1 shows the structure of MAPT in relation to these LCRs. Extensive genotyping across the interval identified 2 haplotypes in almost complete disequilibrium.9 These haplotypes are commonly referred to as the H1 haplotype and H2 haplotype. Recombination in the inverted segment between carriers of the H1 and H2 haplotypes would result in a Robertsonian translocation. The high degree of disequilibrium in this region suggests that recombination has been suppressed or that there was a selection against recombinant chromosomes before the inversion became established in the white population.10 A study11 of tau expression in patients with Alzheimer disease found that 1 variant of the H1 haplotype led to an increase in overall tau levels and specifically in 4R tau levels, creating an imbalance of isoforms. Similar changes in expression could be found in these diseases.
Figure 1.
The structure of the tau gene, indicating the locations of the microtubule-binding domains (asterisks) and the flanking low-copy repeats (LCRs).
There is a locus in or near the MAPT gene that affects susceptibility to PSP and CBD. Conrad et al7 established that common variations in the MAPT gene affect susceptibility to PSP. They reported that the A0 allele of a di-nucleotide repeat marker located in intron 9 of MAPT is observed in 57% of control chromosomes compared with 95.5% of PSP cases.7 The A0 allele was also shown to be overrepresented in CBD chromosomes.12 Other tauopathies have a less certain association with MAPT region polymorphisms.7 The A0 allele is not believed to be biologically relevant to the disease process but is instead in linkage disequilibrium with some other polymorphism.7 The A0 allele is inherited with the H1 haplotype, so it is not surprising that the H1 haplotype is also overrepresented in PSP cases.10 It is uncertain whether the increased risk of PSP and CBD is associated with a specific common variation of the haplotype or with a rare mutation found on some chromosomes with the H1 haplotype.7
Despite the varied clinical features used to categorize the different diseases, there is overlap, suggesting that there could be a shared underlying biochemical abnormality resulting from the altered expression of tau.12 To explore this possibility, we performed a high-density association scan looking for markers that may confer susceptibility to several different tauopathies. In this article, we focus on the markers contained in the region including and surrounding tau. Using the present data, and genotypes imputed using HapMap, it was shown that a significant association exists across the entire inverted interval on chromosome 17 for PSP and CBD cases.
METHODS
SAMPLE COLLECTION AND PREPARATION
The included samples were collected by the Memory and Aging Center at the University of California, San Francisco, or by one of us (K.C.W.). Cases of PSP, CBD, FTD, and FTD with amyotrophy were collected from an unrelated white population. Cases of FTD met the Neary criteria, and cases of PSP met the Litvan criteria. Although all the cases were clinically confirmed, only 46 had pathologic confirmation of disease. None of the cases have known tau or progranulin mutations. Unaffected individuals from the same population were used as controls. DNA was isolated from whole blood using the Puregene kit (Gentra Systems, Minneapolis, Minnesota). The number of patients used for each diagnosis in this study are as follows: FTD, 56 (32 men and 24 women); PSP, 36 (20 men and 16 women); CBD, 22 (9 men and 13 women); amyotrophic lateral sclerosis, 18 (11 men and 7 women); and controls, 98 (42 men and 56 women). The mean age was 73 years for controls and 67 years for cases, with an average age at onset of 60 years. All the participants provided informed consent as approved by the University of California, San Francisco, and The University of North Carolina at Chapel Hill human subjects institutional review boards.
GENOTYPING
Genotyping was performed using the GeneChip 500K Affymetrix SNP arrays using the protocol provided by Affymetrix (Affymetrix, Inc, Santa Clara, California). The BRLMM algorithm was used to make genotyping calls. Acceptable genotypes had a confidence score of less than 0.5. Any call that did not meet this threshold was removed from further analysis.
ANALYSIS
The genotypes of cases vs controls were compared using a Fisher exact test to determine whether the allele frequency in the cases was significantly different from the controls. Markers that were considered to be out of Hardy-Weinberg equilibrium were excluded from analysis. There was no population stratification detected when tested using EIGENSTRAT software (Harvard Medical School, Boston, Massachusetts). Genotype calls made using the BRLMM algorithm were used to infer the remaining known HapMap markers in the area based on correlation using the program IMPUTE from the Chiamo suite (Chiamo Genetics Software Suite, University of Oxford, Oxford, United Kingdom). Imputed genotypes were considered acceptable at a posterior probability greater than 0.8, and markers were included in association tests if the call rate was greater than 80%.
RESULTS
Genomewide, the mean sample call rate was 95%, and the mean single nucleotide polymorphism (SNP) call rate was 92% on the Affymetrix 500K platform. Less than 1% of SNPs were out of Hardy-Weinberg equilibrium, and 1.5% of SNPs were monomorphic.
From the 326 SNPs typed in the MAPT region ranging from approximately 40.4 to 42.5 Mb on chromosome 17, we attempted to impute an additional 4845 Hap-Map SNPs. After eliminating imputed SNPs with a posterior probability lower than 0.8, 1477 SNPs remained. Of these SNPs, 60 were monomorphic and 68 were not in Hardy-Weinberg equilibrium. Any marker with a sample call rate less than 80% was removed. Genotypes for 1169 genotyped and imputed SNPs were used to explore the region near the MAPT gene for allelic association with PSP, CBD, FTD, and FTD with amyotrophy.
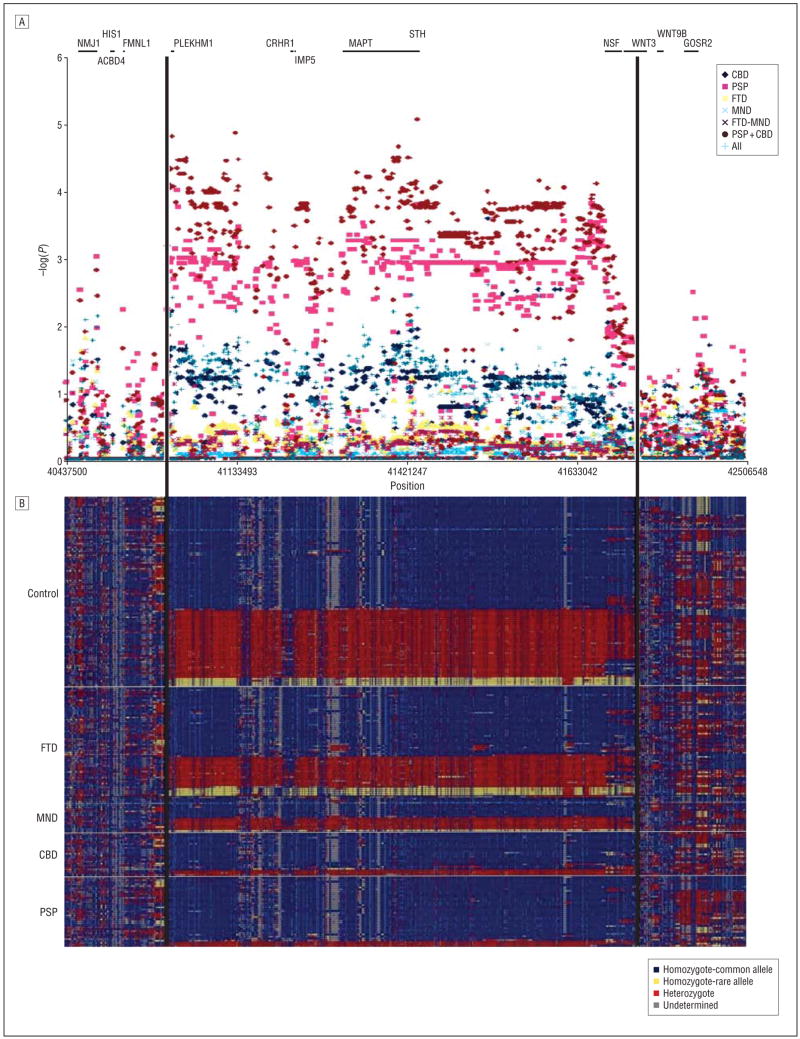
Figure 2A shows a plot of the probability that the cases and controls have equivalent genotype frequencies for each of the typed or imputed SNPs that met the inclusion criteria for each of the disease classification models tested. All of the significant associations observed are within the boundaries of the chromosomal inversion that distinguish the H1 and H2 haplotypes. Although there are some exceptions, most markers across the inversion for any given comparison fall within a constant range of probabilities across the interval. The most striking associations observed are for PSP alone or combined with CBD vs controls across the entire region of the chromosomal inversion. Rarely, a marker from other comparisons will reach a nominally significant association, but these events are rare and are not constant across the inverted interval. Inspection of the raw allele-specific hybridization intensity for these markers does not robustly distinguish between genotype clusters and are not considered to be significant associations. The region where allelic association is detected clearly defines the inversion interval boundaries. Figure 2B shows the genotypes for all samples across the region of interest in the following order: control, FTD, FTD with amyotrophy, CBD, and PSP. Each row represents an individual, and each column represents a marker. Known genes are indicated as lines above the genotypes. The samples were sorted based on diagnosis and haplotype similarity. Two distinct haplotypes can be identified in this figure consistent with previous designation of the H1 and H2 haplotypes.
Figure 2.
Association and haplotypes across the interval. A, A plot of the negative log of the P value from the comparisons between cases and controls. Known genes are represented as lines at the top of the figure. B, The genotypes for all samples across the region of interest. Each row corresponds to a sample. The samples were sorted based on diagnosis and haplotype similarity. Samples with mostly blue, or major, alleles have the H1 haplotype, and samples with mostly yellow, or minor, alleles have the H2 haplotype. Samples with mostly red, or heterozygote, alleles are H1/H2. CBD indicates corticobasal degeneration; FTD, frontotemporal dementia; MND, motor neuron disease; and PSP, progressive supranuclear palsy. The figure was created using the National Institute of Environmental Health Sciences SNPs Visual Genotypes program.13
Table 1 provides the counts for the 3 haplotype combinations (H1/H1, H1/H2, and H2/H2) for each category of diagnosis. Very few heterozygous haplotypes, and no homozygous H2 haplotypes, were seen in either PSP or CBD. Compared with controls, using a Fisher exact test, only PSP and PSP combined with CBD were significantly different. This confirms that the H1 haplotype is overrepresented in PSP and CBD cases compared with controls and that FTD and FTD with amyotrophy have H1 levels in the same proportion as controls (Table 2).
Table 1.
Comparison of Haplotype Combinations by Diagnosis
| Participants, No. |
P Value | Odds Ratio (95% Confidence Interval) | |||
|---|---|---|---|---|---|
| H1/H1 | H1/H2 | H2/H2 | |||
| Control | 59 | 35 | 4 | ||
| PSP | 33 | 3 | 0 | .001 | 7.27 (2.08–25.36) |
| CBD | 19 | 3 | 0 | .06 | 4.19 (1.16–15.10) |
| FTD | 37 | 15 | 4 | .46 | 1.29 (0.65–2.55) |
| MND | 11 | 6 | 1 | >.99 | 1.04 (0.37–2.91) |
| PSP combined with CBD | 52 | 6 | 0 | <.001 | 5.73 (2.24–14.62) |
| All diagnoses | 100 | 27 | 5 | .03 | 2.07 (1.17–3.64) |
Abbreviations: CBD, corticobasal degeneration; FTD, frontotemporal dementia; MND, motor neuron disease; PSP, progressive supranuclear palsy.
Table 2.
Haplotypes by Diagnosis
| Participants, No. |
|||||||
|---|---|---|---|---|---|---|---|
| Control | PSP | CBD | FTD | MND | PSP Combined With CBD | All Diagnoses | |
| H1 | 153 | 69 | 41 | 89 | 28 | 110 | 227 |
| H2 | 43 | 3 | 3 | 23 | 8 | 6 | 37 |
| Total | 196 | 72 | 44 | 112 | 36 | 116 | 264 |
| %H1 | 0.781 | 0.958 | 0.932 | 0.795 | 0.778 | 0.948 | 0.860 |
Abbreviations: CBD, corticobasal degeneration; FTD, frontotemporal dementia; MND, motor neuron disease; PSP, progressive supranuclear palsy.
COMMENT
The association found in these tauopathies across this interval on chromosome 17 further supports the theory that 1 or more loci in this region are affecting susceptibility specifically to PSP and CBD. Because the markers are highly correlated and the association is seen across the whole region, it is difficult to narrow down a disease-causing variant or even a possible candidate gene. However, considering the pathologic features of these diseases and the involvement of tau mutations seen in familial forms, the MAPT gene represents the most likely cause driving the association.
Although all of the diseases in this study are tauopathies, not all of the diseases were highly associated with this region. The hypothesis leading to this study was that similar diseases are caused by mutations in genes controlled by similar biochemical pathways. So, although the mutation causing any particular presentation of 1 of these diseases may be located in different genes, the pathologic outcome is the same. There were other significantly associated regions in the genomewide scan that may be affecting susceptibility to the diseases that were not as highly associated with the tau region. No association was seen between FTD samples and this region, although cases with FTD can have tau inclusions, and mutations of the tau gene have been found in affected families. The lack of association may be affected by the small sample size and the heterogeneity of sporadic FTD. Most cases of FTD do not have tau mutations or inclusion, but cases of PSP and CBD are almost always accompanied by tau inclusions.
The odds ratio was calculated to determine the risk associated with a particular haplotype. Controls have a 7-fold greater odds (95% confidence interval, 2.08–25.36) of having an H2 haplotype, on 1 or both chromosomes, compared with PSP and a 4-fold greater odds (95% confidence interval, 1.16–15.10) compared with CBD. When CBD and PSP are considered together, the odds ratio is in the middle, with a value of 5.7 (95% confidence interval, 2.24–14.62). This suggests that the H2 haplotype provides some protection from the PSP and CBD diseases. This proposed protective allele is considered to be significant for PSP, CBD, and PSP combined with CBD because none of the confidence intervals are less than 1.
Imputation filled in missing genotypes and genotypes for markers not included in the GeneChip 500K Affymetrix sets. This gave a fuller picture of the association in the region. Imputation methods are useful for association studies because they combine information from genotyped markers with existing data sets, such as HapMap. Testing a larger number of markers across the genome provides a finer grid for association. However, in an area of high linkage disequilibrium, as in the chromosome 17 inversion, the true disease-causing variant cannot be distinguished from the surrounding markers even with the extra imputed genotypes. Imputation added little to the association due to the strong linkage disequilibrium in the region that was implicated. The regions flanking this inversion are much more thoroughly evaluated, and there is little reason to investigate these flanking regions further. Filtering the data for call rate and posterior probability removed noise and most false-positives that were detected using unfiltered data. The genotype calling software used to impute genotypes resulted in more stringent and reliable calls.
The results of this association study provide strong evidence that a susceptibility locus in the MAPT gene region is related to certain Pick complex diseases, but the high degree of linkage disequilibrium in the region makes it difficult to draw conclusions about the exact location of the locus. To our knowledge, previous studies have reported results from candidate gene studies focusing on the tau gene. We instead looked at the whole genome and found an association with the entire inversion region, with no evidence that any part of the region is more important than any other. The genotypes are constant across the inversion due to the high level of linkage disequilibrium but, outside the inversion, they become highly variable, with no identifiable pattern. This is also supported by the constant level of association that drops off at the boundaries of the inversion. The inversion is likely a recent event because it is found only in white populations. Although a specific cause cannot be determined, something in the inversion is likely affecting expression of the tau gene and, ultimately, disease status. The inversion, or, more specifically, the H2 haplotype, seems to offer some protection against PSP and CBD.
Acknowledgments
Funding/Support: This work was supported in part by the Schiller family, which has an endowed professorship at The University of North Carolina at Chapel Hill for research into the cause and cure of PSP; grant RO1NS36733 from the National Institute of Neurological Disorders and Stroke (Dr Wilhelmsen); the California State Alzheimer’s Disease and Related Disorders Fund (Drs Wilhelmsen and Miller); The Amyotrophic Lateral Sclerosis Foundation (Dr Wilhelmsen); The French Foundation (Dr Wilhelmsen), program project grant PO1 AG19724 from the National Institute on Aging (Drs Miller and Wilhelmsen); and grant MO1RROOO79 from the National Center for Research Resources (General Clinical Research Center at the University of California, San Francisco).
Footnotes
Author Contributions: All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Miller and Wilhelmsen. Acquisition of data: Webb, Miller, Bonasera, Boxer, and Karydas. Analysis and interpretation of data: Webb and Wilhelmsen. Drafting of the manuscript: Webb. Critical revision of the manuscript for important intellectual content: Miller, Bonasera, Boxer, Karydas, and Wilhelmsen. Statistical analysis: Wilhelmsen. Obtained funding: Miller and Wilhelmsen. Administrative, technical, and material support: Miller, Boxer, and Wilhelmsen. Study supervision: Wilhelmsen.
Financial Disclosure: None reported.
Additional Contributions: We thank the patients for their willingness to participate in this investigation. With her permission, we specifically thank 1 of the patients with PSP, Carol M. Schiller, PhD. Dr Schiller became the first tenured female senior scientist at the National Institute of Environmental Health Sciences, board-certified toxicologist, and graduate of The University of North Carolina at Chapel Hill law school.
References
- 1.Williams DR. Tauopathies: classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau [review] Intern Med J. 2006;36(10):652–660. doi: 10.1111/j.1445-5994.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 2.Neary D, Snowden J, Mann D. Frontotemporal dementia [review] Lancet Neurol. 2005;4(11):771–780. doi: 10.1016/S1474-4422(05)70223-4. [DOI] [PubMed] [Google Scholar]
- 3.Kertesz A. Pick complex: an integrative approach to frontotemporal dementia: primary progressive aphasia, corticobasal degeneration, and progressive supra-nuclear palsy [review] Neurologist. 2003;9(6):311–317. doi: 10.1097/01.nrl.0000094943.84390.cf. [DOI] [PubMed] [Google Scholar]
- 4.Rademakers R, Cruts M, van Broeckhoven C. The role of tau (MAPT) in frontotemporal dementia and related tauopathies [review] Hum Mutat. 2004;24(4):277–295. doi: 10.1002/humu.20086. [DOI] [PubMed] [Google Scholar]
- 5.Rademakers R, Melquist S, Cruts M, et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14(21):3281–3292. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 6.Pittman AM, Fung HC, de Silva R. Untangling the tau gene association with neurodegenerative disorders [review] Hum Mol Genet. 2006;15(spec No 2):R188–R195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- 7.Conrad C, Andreadis A, Trojanowski JQ, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol. 1997;41(2):277–281. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 8.Cruts M, Rademakers R, Gijselinck I, et al. Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the tau region. Hum Mol Genet. 2005;14(13):1753–1762. doi: 10.1093/hmg/ddi182. [DOI] [PubMed] [Google Scholar]
- 9.Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 10.Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8(4):711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 11.Myers AJ, Pittman AM, Zhao AS. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25(3):561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Josephs KA, Petersen RC, Knopman DS, et al. Clinicopathologic analysis of frontotemporal and corticobasal degenerations and PSP. Neurology. 2006;66(1):41–48. doi: 10.1212/01.wnl.0000191307.69661.c3. [DOI] [PubMed] [Google Scholar]
- 13.NIEHS SNPs Program. [Accessed September 2007];NIEHS Environmental Genome Project Web page. http://egp.gs.washington.edu.