Abstract
Silk-elastinlike protein polymers (SELPs) of varying ratios and lengths of silk and elastin blocks capable of hydrogel formation were evaluated as matrices for controlled delivery of plasmid DNA. Influence of polymer structure, ionic strength of the media and gelation time on DNA release from two structurally related hydrogels, SELP-47K and SELP-415K, was evaluated. The influence of elastase-induced degradation on the swelling behavior and DNA release from these hydrogels was investigated. Results indicate that release is a function of polymer structure, concentration and cure time. SELP-415K which has twice the number of elastin units as that of SELP-47K demonstrated higher release than that of SELP-47K. DNA release from these hydrogels is an inverse function of polymer concentration and cure time, with higher release observed at lower polymer concentration and shorter cure time. Results indicate that ionic strength of the media governs the rate of release. An increase in swelling ratio was observed in the presence of elastase at 12wt% composition for both SELP analogs. Release in the presence of elastase was enhanced due to increased swelling ratio and loss of hydrogel integrity. These studies allude to the utility of recombinant techniques to control plasmid DNA release and biodegradation in SELP hydrogels.
INTRODUCTION
Progress made in gene therapy is severely undermined by transient and low transfection efficiency of non-viral vectors and the toxicity and immunogenicity of viral vectors (Verma and Somia, 1997). Several systemic and localized gene delivery platforms have been explored to improve the duration and localization of gene transfer (Shen et al., 2007, Cohen-Sacks et al., 2004, Wang et al., 2004). Encapsulation of naked DNA, non-viral vectors, or viral vectors in polymeric matrices provides several advantages. These include (i) protection from enzymatic degradation, (ii) prolonged delivery, and (iii) efficient localization of transgene expression thereby minimizing toxicity and maximizing therapeutic efficacy (Doukas et al., 2001, Wang et al., 2003, Wang et al., 2005).
Both natural and synthetic polymers have been applied as matrices for controlled gene delivery (Dang and Leong, 2006, Shen et al., 2007, Cohen-Sacks et al., 2004, Wang et al., 2005, Bellocq et al., 2004). Limited control over polymer sequence and structure or issues surrounding the biocompatibility and biodegradability of such polymers has reduced the viability of these delivery platforms for gene delivery applications (van de Weert et al., 2000). Genetically engineered polymers couple the versatility of biocompatibility and biodegradability of natural polymers on one hand with the high degree of control over polymer structure achievable by recombinant synthesis on the other (Haider et al., 2004). Silk-elastinlike protein polymers (SELPs) are a class of genetically engineered polymers composed of repeating units of silk-like (GAGAGS) and elastin-like (GVGVP) peptide blocks in the polymer structure (Cappello et al., 1990). Silk provides cross linking capability and renders mechanical strength, while elastin enhances aqueous solubility. By carefully controlling the ratio and length of silk and elastin units using recombinant techniques, SELPs of diverse functionalities (stimuli-sensitive, biocompatible and biodegradable) can be produced (Megeed et al., 2002, Nagarsekar et al., 2003, Haider et al., 2005).
SELPs with two or more silk units in the monomer repeat and with favorable silk to elastin (S:E) ratio tend to form hydrogels with elevation of temperature from room temperature to 37°C. Solutions of two SELP analogs, 47K and 415K (Fig. 1), which undergo irreversible sol-to-gel transition at 37°C have been developed that are capable of matrix-mediated gene delivery (Megeed et al., 2004, Haider et al., 2005). SELP 47K has a higher S:E and forms hydrogels in the concentration range 4–12 wt% where as SELP 415K forms hydrogels above 10wt% concentration due to its low S:E ratio (Haider et al., 2005). This allows the injection of aqueous polymer-DNA solutions that transform into solid hydrogel matrices within minutes in the body, enabling temporal and spatial control over gene release. Controlled plasmid DNA delivery can be beneficial in tissue engineering, vaccine delivery and a number of other biomedical applications (Doukas et al, 2001, Megeed et al, 2004). Controlled release of DNA from SELP-47K hydrogels is shown to be a function of polymer concentration and cure time (Megeed et al., 2002). SELP-415K which has eight more elastin repeats in its monomer sequence than that of SELP-47K has been shown to have higher swelling ratio (Haider et al., 2005) but its DNA release characteristics are unknown. The influence of length and sequence of elastin units (SELP-415K) on interaction with DNA and elastase-induced biodegradation in these polymers is also unexplored. The presence of elastin blocks in SELPs that can potentially be degraded by the endogenous elastase (released from leukocytes) can alter the network properties of the formed matrix and thereby the release of DNA from these systems. The current study reports the influence of SELP structure, composition, ionic strength of the media, and hydrogel cure time on controlled release of plasmid DNA. Also reported is the influence of elastase incorporation on the swelling behavior and DNA release from these matrices
Fig. 1.
Amino acid sequences of: A) SELP 47K; B) SELP 415K. Lys (K) residues underlined. The elastin blocks are highlighted in gray. Head and tail sequences not shown.
MATERIALS AND METHODS
Preparation of DNA-containing hydrogels
SELP-47K was provided by Protein Polymer Technologies, Inc. (San Diego, CA). SELP-415K was biosynthesized and characterized by procedures described previously (Haider et al., 2005). Leukocyte elastase was purchased from Sigma (St. Louis, MO). Plasmid pRL-CMV-luc 4.08 kbp (Promega, Madison, WI) was propagated in Novablue Singles Competent Cells (Novagen, San Diego, CA), and purified using an EndoFree Giga Kit (QIAGEN Sciences, Germantown, MD) according to manufacturer’s instructions. Syringes containing frozen 12 wt% (%w/w) polymer solutions were thawed in a beaker containing 500 ml of water for 5 min at room temperature. Polymer solutions and plasmid DNA were gently mixed. The volume of the mixture was adjusted by addition of PBS and MilliQ ultrapurified water from Millipore (Billerica, MA) to yield 250 μg/ml plasmid DNA in 11.5 wt% polymer. The mixtures were then transferred to disposable syringes (1ml), incubated at 37°C for 4 hr and 48 hr respectively. After the appropriate cure time, the syringe tip was cut off, and the DNA containing hydrogels were extruded and cut into 50 mm3 cylindrical discs (2.3 mm radius) using razor blade for swelling and release studies. The hydrogels were robust and did not deform.
Influence of ionic strength on DNA release
DNA-containing hydrogel discs were placed in 4 ml glass vials containing 3 ml of the release buffer. PBS (10mM, pH 7.4, 0.01% w/v NaN3) buffer with total ionic strength (μ), adjusted with NaCl to 0.016 M, 0.16 M, and 1.6M respectively were used as release media. Vials containing hydrogels submerged in buffer were incubated at 37 °C in a shaking (120 rpm) incubator for the duration of the experiments. At predetermined time points, the buffer was sampled and replaced with fresh buffer. The amount of DNA in the release media was determined by Picogreen DNA Quantitation Kit (Molecular Probes, Eugene, OR) and cumulative released DNA was calculated by procedures described previously (Megeed et al., 2002).
Ionic interactions of SELP copolymers with plasmid DNA
The interaction of both polymers, 47K and 415K, containing one lysine residue per monomer repeat (Fig. 1) with plasmid DNA at various ionic strengths was evaluated by a turbidity assay (Megeed et al, 2002). DNA: polymer complexes were formed at the charge ratios of 2:1, 1:1, and 1:2 assuming full ionization of the lysine residues and DNA phosphates at pH 7.4 and room temperature. The upper limit concentration of polymer used in the complexes was 0.0094 μg/μL. Polymer solutions were added drop wise to solutions containing 50 μg of plasmid pRL-CMV-luc in PBS with ionic strengths adjusted to 0.016 M, and 0.16 M to a final volume of 137 μL. After incubation at room temperature for 30 min the absorbance at 400 nm was determined by spectrophotometric techniques.
Determination of swelling behavior
SELP-47K and 415K form hydrogels in the concentration range of 4–12 wt%, and 10–12 wt%, respectively (Haider et al., 2005). Hydrogels were prepared and allowed to reach equilibrium in a PBS solution (Table 1). Soluble polymer fractions remaining in the hydrogels post-gelation were removed by extensively washing the hydrogels for 1 week in 1X dPBS under mild agitation (120 rpm) at 37°C. Fresh buffer was replaced daily throughout the washing period. Hydrogels were equilibrated for 24 hr in PBS or PBS with 100ng elastase. The weight equilibrium swelling ratio (q) was experimentally determined as a ratio of the weight of swollen hydrogel (Ws) over the weight of dry hydrogel (Wd) using procedures described previously (Dinerman et al., 2002, Dandu et al., 2008). Briefly, hydrogels were removed from test conditions after 1 week and gently blotted with a lint-free wipe to remove excess solvent from surfaces prior to determining the swollen (wet) weight. Dry weight was determined by weighing the hydrogels after desiccating the wet hydrogels using Drierite dessicant (Xenia, OH) for 5 days. Gel disks were then weighed and placed into a vacuum oven at 30°C for 24 h and then re-weighed to ensure no change in dry hydrogel weight following vacuum drying.
Table 1.
Polymer Composition and Concentration and Elastase Effects on Swelling
| SELP Analog | SELP Concentration | Release Media |
|---|---|---|
| 47K | 4% | PBS |
| 47K | 8% | PBS |
| 47K | 12% | PBS |
| 415K | 12% | PBS |
| 47K | 4% | PBS with 100ng Elastase |
| 47K | 8% | PBS with 100ng Elastase |
| 47K | 12% | PBS with 100ng Elastase |
| 415K | 12% | PBS with 100ng Elastase |
| 47K | 4% | PBS with 10ng Elastase |
| 47K | 4% | PBS with 50ng Elastase |
| 47K | 4% | PBS with 150ng Elastase |
| 47K | 4% | PBS with 200ng Elastase |
Evaluation of polymer degradation and DNA release in the presence of elastase
Hydrogels were prepared as described above. Cylindrical SELP hydrogel analogs with 50 mm3 volume were placed in 1 mL PBS or PBS containing physiological concentration of elastase (0.1μg/mL) (Hind et al., 1991, Morrison et al., 1999). Media was collected at predetermined time points, and replenished with fresh media at each time point. The DNA concentration was then determined using the Quanti-It Picogreen assay kit.
Statistical analysis
Single factor analysis of variance (ANOVA) and student t-test were carried out at α=0.05 to determine statistically significant differences between test samples. All studies were performed in triplicate.
RESULTS AND DISCUSSION
Effect of polymer structure and cure time on DNA release
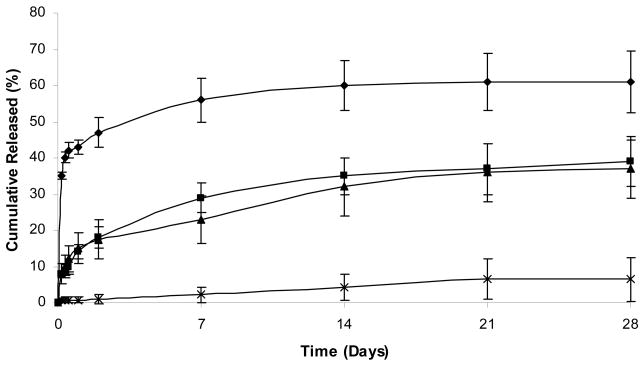
Fig. 2 shows the release profile of plasmid DNA from both polymer constructs at 4 hr and 48 hr cure times respectively. At 4 h cure time cumulative release from 415K hydrogels was higher than that of 47K up to day 28. Since there are an overall lower number of silk units in the polymer backbone than 47K, this is presumed to be due to the lower crosslinking density of 415K (Fig. 1). For both sets of hydrogels cured for 4 hr, there was no significant DNA release beyond day 21 suggesting that the remainder of DNA is trapped in the matrices. An increase in cure time from 4 hr to 48 hr resulted in lower cumulative release for both polymers. This difference in release was more pronounced for hydrogels made from SELP-47K compared to SELP-415K.
Fig. 2.
Influence of polymer structure and cure time (CT) on DNA release from 12 wt% SELP hydrogels at pH=7.4 and μ=0.16 M released over a 28 day period. Symbols represent SELP-415K at 4 hr cure time (CT) (◆) and 48 hr CT (■), and SELP-47K at 4 hr CT (▲) and 48 hr CT (X) respectively. Each symbol represents mean ± standard deviation for n=3 samples.
Previous studies suggest that an increase in cure time for both polymers results in a decrease in their degree of swelling (Haider et al., 2005). These physically cross linked hydrogels are known to have a fraction of soluble polymer that does not participate in the hydrogel network (Dandu et al., 2008, Dinerman et al., 2002). With time there increased chances for more silk units in the vicinity to be recruited at the crosslinking site allowing the formation of a denser network therefore increased crosslinking density, resulting in decreased release. The reason for the pronounced effect of cure time for 47K is presumed to be the higher number of silk units in the polymer backbone that allows a higher degree of inter-polymer interactions compared to 415K.
Influence of ionic strength on DNA release
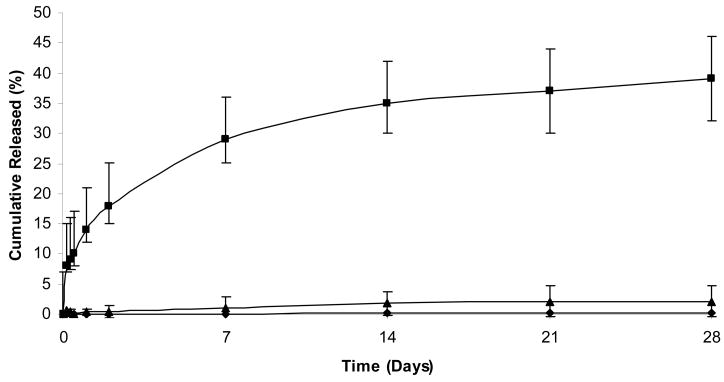
The release of DNA from hydrogels made from SELP 415K showed a strong dependence on the ionic strength of the medium with the highest release observed at 0.16 M (Fig. 3). Previously we had shown that an increase in ionic strength for SELP-415K hydrogels results in a decrease in their degree of swelling (Haider et al., 2005). The lower degree of swelling of SELP-415K at high ionic strength (1.6 M) explains the lower release of plasmid DNA at this ionic concentration compared to the physiologically relevant ionic strength of 0.16 M. At very low ionic strength (0.016M) negatively charged plasmid DNA interacts with lysine residues of the polymer backbone which results in no release consistent with our previous observations (Megeed et al, 2002).
Fig. 3.
Influence of ionic strength on DNA release from SELP 415K hydrogels at pH=7.4. Symbols represent release at 0.016M (◆), 0.16M (■) and 1.6M (▲). Each symbol represents mean ± standard deviation for n=3 samples.
Influence of polymer structure and ionic strength on polymer-DNA interaction
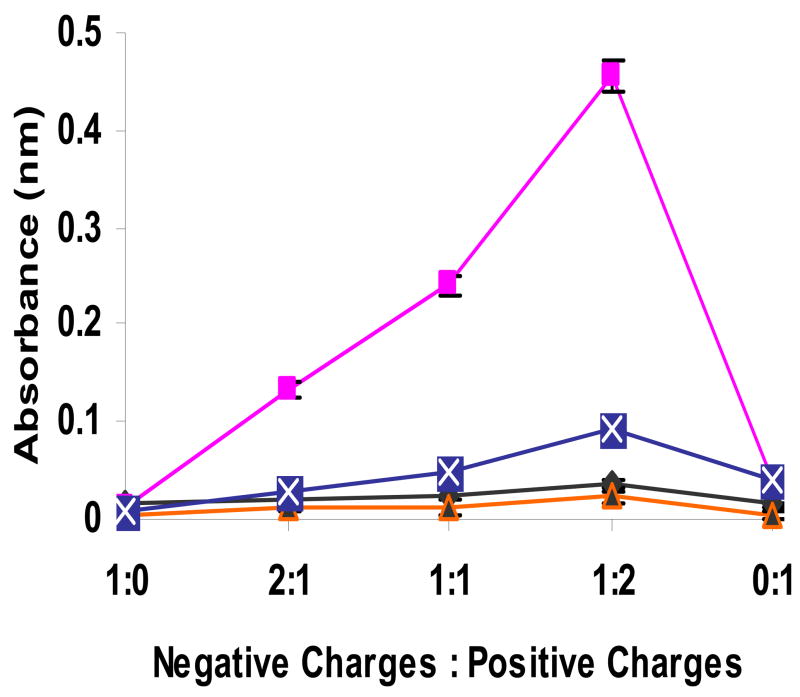
Recombinant techniques allow the introduction of functional amino acid residues at precise locations in the polymer backbone. SELP 415K was designed such that it has a similar molecular weight as SELP 47K but with longer elastin-like units. Consequently there are five more lysine residues in 47K compared to 415K. This could reduce the interaction of negatively charged plasmid DNA with the 415K polymer backbone. To evaluate this phenomenon we measured the absorbance of DNA/polymer complexes at low non-gel forming concentrations. The absorbance of Polymer 47K mixture with plasmid DNA showed a substantial increase when prepared at low ionic strength compared to high ionic strength (Fig. 4). Mixtures of SELP-415K and plasmid DNA showed lower turbidity at both ionic strengths compared to their SELP-47K counterparts due to the presence of lower number of lysine residues.
Fig. 4.
Influence of polymer structure and ionic strength on the turbidity of DNA (at room temperature): Symbols represent absorbance of polymer complexes of SELP-415K at 0.016M (◆) and 0.16M (▲), and SELP-47K at 0.016M (■) and 0.16M ( ). Each symbol represents mean ± standard deviation for n=3 samples.
). Each symbol represents mean ± standard deviation for n=3 samples.
Influence of elastase-induced biodegradation on swelling of SELP hydrogels
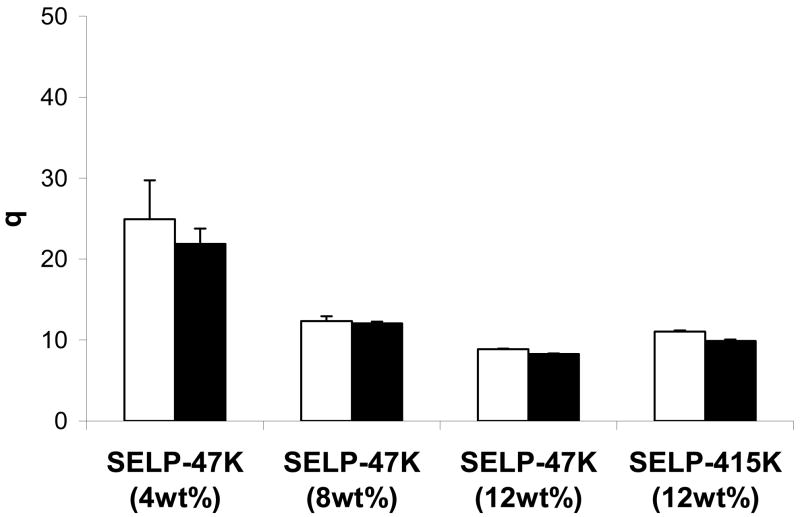
The elastin units in SELPs are susceptible to degradation by endogenous elastases providing a possible mechanism for their biodegradation in vivo. To investigate the nature and extent of biodegradation caused by elastase, studies were conducted in vitro using physiologically relevant concentrations of leukocyte elastase (Hind et al., 1991, Morrison et al., 1999). Results indicate an increase in the observed swelling ratio in the presence of elastase for both SELP analogs at 12wt% (p<0.05) (Fig. 5). However, these differences were not significant at lower polymer concentration (p=0.05). The greater swelling ratio of the elastase treated hydrogels at 12wt% concentration is likely due to a more loosely crosslinked network probably as a result of chain cleavages by the protease. At this stage the extent to which elastase penetrates the hydrogel network is not known. The smaller polymer chains as a result of degradation likely elute from the hydrogel as soluble fraction thus reducing the dry weight (Wd) of the hydrogel and increasing the swelling ratio (q). At low polymer concentration, however, the difference is not significant probably because at low concentration the hydrogel network is already loosely cross linked that additional chain cleavages have lesser effect. It was also observed that in the presence of elastase, 4wt% SELP-47K hydrogels begin to breakdown into pieces and lose their hydrogel integrity within 10 days (data not shown) suggesting the potential utility of this polymer concentration for cases in which it is desirable to degrade the drug carrier within a week.
Fig. 5.
Influence of elastase on the swelling behavior of SELP hydrogels as a function of polymer concentration and structure. The bars represent q of SELP hydrogels (i) in the presence (white) and absence (black) of elastase. Each bar represents mean ± standard deviation for n=3 samples.
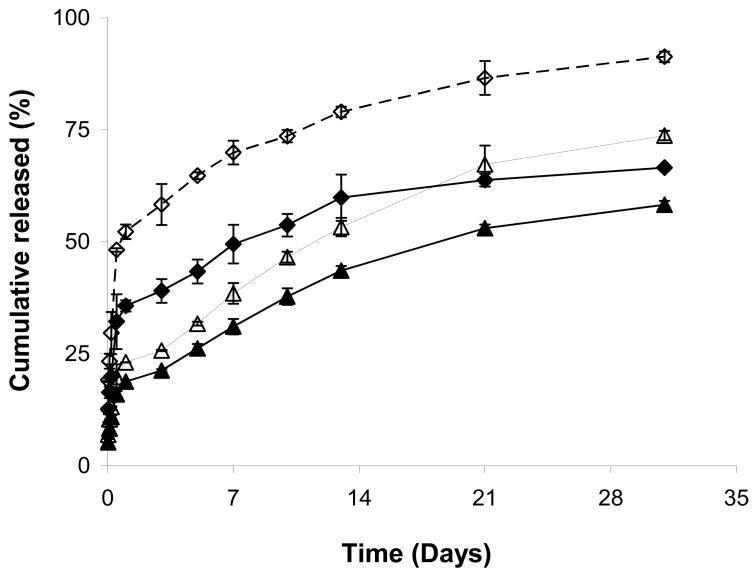
Influence of elastase-induced biodegradation on DNA release
Increased swelling ratio and loss of hydrogel network integrity suggest a larger pore size and greater release rate in the presence of elastase. To understand the extent by which release is enhanced in the presence of elastase, DNA release was evaluated from both SELP hydrogels at similar polymer concentrations (12wt%) in the presence of elastase. As expected in the absence of elastase, SELP-47K, with its shorter elastin units, showed greater release than SELP-415K (p<0.05) (Fig. 6). In the presence of elastase both polymers showed significantly higher release rates than in the absence of elastase (p<0.05) (Fig. 6). For SELP-415K hydrogel, close to 100% DNA release was observed in the presence of elastase compared to 60% in the absence of elastase. These studies indicate that both release and biodegradation are a function of polymer concentration and structure and suggest that by controlling the sequence of the silk and elastin units, polymers of desirable release and biodegradation profiles can be developed for controlled gene delivery applications. Further research is necessary to characterize the pore size and mechanism of DNA release. In the design of polymeric matrices for gene delivery several important parameters need to be considered. These are ease of administration to the site where gene release is desired (e.g., intratumoral or intramuscular), their viscosity, the pore size of the hydrogels, interaction of plasmid DNA with the polymer backbone and its influence on gene release, biodegradation of the matrix and its biocompatibility and elimination from the body. By molecular engineering of SELPs these parameters may be controlled (Dandu and Ghandehari, 2007). In this report by increasing the length of the elastin unit while maintaining molecular weight constant, we demonstrated that polymer-DNA interaction, release and biodegradation can be modulated. Hence by design of polymer structure, matrix mediated delivery of plasmid DNA from SELP hydrogels may be optimized. In addition, these studies can set the stage for utilization of such matrices for controlled release or incorporation of other bioactive agents such as viruses, cells, proteins and peptides.
Fig. 6.
Influence of elastase-induced degradation on release of DNA from SELP hydrogel analogs. Symbols represent cumulative percent release of DNA from 12wt% hydrogels of SELP-47K in the presence (△) and absence (▲) of elastase, and SELP-415K in the presence (◇) and absence (◆) of elastase. Each symbol represents mean ± standard deviation for n=3 samples.
CONCLUSION
Results indicate that polymer structure, concentration, ionic strength and hydrogel cure time govern the rate of DNA release from SELP hydrogels. The influence of the ratio of silk to elastin and the length of elastin units on release is demonstrated by the higher release rate observed in SELP-415K compared to SELP-47K. Treatment with elastase leads to the biodegradation of elastin units in SELPs. These studies provide a framework for the design and development of novel biodegradable and biocompatible SELP-based matrix-mediated delivery systems for gene therapy applications.
Acknowledgments
Funding for this study was provided under NIH R01 CA107621.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bellocq NC, Kang DW, Wang X, Jensen GS, Pun SH, Schluep T, Zepeda ML, Davis ME. Synthetic biocompatible cyclodextrin-based constructs for local gene delivery to improve cutaneous wound healing. Bioconjug Chem. 2004;15:1201–11. doi: 10.1021/bc0498119. [DOI] [PubMed] [Google Scholar]
- Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, Ferrari F. Genetic engineering of structural protein polymers. Biotechnol Prog. 1990;6:198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
- Cohen-Sacks H, Elazar V, Gao J, Golomb A, Adwan H, Korchov N, Levy RJ, Berger MR, Golomb G. Delivery and expression of pDNA embedded in collagen matrices. J Control Release. 2004;95:309–320. doi: 10.1016/j.jconrel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Dandu R, Cappello J, Ghandehari H. Characterization of structurally related adenovirus-laden silk-elastinlike hydrogels. J of Bioact and Comp Poly. 2008;23:5–19. [Google Scholar]
- Dandu R, Ghandehari H. Delivery of bioactive agents from recombinant polymers. Progress in Polymer Science. 2007;32:1008–1030. [Google Scholar]
- Dang JM, Leong KW. Natural polymers for gene delivery and tissue engineering. Gene Delivery for Tissue Engineering. 2006;58:487–499. doi: 10.1016/j.addr.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Dinerman AA, Cappello J, Ghandehari H, Hoag SW. Swelling behavior of a genetically engineered silk-elastinlike protein polymer hydrogel. Biomaterials. 2002;23:4203–10. doi: 10.1016/s0142-9612(02)00164-3. [DOI] [PubMed] [Google Scholar]
- Doukas J, Chandler LA, Gonzalez AM, Gu D, Hoganson DK, Ma C, Nguyen T, Printz MA, Nesbit M, Herlyn M, Crombleholme TM, Aukerman SL, Sosnowski BA, Pierce GF. Matrix immobilization enhances the tissue repair activity of growth factor gene therapy vectors. Hum Gene Ther. 2001;12:783–98. doi: 10.1089/104303401750148720. [DOI] [PubMed] [Google Scholar]
- Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, Ghandehari H. Molecular engineering of silk-elastinlike polymers for matrix-mediated gene delivery: biosynthesis and characterization. Mol Pharm. 2005;2:139–50. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- Haider M, Megeed Z, Ghandehari H. Genetically engineered polymers: status and prospects for controlled release. J Control Release. 2004;95:1–26. doi: 10.1016/j.jconrel.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Hind CR, Joyce H, Tennent GA, Pepys MB, Pride NB. Plasma leukocyte elastase concentrations in smokers. J Clin Pathol. 1991;44:232–5. doi: 10.1136/jcp.44.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeed Z, Cappello J, Ghandehari H. Controlled release of plasmid DNA from a genetically engineered silk-elastinlike hydrogel. Pharm Res. 2002;19:954–9. doi: 10.1023/a:1016406120288. [DOI] [PubMed] [Google Scholar]
- Megeed Z, Haider M, Li D, O’Malley BW, Jr, Cappello J, Ghandehari H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. J Control Release. 2004;94:433–45. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- Morrison HM, Welgus HG, Owen CA, Stockley RA, Campbell EJ. Interaction between leukocyte elastase and elastin: quantitative and catalytic analyses. Biochim Biophys Acta. 1999;1430:179–90. doi: 10.1016/s0167-4838(98)00270-2. [DOI] [PubMed] [Google Scholar]
- Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Genetic engineering of stimuli-sensitive silkelastin-like protein block copolymers. Biomacromolecules. 2003;4:602–7. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- Shen X, Tong H, Jiang T, Zhu Z, Wan P, Hu J. Homogeneous chitosan/carbonate apatite/citric acid nanocomposites prepared through a novel in situ precipitation method. Composites Sci Tech. 2007;67:2238–2245. [Google Scholar]
- Van De Weert M, Hennink WE, Jiskoot W. Protein instability in poly(lactic-co-glycolic acid) microparticles. Pharm Res. 2000;17:1159–67. doi: 10.1023/a:1026498209874. [DOI] [PubMed] [Google Scholar]
- Verma IM, Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389:239–42. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu JK, Krol A, Li YP, Li CY, Yuan F. Systemic dissemination of viral vectors during intratumoral injection. Molecular Cancer Therapeutics. 2003;2:1233–1242. [PubMed] [Google Scholar]
- Wang Y, Li CY, Yuan F. Systemic virus dissemination during local gene delivery in solid tumors and its control with an alginate solution. International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference; United States. 2004. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu S, Li CY, Yuan F. A novel method for viral gene delivery in solid tumors. Cancer Research. 2005;65:7541–7545. doi: 10.1158/0008-5472.CAN-05-1112. [DOI] [PubMed] [Google Scholar]