Abstract
Genome wide association studies can potentially identify the genetic causes behind the majority of human diseases. With the advent of more advanced genotyping techniques, there is now an explosion of data gathered on single nucleotide polymorphisms (SNPs). The need exists for an integrated system that can provide up-to-date functional annotation information on SNPs. We have developed the SNP Integration Tool (SNPit) system to address this need. Built upon a federated data integration system, SNPit provides current information on a comprehensive list of SNP data sources. Additional logical inference analysis was included through an inference engine plug in. The SNPit web servlet is available online for use. SNPit allows users to go to one source for up-to-date information on the functional annotation of SNPs. A tool that can help to integrate and analyze the potential functional significance of SNPs is important for understanding the results from genome wide association studies.
Keywords: Single nucleotide polymorphisms (SNPs), Public health genetics, Biomedical informatics, Data integration, SNP annotation system, SNP integration system
1. Introduction
The majority of leading human diseases have a genetic component [1]. To understand both the genetic component behind human disease as well as to materialize the vision of predictive, preventive, and personalized medicine [2], scientists and researchers need to focus their attentions on the genetic variation within individuals. Genome wide association studies are a powerful epidemiologic method that allows researchers to detect genetic variation, even those variations with subtle effects [3]. With more advanced genotyping techniques, genome wide association studies result in more and more data on single nucleotide polymorphisms.
Single nucleotide polymorphisms (SNPs) are the most common form of genetic variation in the human genome, comprising about 90 percent of the polymorphism in the genome [4]. SNPs play an important role in genome wide association studies because they act as the primary biomarkers. The location of these biomarkers can be tremendously important in terms of predicting functional significance.
The prediction of functional significance or functional annotation of SNPs is complex. There are a limited number of commercial and public projects out there that deal with SNP annotation (PupaSNPFinder [5], SNPer [6], SNPselector [7], LS-SNP [8], SNP Function Portal [9], SNPLims [10]), they look at a certain subset of possible predictors and they use a data warehouse or pipeline approach to storing data. Some of the previous SNP tools are difficult to use and lack current documentation. Furthermore, to our knowledge, none of the previous SNP tools use a federated database approach, and thus, they run the risk of providing out-of-date information.
In addition to making sure that the data is current, a tool that focuses on SNP annotation must address numerous bioinformatics challenges. These include dealing with large numbers of data sources that are involved in SNP annotation, annotation sources of which scientists might not be aware, and the explosion of data due to the combination of all these data sources.
To address all these challenges, we have created a SNP Integration Tool (SNPit) that uses a comprehensive, federated database approach to look at a wide array of possible functional SNP annotation predictors and we seek to provide an easy-to-use integrated interface as well as some expert rules analysis. In contrast to the above mentioned tools, a federated data integration system does not store the information locally, instead it queries, normalizes, and analyzes the data in real time, providing the benefit of real time retrieval [11, 12], though the speed of the query is compromised. In addition, the use of a mediated schema within our data integration system provides a common semantic ontology that is not specific to the individual data sources, thus enabling flexibility when new data sources are added.
2. Implementation
SNP Integration Tool (SNPit) is built upon a pre-existing open source federated data integration system called BioMediator [13]. BioMediator is a data integration system developed by the University of Washington. We decided that BioMediator was the optimal choice for the domain of SNP annotation because it is a federated data integration system, it is easy to query, it is XML-based, and it uses a mediated schema which allows for more flexible data modelling.
BioMediator’s main objective is to take information from disparate sources and integrate it together through a multi-tiered system (Figure 1) [12, 14]. The researcher issues a single query, and the system gathers the information from the relevant sources. The central component to BioMediator’s system is its source knowledge base, which consists of descriptions of the various data sources, mappings from the source to the mediated schema, and the mediated schema itself. The mediated schema is an outline of the data sources’ objects and mappings, it can be thought of as an ontology of the entities and relationships in a domain [13]. In addition to the source knowledge base, components of the BioMediator system include wrappers that conduct syntactic translations by translating the returned data results into an XML document, a metawrapper that conducts semantic translations by mapping the returned XML document onto the mediated schema, and a query processor that queries against the mediated schema.
Figure 1. System architecture of BioMediator.
The various components of BioMediator. Includes the multi-tiered components of the system – the query processor, mediated schema, and wrappers to the different data sources.
The BioMediator system has a modular architecture; thus, it is easily extendable to additional analytical tools. One additional component that was added for the SNPit tool was the Java Expert System Shell (Jess) [15] rule-based system plug-in, which permitted us to add logical inference [16]. Jess allowed us to include a declarative programming approach and create new knowledge from a set of rules [17]. The inference engine allowed us to formulate sets of rules over the returned result sets in order to conduct various analyses and filtering of the data.
Much of the challenge when it comes to integrating a large number of data sources is that sometimes the result sets become overwhelming and difficult to understand to a human annotator. Owing to this unsatisfactory situation, we have also begun incorporating expert rules into the BioMediator system to enable proper filtering of data as well as improving the repeatability of the results. For example, it is difficult for a human to continually apply sets of expert-defined rules in a consistent fashion, especially when there are dependencies among rules. Additionally, given the proper framework, rules in the BioMediator system could be easily modified, shared, and applied repeatedly for iterative refinement.
Thus, SNPit builds upon both BioMediator and the Jess plug-in. A mediated schema and wrappers were created to model the different data sources that help to reveal important SNP information. Various data sources have been included such as dbSNP [18], EntrezGene [19], HGMD [20], Haplotter [21], GVS [22], SIFT [23], UCSC [24, 25, 26], Transfac [27], BDGP [28]. Table 1 includes the list of data sources from which SNPit currently pulls information (Table 1). SNPit is currently available in two different user interfaces: a graph-based visualization tool entitled Touchgraph which is available over the BioMediator website and a web servlet available over the SNPit website. The speed of the queries depends on both the number of SNPs submitted as well as the speed of the individual data sources that SNPit integrates with. Figure 2 demonstrates the components of SNPit with the data sources section, BioMediator section, and interfaces section.
Table 1. Data sources that are integrated into SNPit.
Different data sources that are being included in SNPit.
| dbSNP | central repository database for both single base nucleotide substitutions and short deletion and insertion polymorphisms [18] |
| EntrezGene | gene-centric database that includes information on nomenclature, chromosomal localization, gene products and their attributes [19] |
| HGMD | provides publication evidence to genes responsible for human inherited diseases [20] |
| Haplotter | web tool that includes evidence for positive selection within the human genome, two statistical measures that look at the linkage disequilibrium of positively selected alleles and frequencies of polymorphisms are provided [21] |
| GVS | local database that provides access to information in dbSNP and includes tag SNP and linkage disequilibrium analysis [22] |
| SIFT | resource for predicting the functional impact of nonsynonymous coding SNPs, algorithm sorts tolerant from intolerant polymorphisms [23] |
| UCSC Phastcons | predicts evolutionary conservation of noncoding SNPs based on aligning genomic sequences of different species [26] |
| UCSC Genscan | predicts whether the SNP lies in a region that is likely to be gene region, based on transcriptional, translational, and donor/acceptor splicing signals [24] |
| UCSC GNFAtlas2 | displays gene-centric expression of polymorphisms over 79 human tissues [25] |
Figure 2. SNPit diagram of the various system components.

SNPit’s various components, including the query layer, common data model, translation and interface between the different data sources and interfaces of SNPit.
The current version of SNPit includes a query interface, query processor, semantic translation engine, and interfaces to each of the data sources. By collaborating with different experts in both public health genetics and as well as genome sciences, various data sources have been identified that provide functional predictions for SNP annotation. Wrappers have been created to interface with these disparate data sources. A central data model (mediated schema) has been created and the necessary translational rules that translate the source specific data models into the common data model have been written. Implemented in the Protégé [29] ontology editor, Figure 3 is a graphical representation of a subset of the mediated schema, demonstrating the SNP, gene, evolutionary conservation, and splice site entities and the relationships between those entities. The SNPit system comes in two different user query interfaces that allow the users to access the common data model with a particular query: the graphical user interface implemented through TouchGraph [30] as well as a text-based web servlet. Most of our focus was on the development of the web servlet’s Internet accessibility; in particular, we designed it for simplicity and ease-of-use. Using the left-hand-side of the interface, a user can query a number of different entities. The user can also examine details of each entity, in text-form, by clicking on the resource.
Figure 3. Visio diagram of the mediated schema.

Subset of the mediated schema, demonstrating the entities, relationships, and attributes of the entities: SNP, gene, evolutionary conservation, splice site, and human gene mutations.
Data has been collected on a variety of possible prioritization attributes. The Human Gene Mutation Database (HGMD) provides information on whether a particular SNP might be within a gene known to cause an inherited disease. Genomic context, which is defined as whether a SNP is in an intron, exon, promoter, or coding region, is provided by the UCSC Browser’s Genscan Gene Prediction track. The UCSC’s Tissue Expression track contains information on a polymorphism’s transcriptional expression over 79 human tissues. The ECR Browser captures the evolutionary relationships between the genomes of vertebrates and non-vertebrates. Haplotter tells us about recent positive selection within the human genome of certain SNPs. SIFT returns predictions for which SNPs will affect protein function. Genome Variation Server (GVS) provides information on linkage disequilibrium (LD) between two SNPs, LD is calculated using the Hill method [31]; users can select LD information specific to the population they are studying when querying the LD component of SNPit, HapMap provides the source of the genotype data. SNPit was created for the purpose of human SNP annotation integration and analysis; however, SNP data from other genomes can be queried through SNPit, depending on whether or not those genomes are supported by the data sources that SNPit integrates with. All of the collected functional annotation data enabled us to implement some preliminary inference rules which provide expert system decision ability.
Various publications have confirmed the predictive potential of in silico tools for SNPs in coding regions and evolutionarily conserved regions [32, 33]. Prioritization schemes for SNPs based on biological plausibility can already be determined [34]; for example, SNPs in coding regions would be given higher priority over non-coding SNPs. Researchers who manually created and used a similar decision tree with rankings 1, 2, 3, and 4 were able to identify 119 polymorphisms with low or neutral predictive power in a test set of 140 SNPs [35], demonstrating the potential value of prioritization strategies either before or after an association study. In our research, we used a similar approach, though we acknowledge that variations in both the prioritization scheme and heuristic weights are possible.
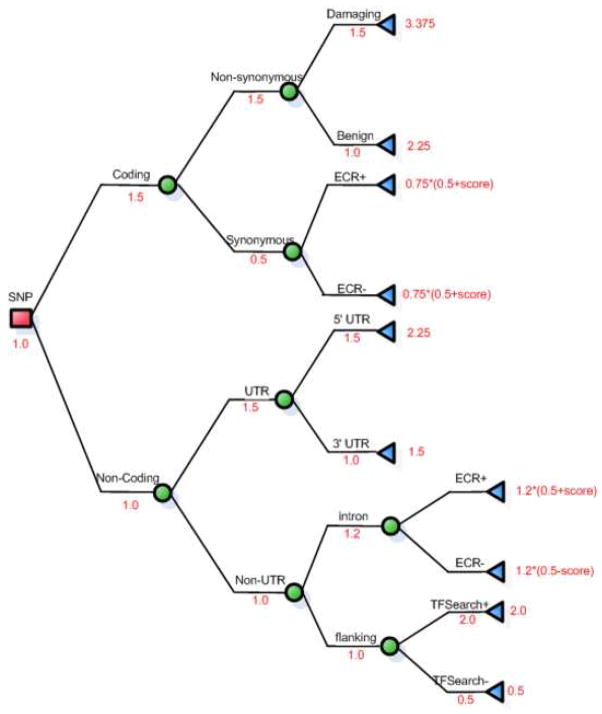
Preliminary logical inference rules were applied to the returned SNP annotation data by first creating a decision tree building upon biological knowledge [35, 34]. The initial decision tree was then vetted and revised as appropriate with the input of two experts, one of whom co-authored this paper; the experts also assigned weighted scores to the functional categories of different SNPs. The scores ranged from 1 to 4, with higher scores indicating a higher predicted relative risk of developing the phenotype. Potentially functional SNPs can be prioritized using different criteria; for example, the type of variant (nonsense, missense, splice site, promoter) can help to provide possible clues to the SNP’s biological relevance [34]. Currently, numerous bioinformatics tools access online biological databases and return information on both experimental and epidemiologic evidence; a decision tree was created to reflect the prioritization rankings of SNPS. Figure 4 demonstrates the preliminary rules and heuristic weights assigned to different SNP characteristics; heuristic weights were assigned to each node in the decision tree, with the score of the final node calculated by multiplying the previous nodes in its’ path. For example, SNPs that are in a coding region, that are non-synonymous and damaging, have a risk of moderate to very high; a heuristic weight of 3.375 was assigned to this branch of the tree. Using these expert heuristic weights, we were able to create some preliminary rules in Jess that capture the relative rankings of SNPs based on the evidence integrated together through the BioMediator federated integration system (Figure 5). The results of the rankings are then displayed in text-form through the web servlet interface.
Figure 4. Decision tree with heuristic weights.
Decision tree with paths and weights, heuristic weights were assigned to each node in the decision tree to reflect the prioritization of SNPs based on functional annotation. TFSearch refers to transcription factor binding and ECR refers to evolutionary conservation.
Figure 5. Example of Jess inference rule.
Pseudo-code demonstrating how Jess rules can be used to filter out SNPs that are nonsynonymous, (top) shows the pseudo-code and (bottom) demonstrates the actual Jess syntax.
3. Results
Using the SNP rs405509 as a user case scenario, the following results can be observed from the different SNPit interfaces.
SNPit Touchgraph Interface
The graphical user interface (GUI) component to SNPit can be accessed as a separate download from the web servlet component. Starting from an initial seed query of rs405509, the GUI expands out to reveal all the different pieces of information collected on rs405509. The different pieces of information are displayed in the form of nodes - which represent the entities - and edges - which represent the relationships – of the queried SNP. In the example, Figure 6, linkage disequilibrium information is displayed for rs405509. For SNP rs405509, the results demonstrate that the initial queried SNP is in linkage disequilibrium with SNPs rs10119, rs8106922, rs225925, rs439401, and rs405697. This result indicates that rs405509 is correlated with rs10119, rs8106922, rs225925, rs439401, and rs405697; this correlation can be an indication of which additional SNPs need to be studied.
Figure 6. Touchgraph interface for SNPit.

Touchgraph demonstrating how two expansions out from the central SNP leads to numerous SNPs being in linkage disequilibrium with the queried SNP.
SNPit Web Interface
The main website to the SNPit system was designed for simplicity and ease of use (Figure 7). The left side menu offers the ability to search by SNP or search by Gene. SNP input to the system can come from a variety of places including genome wide association study results. Within these categories, users can select either an entity to search by (Allele Frequency, Evolutionary Conservation, Gene Prediction, Linkage Disequilibrium, Population, Positive Selection, Protein Function Prediction, SNP, Tissue Expression, Gene, and Human Gene Mutation). Users can also select the Main Search option to search all the data sources together or the Rank SNPs option to view a list of queried SNPs ranked by potential functionality.
Figure 7. SNPit Web Interface.
SNPit web servlet, left menu includes links to various data sources.
Results from the Main Search option using the user case scenario rs405509 demonstrates a portion of the information that is returned from both dbSNP and HGMD (Figure 8). Background information from dbSNP reveals that rs405509 has the chromosome location chr19: 50100675–50100676, resides in the APOE gene, and has a predicted functional class (defined by dbSNP as the classification of the polymorphism and its nearby gene features [18]) of being in a locus region, meaning that the variation is 2 kb 5′ or 500 bp 3′ of a gene. Results from the Rank SNPs option using a collection of other SNPs demonstrate how the inference rules and heuristic weights are displayed to the user (Figure 9). Depending on the potential function of a SNP, a ranking and determination is assigned and displayed.
Figure 8. Results from SNPit web interface.
Subset of results returned when the user queries on rs405509.
Figure 9. Decision tree with heuristic weights.
Ranked SNPs based on decision tree.
4. Discussion
The SNPit system’s federated integration approach and its focus on a comprehensive set of data sources differentiates it from existing SNP annotation tools. Additional functional annotation considerations will be added during future developments, and we plan on incorporating these information sources as they become available and are deemed feasible. Additional features and functionality to the SNPit website will also be pursued as deemed appropriate. This includes providing the results in tab-delimited table format as well as the present flat file format. There are currently plans on incorporating uncertainty measurements into the SNPit system, which will help to address the issue of the accuracy in different data sources. That way, we’d be able to parameterize the confidence of the results reported. Our primary objective continues to be the creation of a quality system that is actively used by population geneticists, and we hope to accomplish this goal by continuing to include as many users as we can in both the design and development stages. IRB approval has been attained to develop a survey that can measure the usability and usefulness of the SNPit website. This utility survey will eventually be distributed over the SNPit website, allowing us to continue refining the design of the system.
5. Conclusions
Genome wide association studies hold enormous potential in terms of uncovering the genetic mechanisms behind human diseases. Advances in genotyping result in ever increasing amounts of information on SNPs. The ability to integrate and analyze SNP data is instrumental to understanding the results of genome wide association studies. SNPit is a tool that attempts to use a federated data integration system to predict the functional annotation of SNPs.
Acknowledgments
This work was supported with the following grants: NIH NLM T15LM07442, NSF IIS-0513877, NIH NHGRI/NLM R01 HG02288. The authors would like to extend thanks to the BioDIAG and UII group for their contributions to the SNPit project.
Footnotes
Authors’ contributions
TS designed the mediated schema for the SNPit system, coded the wrappers to the different data sources and the web servlet, and wrote the first draft of this paper. CC provided expertise on SNP annotation, provided the heuristic weights for the decision tree, and guided the development of the project. PTH provided biomedical informatics expertise and guided the development of the project. All authors participated in the drafting of this paper and approved the final draft for submission.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miniño HMA, Smith B. National Center for Health Statistics. Vol. 54. 2006. Deaths: Preliminary Data for 2004 National vital statistics reports. [PubMed] [Google Scholar]
- 2.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–3. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 3.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 4.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–6. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 5.J. M. Conde L Fau - Vaquerizas, J. Vaquerizas Jm Fau - Santoyo, F. Santoyo J Fau - Al-Shahrour, S. Al-Shahrour F Fau - Ruiz-Llorente, M. Ruiz-Llorente S Fau - Robledo, J. Robledo M Fau - Dopazo and J. Dopazo, PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level.
- 6.Riva A, Kohane IS. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–5. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 7.S. G. Xu H Fau - Gregory, E. R. Gregory Sg Fau - Hauser, J. E. Hauser Er Fau -Stenger, M. A. Stenger Je Fau - Pericak-Vance, J. M. Pericak-Vance Ma Fau - Vance, S. Vance Jm Fau - Zuchner, M. A. Zuchner S Fau - Hauser and M. A. Hauser, SNPselector: a web tool for selecting SNPs for genetic association studies.
- 8.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Dai M, Xuan W, McEachin RC, Jackson AU, Scott LJ, Athey B, Watson SJ, Meng F. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–9. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 10.Orro A, Guffanti G, Salvi E, Macciardi F, Milanesi L. SNPLims: a data management system for genome wide association studies. BMC Bioinformatics. 2008;9(Suppl 2):S13. doi: 10.1186/1471-2105-9-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadag E, Louie B, Myler PJ, Tarczy-Hornoch P. Biomediator data integration and inference for functional annotation of anonymous sequences. Pac Symp Biocomput. 2007:343–54. [PubMed] [Google Scholar]
- 12.Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine. J Biomed Inform. 2007;40:5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Mork P, Halevy AY, Tarczy-Hornoch P. A Model for Data Integration Systems of BioMedical Data Applied to Online Genetic Databases; Proceedings of the American Medical Informatics Annual Fall Symposium; Washington, D.C. 2001. pp. 473–77. [PMC free article] [PubMed] [Google Scholar]
- 14.Louie B, Mork P, Shaker R, Kolker N, Kolker E, Tarczy-Hornoch P. Integration of Gene Annotation Data Using the BioMediator System (Poster). AMIA Fall Symposium 2005; Washington, D.C.. 2005. [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman-Hill E. Jess (Java Expert Systems Shell) Sandia National Laboratories; 2008. [Google Scholar]
- 16.Cadag E, Louie B, Myler P, Tarczy-Hornoch P. BioMediator Data Integration and Inference for Function Annotation of Anonymous Sequences. Pacific Symposium on Biocomputing; Maui, Hawaii. 2007. [PubMed] [Google Scholar]
- 17.Friedman-Hill E. Jess In Action: Rule-Based Systems in Java. Manning Publications Co.; CT: 2003. [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–8. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 21.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SeattleSNPs. NHLBI Program for Genomic Applications, SeattleSNPs. Seattle, WA: [Google Scholar]
- 23.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karolchik D, Kuhn R, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, WJ K. The UCSC Genome Browser Database: Genscan Gene Predictions. Nucleic Acids Res. 2008 Jan;:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karolchik D, Kuhn R, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, WJ K. The UCSC Genome Browser Database: GNF Expression Atlas 2. Nucleic Acids Res. 2008 Jan;:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karolchik D, Kuhn R, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, WJ K. The UCSC Genome Browser Database: Vertebrate Multiz Alignment & PhastCons Conservation. Nucleic Acids Res. 2008 Jan;:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–7. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 29.Musen M, Crubézy M, Fergerson R, Noy NF, Tu S, Vendetti J. Protégé-2000. Stanford Medical Informatics; Stanford, CA: [Google Scholar]
- 30.Shapiro A. TouchGraph: open source software for graph visualization using spring-layout and focus+context techniques. 2006 [Google Scholar]
- 31.Hill WG. Estimation of linkage disequilibrium in randomly mating populations. Heredity. 1974;33:229–39. doi: 10.1038/hdy.1974.89. [DOI] [PubMed] [Google Scholar]
- 32.Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–9. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Spitz MR, Amos CI, Lin J, Schabath MB, Wu X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004;64:2251–7. doi: 10.1158/0008-5472.can-03-2800. [DOI] [PubMed] [Google Scholar]
- 34.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–7. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 35.Bhatti P, Church DM, Rutter JL, Struewing JP, Sigurdson AJ. Candidate single nucleotide polymorphism selection using publicly available tools: a guide for epidemiologists. Am J Epidemiol. 2006;164:794–804. doi: 10.1093/aje/kwj269. [DOI] [PubMed] [Google Scholar]