Summary
The structural origin underlying differential nucleotide excision repair (NER) susceptibilities of bulky DNA lesions remains a challenging problem. As a model system, we investigate the 10S (+)-trans-anti-[BP]-N2-dG (G*) adduct in double-stranded DNA. This adduct arises from the reaction, in vitro and in vivo, of a major genotoxic metabolite of benzo[a]pyrene (BP), (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene, with the exocyclic amino group of guanine. The removal of this lesion by the NER apparatus in cell-free extracts has been found to depend on the base sequence context in which the lesion is embedded, providing an excellent opportunity for elucidating the properties of the damaged DNA duplexes that favor NER. While the BP ring system is in the B-DNA minor groove, 5′-directed along the modified strand, there are orientational distinctions that are sequence-dependent and are governed by flanking amino groups (Nucleic Acids Res. 35 (2007) 1555-68). To elucidate the sequence-governed NER susceptibility, we have conducted MD simulations for the 5′-…CG*GC…, 5′-…CGG*C…, and 5′-…TCG*CT… adduct-containing duplexes. We also investigated the 5′-…CG*IC… and 5′-…CIG*C…. sequences, which contain “I”, 2′-deoxyinosine with hydrogen replacing the amino group in 2′-deoxyguanosine, to further characterize the structural and dynamic roles of the flanking amino groups in the damaged duplexes. Our results pinpoint explicit roles for the amino groups in tandem GG sequences on the efficiency of NER, and suggest a hierarchy of destabilizing structural features that differentially facilitate NER of the BP lesion in the sequence contexts investigated. Furthermore, combinations of several locally destabilizing features in the hierarchy, consistent with a multipartite model, may provide a relatively strong recognition signal.
Keywords: benzo[a]pyrenyl-guanine lesion, GG mutation hotspot, guanine amino group, sequence-dependent conformational variability, nucleotide excision repair susceptibility
Introduction
The factors that invoke nucleotide excision repair (NER) of bulky DNA adducts have been of intense interest for a number of years.1–9 Various experimental studies have suggested the importance of distortions such as kinks in the damaged DNA,10 impaired Watson-Crick hydrogen bonding,4 flipped out nucleotides on the partner strand,9 enhanced local dynamics,11–14 and thermodynamic destabilization,5, 15 including a multipartite model involving combinations of various local thermodynamically destabilizing DNA distortions.14, 16 Bulky lesions derived from the binding of benzo[a]pyrene (BP) metabolites to DNA which do not cause thermal destabilization17 have been found to be resistant to repair by human NER in cell-free extract assays.15, 16, 18
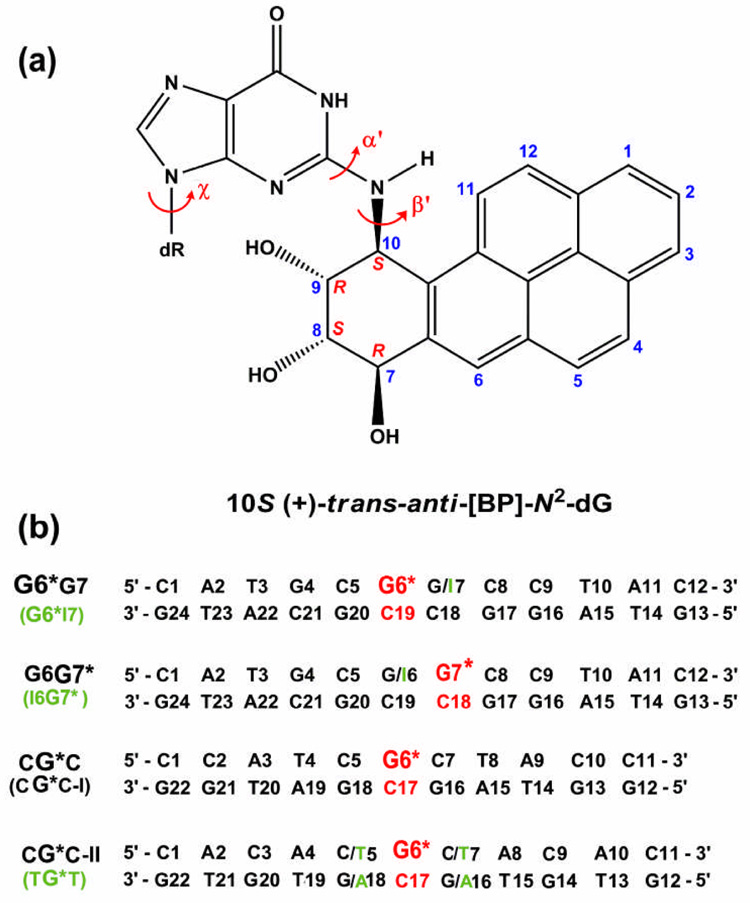
A set of DNA duplexes all containing the same BP-derived lesion but in different sequence contexts, and with differing susceptibilities to human NER, provide a fascinating opportunity to explore the structural features that produce observed differences in NER. These duplexes all contain the highly mutagenic major 10S (+)-trans-anti-[BP]-N2-dG adduct (Figure 1a) produced from the reaction of the strongly tumorigenic metabolite of BP, (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene ((+)-anti-BPDE),19 with the exocyclic amino group of guanine in DNA.20–22 These duplexes differ in sequence contexts surrounding the lesion: 5′-…GCG*GC… (G6*G7), 5′-…CGG*CC… (G6G7*), 5′-…TCG*CT… (CG*C-I), 5′-…ACG*CA… (CG*C-II), 5′-…ATG*TA… (TG*T) and 5′-…CIG*CC… (I6G7*), with asterisks designating the modified bases, and “I”, 2′-deoxyinosine (Figure 1b). Relative repair susceptibilities in a eukaryotic NER assay are in the ratio of 4.1 ± 0.3 (G6*G7): 2.4 ± 0.2 (TG*T): 1.7 ± 0.2 (G6G7*): 1.5 ± 0.2 (CG*C-II): 1.3 ± 0.2 (I6G7*):1.0 (CG*C-I).23 The GG tandem guanine sequence has been of unusual interest because it is a mutational hotspot in mouse embryonic NIH 3T3 cells,24 mammalian V-79 cells25 and the SupF gene of an E. coli plasmid.26, 27
Figure 1.
(a) Structure of the 10S (+)-trans-anti-[BP]-N2-dG adduct. The BP-DNA linkage site torsion angles α′ and β′ are defined as follows: α′, N1-C2-N2-C10(BP); β′, C2-N2-C10(BP)-C9(BP). χ is the glycosidic torsion angle, defined as O4′-C1′-N9-C4. (b) Sequence contexts, where G6* and G7* (red) represent the lesion modified guanines. The duplex G6*I7 is identical to G6*G7 except for the designated 2′-deoxyinosine (green). Similarly, the duplex I6G7* is identical to G6G7* except for the designated 2′-deoxyinosine (green. Note that the CG*C-II duplex differs in the next nearest neighbors and beyond from the CG*C-I sequence context of the current study. In the following text, “CG*C” refers to CG*C-I unless specified otherwise.
Structural studies by high resolution NMR methods have been carried out for the G6*G7, G6G7* and CG*C-I duplexes.28, 29 These reveal that the BP ring system in all cases is located in the minor groove, oriented towards the 5′-direction of the modified strand. However, subtle structural differences are observed in the three sequence contexts because of sequence-dependent steric competition between the bulky amino groups of the guanines flanking the modified G* and the BP residue, which is positioned in the minor groove in all cases.28, 29 Additionally, MD studies for the CG*C-II and TG*T sequence contexts (Figure 1b) indicated that the BP ring system also resides in the minor groove, 5′-directed along the modified strand but with more dynamic flexibility in the TG*T case.14, 30
Our goal in the present study is to determine how the 10S (+)-trans-anti-[BP]-N2-dG adduct impacts the dynamic structural properties of these damaged duplexes in a sequence-dependent manner; we aim to gain deeper understanding of the properties that favor NER by comparing our findings to the experimental NER results. We have employed 10.0 ns unrestrained molecular dynamics (MD) simulations to provide ensembles of structures for the G6*G7, G6G7* and CG*C-I duplexes; these ensembles, based on NMR solution structures, yielded new insights into the local dynamic properties of the duplexes. Our results delineated the roles of the exocyclic amino groups of guanines flanking the lesion on the duplex dynamic properties. We further substantiated these findings by replacing the flanking G’s by I’s (2′-deoxyinosine, with hydrogen replacing the amino group in 2′-deoxyguanosine) for the GG sequence contexts. Coupled with previous MD results for the CG*C-II and TG*T sequence contexts,14 our investigations suggest a hierarchy of destabilizing structural features that thermodynamically facilitate the recognition of the BP-modified duplexes by the NER apparatus for the sequences we considered. Dynamic, episodically denatured Watson-Crick hydrogen bonding at the base pair on the 5′-side of the lesion provides the strongest signal, followed by dynamic local untwisting accompanied by a large Roll/bend. Enlarged minor groove widths and modest perturbations of hydrogen bonding at and adjacent to the lesion site are common to all of the damaged duplexes, with only slight sequence-dependent dynamic differences, and are weaker recognition factors. Furthermore, various elements from the hierarchy can be combined in a multipartite model to produce a relatively strong NER recognition signal. We propose that the varied destabilizing features differentially facilitate the insertion of a β-hairpin from the human recognition factor XPC (the homolog of yeast Rad431) between the two strands of the BP-modified duplexes at or near the lesion site.
Results
In this study, we utilized the AMBER 8.0 simulation package32 to produce 10.0 ns MD ensembles for the 10S (+)-trans-anti-[BP]-N2-dG adduct in the G6*G7, G6G7*, and CG*C-I duplexes, as well as their unmodified control duplexes G6G7 and CGC-I. We used the high resolution NMR solution structures28, 29 as initial models, and generated unrestrained MD ensembles that preserved the NMR structural features for each lesion-containing duplex, yet provided fuller views of their dynamic nature. We also investigated the 2′-deoxyinosine sequences G6*I7 and I6G7* to further substantiate our understanding of the roles of the lesion-flanking amino groups.
Unrestrained MD Simulations Reproduce NMR-Derived Interproton Distances and Other Structural Features of the NMR Solution Structures
The unrestrained MD simulations provide structures consistent with the NMR experimental data.28, 29 To validate our MD simulation results, we computed key interproton distances that govern the positioning of the BP rings with respect to the DNA, and compared these computed distances to the experimentally observed ranges for the G6*G7, G6G7* and CG*C-I duplexes (Table S1, Supplementary Materials).28, 29 Specifically, for the 10S (+)-trans-anti-[BP]-N2-dG adduct in the G6*G7 sequence context, a total of 12 interproton NOE cross-peaks between BP rings and DNA were identified. Only two computed distances deviate from the experimental NOE-based distance ranges, and these by only 0.4 Å when the standard deviations in the NOE distances taken from the MD trajectory are included (Table S1a, Supplementary Materials). For the G6G7* case, a total of 14 intermolecular NOE cross-peaks between the BP rings and DNA were identified, and all computed values are within the experimentally observed NOE-based distance ranges (Table S1b, Supplementary Materials). For the CG*C duplex, 17 interproton NOEs were assigned with the greatest confidence (S. Broyde, unpublished data), and the computed and experimental values are in good agreement in all but one case (Table S1c, Supplementary Materials). Thus, our MD simulations reproduce the experimentally observed NOE distances reasonably well for all three cases. In addition, our MD simulations also reproduce, within standard deviations, the carcinogen-DNA linkage site and glycosidic torsion angles α′, β′, and χ of the NMR models (Table S2, Supplementary Materials).
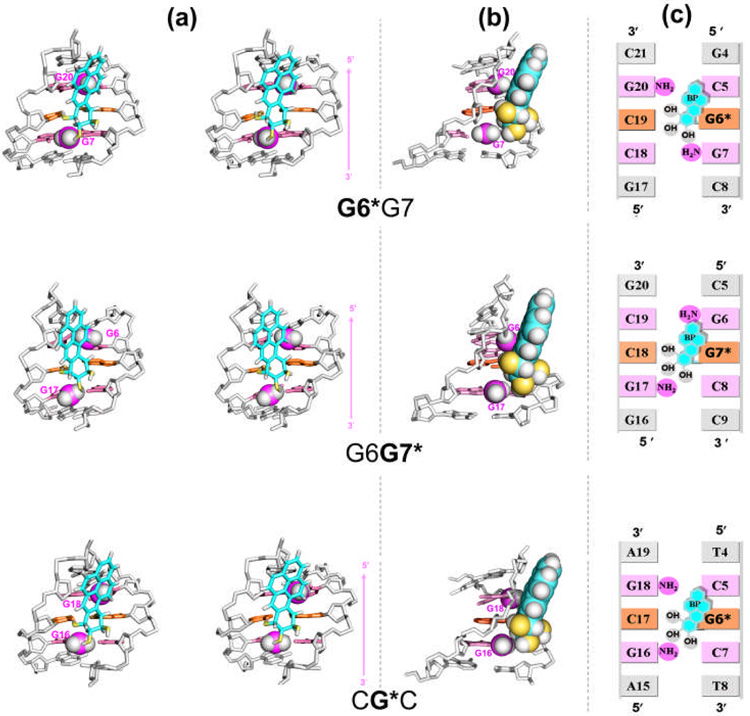
The best representative structures of the 10S (+)-trans-anti-[BP]-N2-dG adduct in the G6*G7, G6G7* and CG*C sequence contexts are shown in Figure 2a. The best representative structure33 is a real frame from the ensemble, and represents the most populated conformation, or the conformation that is the closest to all other snapshots in the ensemble, as determined by a cluster analysis. The views in Figure 2a are looking into the minor groove of each duplex, with the backbones of the duplexes aligned to optimally superimpose each other, so as to highlight the differential orientations of the BP moieties. In each case, the BP rings are 5′-directed along the modified strand in the minor groove of the duplex but with distinctions in the orientations, which are governed by the amino groups in the vicinity of the lesions (Figure 2b). Supplementary materials movie_1, Supplementary materials movie_2, and Supplementary materials movie_3 show rotating views of these structures. Particularly in the G6G7* case, the BP aromatic rings are restrained by the unmodified G6 exocyclic amino group from aligning within the contours of the minor groove, and the BP rings lean on the backbone of the complementary strand (Figure 2a). This positioning of the BP residue for the G6G7* duplex was also manifested by NMR observed interproton NOEs between H6 of the BP aromatic ring and the sugar of C18, the Watson-Crick partner of the modified G* in G6G7* (Table S1b, Supplementary Materials). However, analogous connectivities were not observed between BP and C19 in the G6*G7 duplex (Table S1a, Supplementary Materials).28 As a consequence of the more poorly aligned positioning of the BP ring system, the minor groove is wider at the lesion site in G6G7* than in G6*G7 (Figure S1 and Table S3, Supplementary Materials), also noted in the NMR solution models.28 On the other hand, G6*G7 and CG*C resemble each other in that the orientation of the BP moieties is aligned with the natural contours of the minor grooves, because the lesions are flanked by the same 5′-neighboring C:G base pairs in both duplexes (Figures 2b and 2c).
Figure 2.
(a) Orientational differences of BP rings in the G6*G7, G6G7* and CG*C sequence contexts in stereo. Views are looking into the minor grooves. Arrows on the right indicate the orientation of the modified strands. The structures shown are the best representative conformations33 for the last 7.0 ns of the MD simulations. Only the central 5-mers are shown. The color codes are as follows: for the BP moiety, the carbon atoms are cyan, oxygen atoms yellow and hydrogen atoms white; the modified guanine and its partner cytosine are orange. The guanine amino groups are violet (N) and white (H). The flanking base pairs are pink. The DNA duplexes are white. Hydrogen atoms and pendant phosphate oxygen atoms in the DNA duplexes are not displayed for clarity. The amino groups are in CPK representation. The backbones of the duplexes were aligned to be optimally superimposed in order to highlight the different orientations of the BP rings. Supplementary materials movie_1, Supplementary materials movie_2, and Supplementary materials movie_3 show rotating views of these structures (temporarily available on http://broyde.nyu.edu/~caiy/JMB_March_2008/). (b) A view highlighting the differential steric effects of guanine amino groups in the different sequence contexts. The BP moiety and relevant guanine amino groups are in CPK representation. (c) Schemes illustrating the relative positioning of the lesion and guanine amino groups in the different sequence contexts.
The differences in the positioning of the BP rings in the three sequence contexts are manifested by differences in the carcinogen–DNA linkage site torsion angles α′, β′ and the glycosidic torsion angle χ (Figure 1a). The torsion angle β′ in the G6*G7 and CG*C duplexes is somewhat more flexible, as shown by the broader population distribution and greater standard deviation from the mean value (Figures S2 and S3); this difference reflects a more rigid placement of the BP moiety in G6G7*, which is due to the steric hindrance arising from the 5′-flanking amino group of G6 (Figure 2b).
Hydrogen bonding between the BP hydroxyl groups and the adjacent bases/backbones, suggested as well in the NMR solution structures,28 also reflect the differences in the orientations of the BP aromatic ring system (Table S4, Supplementary Materials). These additional hydrogen bonding interactions further anchor the positions of the BP residue in each duplex.
The 3′-Flanking Amino Group of G7 Only in the G6*G7 Duplex Causes Dynamic Flexibility with Episodic Denaturation of Watson-Crick Hydrogen Bonding at the 5′-Neighboring Base Pair
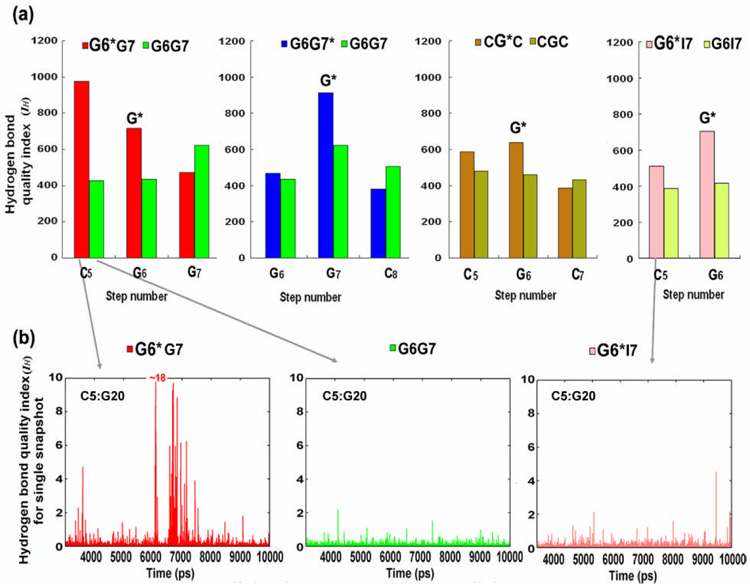
A pronounced dynamic disturbance of Watson-Crick hydrogen bonding is evident at the base pair 5′-flanking the lesion in the G6*G7 duplex. We use our hydrogen bond quality index IH (see Methods) to assess the impact of the 10S (+)-trans-anti-[BP]-N2-dG adduct on the quality of Watson-Crick hydrogen bonding in the G6*G7, G6G7* and CG*C duplexes. The lower the value of IH, the better the quality of the Watson-Crick hydrogen bonding. With ideal Watson-Crick hydrogen bonding, the IH value is zero. However, in real, even unmodified DNA, sequence-dependent deviations from ideal Watson-Crick hydrogen bonding are normal.34 Thus, the IH values in the unmodified control G6G7 and CGC duplexes provide an estimate of their Watson-Crick hydrogen bond quality.
In G6*G7, the unusually high IH value at C5:G20 (Figure 5) indicates that the base pair 5′ to the lesion is markedly disturbed. Furthermore, the time-dependence of the IH value of each snapshot for this C5:G20 base pair reveals that the disturbance is highly dynamic (Figure 5b); that is, the rate of change of IH with time is much greater for the G6*G7 duplex than for the unmodified control. This is unique to the G6*G7 duplex. A similar extent of perturbation is not observed in either the G6G7* or the CG*C case; the analogous positions, G6:C19 in the G6G7* duplex or C5:G18 in the CG*C duplex are only modestly disturbed, and yield IH values close to those of their respective unmodified control steps (Figure 5a, S4 and S5). However, at the lesion-containing base pair, all three sequences show more disturbed (Figure 5a) and dynamic Watson-Crick pairing than in their respective unmodified controls, particularly the N-H⋯O hydrogen bond on the minor groove side of the base pair (Figure S6b, Supplementary Materials, shows the Watson-Crick pairs with groove designations). The great disturbance of the base pair 5′ to the lesion in the G6*G7 duplex is in agreement with NMR experiments;28 these show that pronounced up-field shifts and broadening of the imino proton resonances occur at base pair C5:G20 only in the G6*G7 duplex, indicative of increased frequencies of duplex opening events.35, 36
Figure 5.
(a) Hydrogen bond quality indexes for the 10S (+)-trans-anti-[BP]-N2-dG adduct in the G6*G7(red), G6G7* (blue), CG*C (brown) and G6*I7 (pink) duplexes compared with their respective unmodified controls. Each lesion-modification site is designated with an asterisk. The sum of the hydrogen bond quality was taken over 7000 frames from the last 7.0 ns of the MD simulations. Only the C:G pairs at the central 3-mer or 2-mer are shown because A:T or I:C pairs, with only two hydrogen bonds, are not comparable to C:G pairs with three hydrogen bonds in terms of hydrogen bond quality index. (b) Time-dependence of the hydrogen bond quality index for each snapshot at the most disturbed step 5 (C5:G20) in G6*G7 (left), and its analogous step in the unmodified control G6G7 (middle) and the G6*I7 duplex (right).
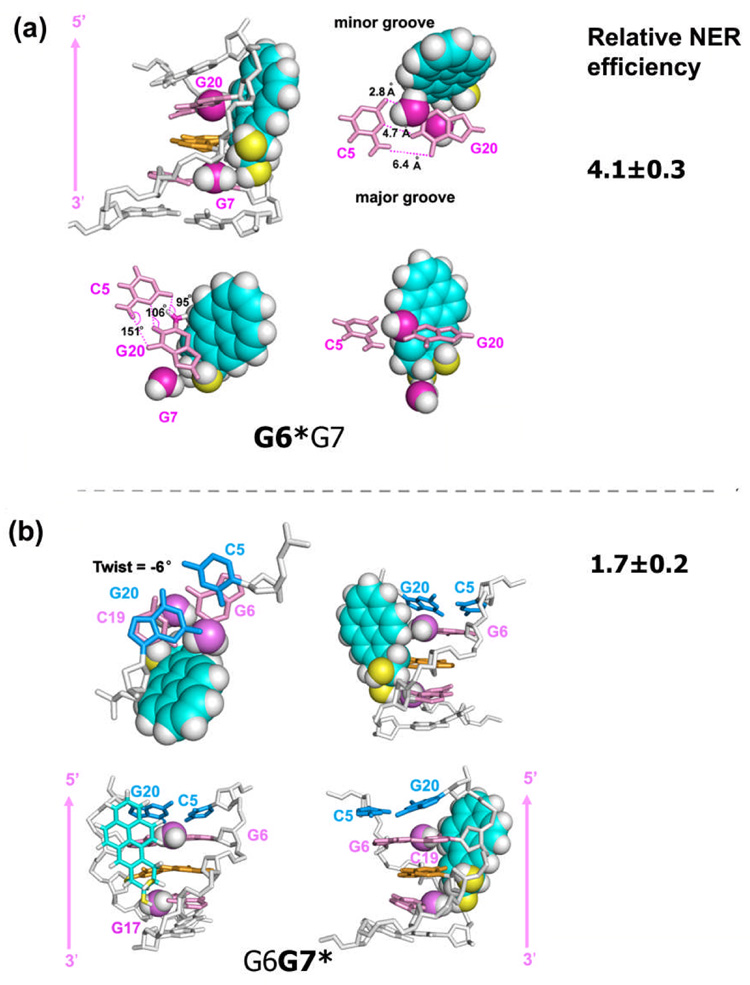
The highly dynamic nature of the 5′-flanking base pair in the G6*G7 duplex originates in the steric hindrance between the BP rings and the amino group flanking the lesion on the 3′-side (G7). During the dynamics simulations, this 3′-side flanking amino group, which is unique to the G6*G7 case, crowds the BP benzylic ring, forcing the distal BP rings into a position where they disturb the exocyclic amino group of G20, on the complementary strand 5′-side to the lesion; as a result, the distal BP rings intrude into the space normally occupied by the Watson-Crick base pair C5:G20 (Figure 3a). The time-dependence studies of the hydrogen bonding distances and angles for the C5:G20 pair (Figure S6, Supplementary Materials) suggest that all three hydrogen bonds are in fact transiently broken during the dynamics. Figure 3a shows details of the most disrupted structure taken from the G6*G7 ensemble. In this structure, the amino groups from both G7 and G20 contact the BP ring system: G20 contacts the distal aromatic rings on the 5′-side, and G7 contacts the benzylic ring on the 3′-side. The donor-acceptor atom distances in this Watson-Crick base pair are 2.8 Å (on the minor groove side), 4.7 Å(in the middle), and 6.4 Å (on the major groove side) (Figure S6, Supplementary Materials), while the hydrogen bond angles are 95°, 106°, and 151°, respectively (Figure 3a). Thus all three hydrogen bonds are essentially broken, based on commonly accepted minimal criteria for Watson-Crick pairs of donor-hydrogen-acceptor angles approximately larger than ~120° and donor-acceptor distances lower than ~ 3.4 Å. Such disruption occurs frequently during the 6,000–8,000 ps time frame as indicated by the time-dependence and population distribution of the hydrogen bond distances and angles (Figure S6, Supplementary Materials). By contrast, the G16 amino group in the CG*C duplex or the G17 in the G6G7* duplex, on the partner strand, does not crowd the BP as much as does G7 in G6*G7; hence, the 5′-flanking base pair C5:G18 for CG*C or G6:C19 for G6G7* is not significantly perturbed and there is no episodic denaturing effect as in the G6*G7 duplex case (Figure S7, Supplementary Materials).
Figure 3.
(a) Four views of the most denatured structure in G6*G7. Supplementary materials movie_4 shows the structure with the most denatured 5′-flanking Watson-Crick base pair (at 7319 ps) and the structure with least perturbed 5′-flanking neighbor (at 6103 ps). (b) Four views of the most untwisted structure in G6G7*. Supplementary materials movie_5 shows the most untwisted structure (at 4115 ps) and the most overtwisted structure (at 5241 ps). Frames were selected from the last 7.0 ns of the MD simulations for both cases. Pink arrows indicate the orientation of the modified strands. Relative NER efficiencies23 are with respect to the least efficiently repaired 10S (+)-trans-anti-[BP]-N2-dG adduct in the CG*C sequence context.
Simulations with the G6*I7 duplex further substantiate the role of the G7 amino group in the G6*G7 duplex. In the G6*I7 duplex study (Figure 1b) the lesion site and sequence context are the same as in the G6*G7 duplex, except that the G7 deoxynucleoside is replaced by 2′-deoxyinosine, I7. The absence of the G7 amino group in G6*I7 results in the disappearance of the episodic denaturing effect: with the 3′-flanking exocyclic amino group absent, the Watson-Crick pairing of the 5′-flanking base pair is normal (Figure 5) (Figure S6, Supplementary Materials).
The 5′-Flanking Amino Group of G6 in G6G7* Causes Dynamic Local Untwisting Concomitant with a Large Roll/Bend
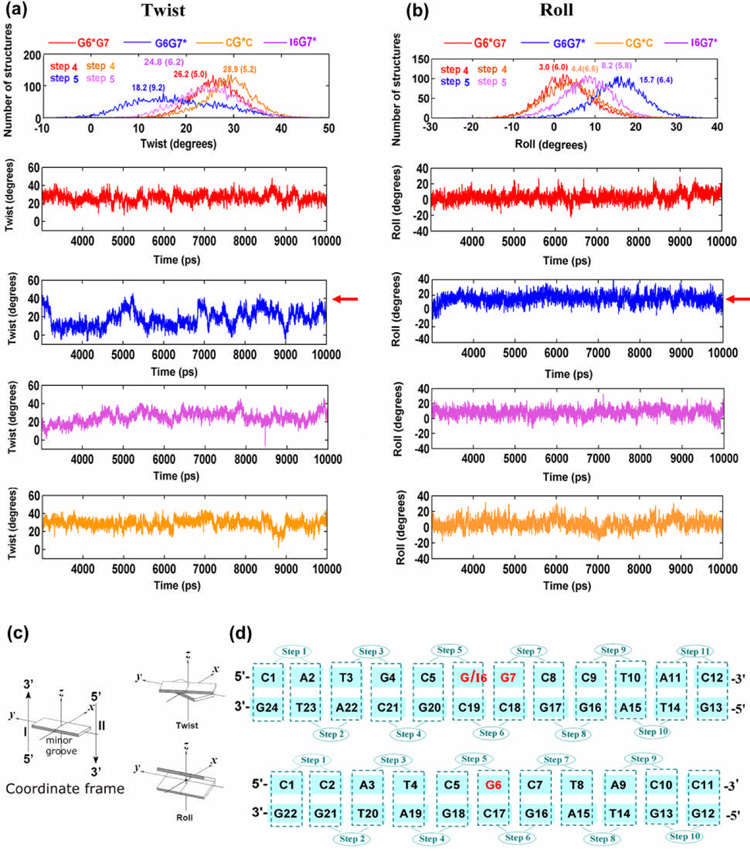
The twist angle is lower and fluctuates more rapidly at step 5 in the G6G7* duplex than in the analogous step 4 in the G6*G7 duplex; ensemble average values, with standard deviations are 18 ± 9° for G6G7* at step 5, and 26 ± 5° for G6*G7 at step 4 (Figure 6). Note that for Roll and Twist, G4:C21 to C5:G20 is step 4, and C5:G20 to G6:C19 is step 5 as shown in Figure 6d. The wider range in the population distribution plot of Twist, −6° to 45° for G6G7* versus 7° to 26° for G6*G7, as well as the higher standard deviation in the mean value for G6G7*, reflect the locally greater dynamic flexibility in the latter (Figure 6a). This dynamic untwisting at the G7*:C18 base pair in the G6G7* duplex is accompanied by a large local Roll/bend (Figure 6b); (Twist and Roll are anti-correlated and DNA bending occurs primarily via Roll.37–39) These structural features were also inferred from the NMR solution structure of the G6G7* duplex.28 In contrast, at the analogous base pair adjacent to the lesion, step 4 in the G6*G7 or the CG*C duplex, the Twist is less flexible and is accompanied by a smaller Roll angle, with both resembling the values and dynamics in the unmodified control duplexes. The population distributions of Twist and Roll angles for all the local base steps are shown in Figure S8, Supplementary Materials.
Figure 6.
Population distribution and time-dependence of duplex helicoidal parameter Twist (a) and Roll (b) at the untwisted step for G6G7* (blue) and at the analogous steps for I6G7* (magenta), G6*G7 (red) and CG*C (brown) duplexes. Ensemble averages and standard deviations (in parentheses) are given. Note that in the population distribution plots in (a) and (b), G6G7* (blue) is plotted against G6*G7 (red) and CG*C (brown) with one step shifted ahead, (e.g. step 5 in G6G7* vs. step 4 in G6*G7 and CG*C, as labeled), in order to permit comparison of the structural properties for the same position in relation to the lesion site. (c) Illustrations of the parameter definitions are adapted from Lu and Olson.57 (d) Numbering schemes for the G6G7/I6G7 and CGC sequence contexts. For the GG sequence contexts, G4:C21 to C5:G20 is step 4, C5:G20 to G6:C19 is step 5.
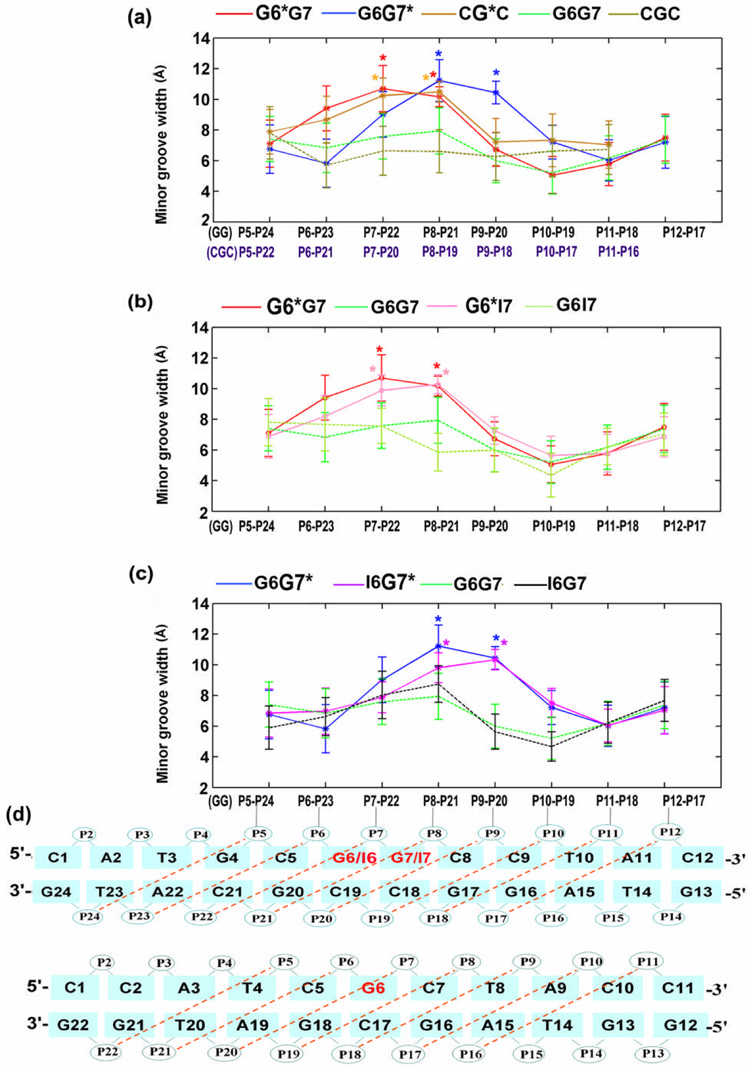
The cause of this unusual untwisting in the G6G7* duplex is primarily the 5′-flanking amino group of G6, and to a lesser extent, the 3′-side amino group of G17 (Figure 2b, discussed subsequently). The exocyclic amino group of G6 flanking the modified base G7* in the G6G7* duplex has the greatest impact on the orientation of the BP ring system and on the flexible local Roll/bend at the lesion site, among the sequence contexts of CG*C, G6*G7 and G6G7*. The G6 amino group prevents the BP ring system from entering the Watson-Crick base pairing territory of the G6:C19 base pair, and thus preserves the integrity of this base pair flanking the lesion site on the 5′-side. However, this occurs at the price of distorting the duplex and, as a result, the minor groove is enlarged locally at the lesion site (ensemble average values are: 11.2 ± 1.4 Å for G6G7* (P8–P21); 10.7 ± 1.5 Å for G6*G7 (P7–P22); and 10.2 ± 1.1 Å for CG*C (P7–P20) (Figure S1 and Table S3, Supplementary Materials)), the duplex unwinds locally, and Roll is increased. The BP rings lean upon the backbone of the complementary strand (Figure 2a), also manifested in the relatively larger and less flexible β′ (Figure S2, Supplementary Materials). Figure 3b shows details of the most untwisted structure. In this structure, the amino group of G6 almost touches the BP aromatic ring system which is in contact with the backbone of the complementary strand; this causes the duplex to untwist (Figure 3b and Figure S8) and the minor groove width at the lesion site to open to 13.1 Å.
These structural insights are further substantiated by studies on the duplex I6G7*, in which guanine G6 is replaced by I6. Replacing the G6 in G6G7* with an I6, without the amino group, abolishes the unusual untwisting of the G7*:C18 base pair in the G6G7* duplex (Figure 6a). Consequently, the population distribution of Twist at step 5 (Figures 6a and 6d) shifts toward higher values, closer to that of the G6*G7 case (Figure 6a), and the average Roll angle is also diminished (Figure 6b). This is reflected by the ensemble average values; the values with standard deviations for Twist at step 5 are 18.2 ± 9.2°, 24.8 ± 6.2° and 26.2 ± 5.0°, for G6G7*, I6G7*, and G6*G7, respectively; for Roll, the values are 3.0 ± 6.0°, 8.2 ± 5.8° and 15.7 ± 6.4°, respectively (Figure 6b) (A full analysis of the helicoidal parameters is provided in Table S5 and Figure S9, Supplementary Materials). However, distinctions remain between I6G7* and G6*G7 in both Twist and Roll, particularly in Roll (Figure 6b). This suggests a secondary role for the amino group of G17 on the 3′-side in determining these properties: specifically, the BP benzylic ring must position itself to avoid collision with the G17 amino group (Figure 3b); consequently, the distal BP aromatic rings become oriented to disturb the C5:G20 and G6:C19 base pairs (step 5), which somewhat impacts the local Twist and Roll.
The untwisting and enlarged Roll/bend is correlated with diminished van der Waals interactions between adjacent base pairs near the lesion site. These interactions are reduced more, relative to the unmodified sequence, in the G6G7* than in the G6*G7 duplex (Table S5, Supplementary Materials). Concomitant with the flexible untwisting at step 5 in the G6G7* duplex, the van der Waals stacking interactions between base pairs C5:G20 and G6:C19 are diminished by 0.8 kcal·mol−1, from −15.0 ± 1.1 kcal·mol−1 to −14.2 ± 1.3 kcal·mol−1, and this step is dynamically most flexible as shown by the largest standard deviation (1.3 kcal·mol−1) (Table S6, Supplementary Materials). The next two steps, (G6:C19)/(G7*:C18) and (G7*:C18)/(C8:G17), also feature reductions in these stacking interaction energies by 1.0, and 0.9 kcal·mol−1, respectively (Table S6, Supplementary Materials). In contrast, the greatest relative destabilization in the G6*G7 duplex is 0.3 kcal·mol−1 ((G7:C18)/(C8:G17)) (Table S6, Supplementary Materials). The sum of these interactions within the central seven-base pair sequence is reduced by 2.7 kcal·mol−1 in G6G7*, 0.4 kcal·mol−1 in G6*G7, and 0.6 kcal·mol−1 in CG*C (Table S6, Supplementary Materials).
The Minor Grooves Are Widened on the 5′-Side of the Lesion in All Cases
To accommodate the bulky BP lesion, 5′-directed in the minor groove, the minor groove at the lesion site for each damaged duplex is enlarged: the greatest effect is when there is a 5′-lesion-flanking guanine exocyclic amino group present on the same strand as the lesion (Figure 4 and Figure S1) Thus, in the G6G7* duplex, the minor groove is enlarged most at the lesion (P8–P21), because of steric hindrance from the 5′-flanking amino group of G6 on the same strand (Figure 4a) (Table S3, Supplementary Materials). Table 2 summarizes minor groove width differences between sequence contexts near the lesion site and their underlying steric origins. However, dynamic analyses show that the mobilities of the grooves differ little in the G6*G7, G6*I7, G6G7*, I6G7* and CG*C duplexes (Figure S1, Supplementary Materials).
Figure 4.
(a), (b) and (c), computed trajectory average minor groove widths (Å) of the G6*G7, G6G7*, CG*C, G6*I7 and I6G7* and their respective unmodified duplexes. (d) minor groove widths for GG sequences are the distance between P5 and P24, P6 and P23, P7 and P22 and so on; for CGC sequences, they are between P5 and P22, P6 and P21, P7 and P20 and so on. For each computed minor groove width, 5.8 Å is deducted to account for the van der Waals radius of the P atoms.54 Lesion-modification sites are designated with asterisks. Error bars indicate standard deviations.
Table 2.
| Sequence contexts | Minor groove width difference at the lesion site b | Guanine amino group providing responsible for enlargement | |
|---|---|---|---|
| G6G7* | P8–P21 | ~0.5 Å | G6 |
| G6*G7 | P7–P22 | 0 | |
| G6*G7 | P7–P22 | ~0.5 Å | G7 |
| CG*C | P7–P20 | 0 | |
| G6G7* | P8–P21 | ~1.4 Å | G6 |
| I6G7* | P8–P21 | 0 | |
| G6*G7 | P7–P22 | ~0.8 Å | G7 |
| G6*I7 | P7–P22 | 0 | |
See Figure 4 for definitions of minor groove widths.
For each comparison, the sequence context with the smaller minor groove width is assigned a value of 0.
The G6*G7 Full Duplex Is Slightly Less Stable by Thermodynamic Analyses
Thermodynamic analyses based on the MD ensembles indicate that the G6*G7 duplex is very modestly less stable on energetic grounds than the G6G7* duplex. We utilize the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) methodology40 in the AMBER 8.0 simulation package32 to carry out thermodynamic analyses, employing the ensemble of structures derived from the last 7.0 ns of the MD simulations. The relative free energies and enthalpies of the two sequence contexts were computed to estimate the relative stabilities of the G6*G7 and G6G7* duplexes. These two duplexes contain the same 10S (+)-trans-anti-[BP]-N2-dG adduct and the same base pair sequences, but different modification sites, and thus are comparable in terms of free energy using the MM-PBSA method. However, the CG*C case could not be included in this analysis due to the sequence difference. Our calculated results indicate that the G6*G7 duplex is less stable than the G6G7* duplex (Table 1) by 0.9 ± 2.9 kcal·mol−1 in total free energy, and by 1.2 ± 1.0 kcal·mol−1 in enthalpy; the contribution from entropy is less certain than the enthalpy.40 Full thermodynamic analyses are provided in Table S7, Supplementary Materials. The computed results showing very modest differences, are, however, consistent with the thermal melting studies of these two duplexes, which showed the same trend, with Tm values of 55 ± 1 °C for the G6*G7 duplex, and 57 ± 1 °C for the G6G7* duplex.28 However, local thermodynamic destabilization, in the lesion vicinity, is much greater for the G6*G7 duplex, as discussed below.
Table 1.
Relative Enthalpies and Free Energies for the G6*G7 and G6G7* Duplexes a
| ΔH (kcal·mol−1) | ΔG (kcal·mol−1) | ||
|---|---|---|---|
| G6*G7 | G6G7* | G6*G7 | G6G7* |
| 1.2 (1.0) | 0 | 0.9 (2.9) | 0 |
For each sequence context, the conformation with the lower enthalpy or free energy is assigned a ΔH or ΔG of 0, respectively. See Table S7, Supplementary Material;, for full MM-PBSA analyses over the 3.0 – 10.0 ns windows.
Discussion
Differences in steric hindrance from exocyclic amino groups of guanines flanking the lesion on either side explain distinct sequence-dependent structural characteristics and dynamic phenomena
In the G6*G7 duplex the amino group of the 3′-flanking G7 leads to episodic unpairing of the 5′-flanking C5:G20 base pair, while in the G6G7* duplex the amino group of the 5′-flanking G6 causes unusual flexible, local untwisting concomitant with an increased local Roll/bend. The importance of the guanine amino group is substantiated by studies in which 2′-deoxyinosine replaces the 2′-deoxyguanosine adjacent to the lesion; this relieves the steric hindrance in the G6*G7 and G6G7* duplexes by replacing the guanine amino groups with hydrogen atoms. In the 2′-deoxyinosine-containing duplex G6*I7, the denaturation effect at the 5′-flanking base pair is eliminated, while in the I6G7* duplex the untwisting is significantly diminished. In the latter case, the amino group on G17 in the complementary strand also plays a role in the untwisting effect. The highly dynamic local untwisting concomitant with a greater local Roll/bend, observed only in the G6G7* duplex, is a plausible explanation on a molecular level to the experimental observation that the same 10S (+)-trans-anti-[BP]-N2-dG adduct in a 5′-…GG*… sequence context exhibits anomalously slow electrophoretic mobility41 and unusually high flexibility,42 while the 5′-…G*G… and CG*C duplexes behave normally.41 Furthermore, the observation that abolishing the G6 amino group abolishes the flexible untwisting and enlarged Roll/bend is also consistent with the electrophoretic mobility study showing that replacing the G 5′ to the lesion with I in a 5′…GG*… sequence context abolishes the unusually slow electrophoretic mobilities.42
Sequence-dependent structural and dynamic differences produce differential NER excision efficiencies
The structural factors that provoke the NER response have been intensely studied.1–9, 16 It is now widely believed that the first and rate-limiting step is the recognition of the bulky lesions by the XPC-hHR23B protein heterodimer complex.1, 43 Within a similar family of bulky polycyclic aromatic hydrocarbon lesions, the human NER response has been correlated with the extent of thermodynamic destabilization of double-stranded DNA associated with the lesions in certain cases; a multipartite model of NER recognition was proposed based on observations that bulky lesions disturb a variety of DNA structural parameters that are detected by the NER recognition machinery in a manner that depends on base sequence context as well as the nature of the bulky lesion.16 Mocquet et al reported that XPC-HR23B distinguishes the stereoisomeric 10S (+)-trans, 10R (−)-trans, and 10R (+)-cis-anti-[BP]-N2-dG lesions by differential strand opening facilitated by local weakening of Watson-Crick hydrogen bonding and base pair stacking interactions, and minor groove widening.44 Our previous studies on the same 10S (+)-trans-anti-[BP]-N2-dG lesion in the CG*C-II and TG*T (Figure 1b) sequence contexts14 suggested an important role for enhanced dynamics in the lesion-recognition process based on experimental results with the prokaryotic UvrABC system.45 Furthermore, the relative repair efficiencies determined for this prokaryotic system45 follow the same sequence-dependent trends as in the mammalian system reported here (about 2 fold higher efficiency for TG*T).
A recent crystal structure of the yeast XPC orthologue Rad4 bound to DNA containing a cyclobutane pyrimidine dimer lesion shows that Rad4 inserts a β-hairpin between the DNA strands and expels two mismatched thymine nucleotides out of the duplex on the unmodified strand to interact with protein residues.31 Broad initial substrate specificity in damage recognition may be gained by Rad4 or XPC binding to the undamaged strand.9, 31 The structure of this Rad4-damaged DNA complex is in accord with the concept that lesions which thermodynamically destabilize the Watson-Crick double helix and facilitate the flipping-out of base pairs and intrusion of the β-hairpin are likely to be detected more efficiently.31 The underlying hypothesis is that, in combination, as in the multipartite model,16 the various distortions in a specific lesion/sequence context cause the needed local destabilization to allow β-hairpin insertion. Moreover, while local thermodynamic destabilization appears to be an important factor for human NER, this factor alone may not be sufficient, an issue requiring further study.
However, the relative importance of the various types of structural distortions in governing the recognition and relative incision efficiencies remains unclear and uncharted. Our series of studies involving the same lesion in various sequence contexts here and in earlier work14 suggest a hierarchy of structural and dynamic features that help us understand the relative NER excision efficiencies for these cases. These are summarized in Table 3. We note that these differences are modest and comparable among the less well excised sequences. The following sequence contexts with the same minor groove-aligned 10S (+)-trans-anti-[BP]-N2-dG adduct have been investigated by us so far in the present and prior work:14, 45 G6*G7, G6*I7, G6G7*, CG*C-I, CG*C-II and TG*T. It should be noted that the previously investigated TG*T sequence14, 45 is the same as CG*C-II except for the bases immediately flanking G* (Figure 1b). Their distinct structural and dynamic properties and differential NER susceptibilities of all sequences investigated are summarized in Table 3.
Table 3.
| Lesion-containing duplexes | Local dynamic structural destabilization | Relative NER dual incision efficiency | ||||
|---|---|---|---|---|---|---|
| W-C base pairing 5′-to the lesion | Twist and Roll/bend | Minor groove width | W-C base pairing at the lesion | Local base stacking interaction | ||
| G6*G7 | Dynamic flexibility and episodic denaturation for all three HBs | Normal | Enlarged | Modestly impaired | Normal | 4.1±0.3 |
| TG*T | Dynamic flexibility and episodic denaturation for only one HB | Dynamically flexible Twist and Roll/flexible bend | Enlarged and dynamically flexible | Modestly impaired | Impaired | 2.4±0.2 |
| G6G7* | Slightly impaired | Dynamic untwisting and enlarged Roll/flexible bend | Enlarged | Modestly impaired | Impaired | 1.7±0.2 |
| I6G7* | Dynamic flexibility and episodic denaturation for only one HB | Normal | Enlarged | Modestly impaired | Impaired | 1.3±0.2 |
| CG*C | Slightly impaired | Modestly enlarged Roll/rigid bend | Enlarged | Modestly impaired | Normal | CG*C-II 1.5±0.2 |
| CG*C-I 1.0 | ||||||
Relative NER efficiency1 is with respect to the least efficiently repaired 10S (+)-trans-anti-[BP]-N2-dG adduct in the CG*C sequence context
Structural analyses for the CG*C-II, and TG*T duplexes are from our prior work2
Experimental data for TG*T and CG*C-II are from Kropachev, K. & Geacintov, N. E. et al., manuscript in preparation.
Specifically, the dynamic local destabilization in Watson-Crick pairing at C5:G20, flanking the lesion from the 5′-side in G6*G7 (Figure 5), with all three hydrogen bonds episodically ruptured, appears to provide a relatively strong NER recognition signal. This G6*G7 duplex is processed by NER assays with the greatest efficiency of the six cases investigated experimentally. While the thermal melting temperatures of G6*G7 and G6G7* differ only slightly, with G6*G7 minimally less stable, consistent with our thermodynamic analyses here, NMR studies have revealed more significant local destabilization for the G6*G7 duplex. Particularly, the G6*G7 duplex manifests significant line-width broadening for the imino proton resonances of G6*, G7 and G20,28 indicative of increased frequencies of local duplex opening events;35, 36 it also features pronounced temperature-dependent up-field chemical shifts at 20–30 °C for the imino proton of G20 and G6*, and to a lesser extent, G7, suggesting local pre-melting in the immediate vicinity of the lesion at a much lower temperature than the global melting point (55 ± 1 °C).28 By contrast, the G6G7* duplex lacks these NMR properties, indicating that the local base pairs are not destabilized at 20–30 °C.28
The TG*T and I6G7* duplexes share with G6*G7 the feature of dynamically denatured Watson-Crick pairing on the 5′-side of the lesion. However, there are distinctions in the degree of destabilization; while all three hydrogen bonds are episodically denatured in the G6*G7 case (Figure S10, Supplementary Materials), providing a stronger recognition signal detected by the NER assays, only one hydrogen bond is episodically disrupted in the TG*T and I6G7* duplexes (Figure S10, Supplementary Materials). Specifically, the “breathing” amplitudes of the 5′-flanking T5:A18 pair in TG*T and the I6:C19 pair in I6G7* are less than in the C5:G20 pair of the G6*G7 duplex (Figure S10, Supplementary Materials). Only one hydrogen bond is episodically totally ruptured (23% of the population) in the T5:A18 base pair14, namely, the (A18) N6-H6⋯O4 (T5) bond, which is on the major groove side of the T:A pair. The NMR study of the TG*T duplex revealed a loss of the imino proton resonance signals in the central 5′-d(…ATG*…)·d(…CAT…) region,30 which is associated with rapid base pair opening rates.35, 36 Similarly, only one hydrogen bond, namely, (C19) N4-H4⋯O6 (I6), at base pair I6:C19, 5′-flanking the lesion in the I6G7* duplex is ruptured in 21% of the population during the MD simulations (Figure S6, Supplementary Materials).
Dynamic untwisting accompanied by a large Roll/bend, which is unique to the G6G7* duplex (Figure 6), appears to provide a lesser recognition signal than hydrogen bond disruption. The TG*T sequence also features dynamic flexibility in Twist and Roll, but without untwisting and enlargement in Roll; the enhanced dynamics in Twist and Roll in the TG*T case could contribute to distinguishing its repair efficiency from that of the I6G7* case, which lacks this feature (Table 3).
Dynamically mobile minor groove widths and diminished base stacking appear to be a third contributing factor to the recognition hierarchies. The TG*T sequence has unique dynamically flexible minor groove widths; this stems from the intrinsically weaker 5′-flanking base pair T5:A18 with only two hydrogen bonds, weaker T/G stacking, and absence of guanine amino groups in the minor groove.14 The TG*T duplex also features impaired base stacking, which is exhibited in the G6G7* and I6G7* sequences as well. The importance of impaired base stacking in lesion recognition was emphasized by Yang in a recent review.46
It is worth noting that the TG*T duplex manifests various elements in the hierarchy of dynamically destabilizing structural features, as in the multipartite model:16 one episodically denatured Watson-Crick hydrogen bond flanking the lesion, dynamically flexible Twist and Roll/bend, dynamic mobility of the minor groove, and impaired stacking interactions. In combination, these destabilizing features can explain the relatively high NER efficiencies in the TG*T case (Table 3).
However, when there is just an enlarged minor groove44, modestly enlarged Roll/rigid bend,47 and only modestly distorted Watson-Crick hydrogen bonding at the lesion and on its 5′-side (Figure 5), NER efficiencies are less. The relative NER efficiencies in the CG*C cases are modest (Table 3); these appear to rely only on the modestly disturbed Watson-Crick hydrogen bonds at and adjacent to the lesion and the widened minor groove. Repair differences between CG*C-I and CG*C-II may reflect next-nearest-neighbor effects on structure and merit further study.
Conclusions
We have employed MD simulations to uncover dynamic structural correlations with repair susceptibilities for the 10S (+)-trans-anti-[BP]-N2-dG adduct in several sequence contexts. By utilizing the identical minor groove-aligned lesion in a number of sequence contexts that exhibit varying susceptibilities to removal by the human NER system in cell-free extracts, we have proposed hierarchies of structural signals for invoking NER with differential efficiencies in these sequence contexts. We find that dynamic episodic denaturation of Watson-Crick base pairing 5′-flanking the lesion correlates with greatest susceptibility, and therefore appears to provide the relatively strongest recognition signal in these sequences. NMR data showing pronounced local thermodynamic destabilization is consistent with this concept.28 Dynamic flexible untwisting concomitant with a significantly enlarged Roll/bend at the lesion site provides a lesser contributing factor. Diminished base stacking interactions and flexibility in the minor groove can contribute additional destabilizing factors to NER recognition. The modest perturbation of Watson-Crick hydrogen bonding at and on the 5′-side of the lesion, and enlarged, but not unusually dynamic minor grooves, observed in all the sequence contexts considered here, apparently offer only moderate recognition signals. While significant episodic hydrogen bond denaturation and untwisting themselves seem more prominent NER signals, a multipartite model16 combining various destabilizing features, which may be correlated, may also produce a relatively strong recognition signal, facilitating β-hairpin insertion by the XPC recognition factor.31 However, much future work remains in order to gain thorough understanding of the hallmarks for NER recognition that successfully initiate the complex processing of bulky lesions by the human NER machinery. Our library of structurally characterized bulky lesions in different sequence contexts offers opportunities to approach this challenging problem in a systematic manner.
Methods
Starting Structures
The starting models were the minor groove conformations of the NMR solution structures for the 10S (+)-trans-anti-[BP]-N2-dG adduct in the G6*G7, G6G7*28 and CG*C sequence contexts.29 The starting models for the unmodified DNA duplexes were built using standard B-DNA in INSIGHTII 2005 (Accelrys, Inc.), and were subjected to energy minimization in SANDER in the AMBER 8.0 simulation package.32 These initial models are shown in Figure S11, Supplementary Materials.
Force Field
MD simulations were carried out employing SANDER in the AMBER 8.0 simulation package,32 with the Cornell et al. force field,48 with parm99.dat parameter set.49 Additional parameters for the 10S (+)-trans-anti-[BP]-N2-dG adduct were the same as detailed previously.50
MD Computation Protocols
Details of the MD protocols are given in Supplementary Materials. The stability of the molecular dynamics simulation was evaluated for each model. For each sequence context, the root mean square deviation (RMSD) of each snapshot in the trajectory relative to its respective NMR solution starting structure was plotted as a function of time and is shown in Figure S12, Supplementary Materials. The average RMSD of all atoms, excluding two base pairs at each end, in the current structure against the initial model for the 10.0 ns simulation is given in Figure S12, Supplementary Materials. For all cases, MD achieved good stability, fluctuating around the mean after 3.0 ns, and we have employed the structural ensembles from the 3.0–10.0 ns time frame for further analyses.
Free Energy Analyses
We utilize the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA)40 method in the AMBER 8.0 simulation package32 to perform the thermodynamics analyses. Details of this method and the protocols are available in Supplementary Materials.
Structural Analyses
The PTRAJ module of the AMBER 8.0 package32 and the CARNAL module of the AMBER7.0 package51 were employed for structural analyses. Frames were selected at 1 ps intervals from the last 7.0 ns of simulation. DNA duplex groove dimensions were analyzed using MD Toolchest 2.052, 53 and 5.8 Å was subtracted from the pairwise phosphorous-phosphorous distances to account for the van der Waals radii of the P atoms.54 To estimate stacking interactions, we computed the van der Waals interaction energies between all adjacent base pairs. The ANAL module of the AMBER 8.0 package32 was employed. INSIGHTII 2005 was employed for visualization and model building. PyMOL (Delano Scientific, LLC.)55 was utilized to make molecular images and movies. Computations were carried out on our own cluster of Silicon Graphics Origin and Altix high-performance computers.
We employed our hydrogen bond quality index,56 to quantitatively assess the quality of Watson-Crick hydrogen bonding, in terms of the deviation from ideal Watson-Crick hydrogen bond distances and angles:
where dDA is the instantaneous donor-acceptor distance, is an ideal donor-acceptor distance (Saenger, 1984) [O6 (G) to N4 (C) is 2.91 Å N1 (G) to N3 (C) is 2.95 Å and N2 (G) to O2 (C) is 2.86 Å] and γ is the instantaneous donor-hydrogen⋯acceptor (D-H⋯A) hydrogen bond angle with an ideal value of 180°. The summation is over all the Watson-Crick hydrogen bonds in a base pair over the trajectory. IH is zero when the Watson-Crick hydrogen bonding is ideal during the dynamics.
Supplementary Material
Acknowledgements
This work was supported by the National Cancer Institute, National Institutes of Health, through grants CA28038 (S.B.), CA099194 (N.E.G.), and CA046533 (D.J.P). Partial support for computational infrastructure and systems management was also provided by grant CA75449 (S.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations
- BP
benzo[a]pyrene
- (+)-anti-BPDE (benzo[a]pyrene diol epoxide)
(+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- dG
2′-deoxyguanosine
- NER
nucleotide excision repair
- MD
molecular dynamics
- NOE
Nuclear Overhauser Effect
- RMSD
root mean square deviation
Footnotes
Kropachev, K., Kolbanovskii, M., Cai, Y., Rodríguez, F., Kolbanovskii, A., Liu, Y., Zhang, L., Amin, S., Patel, D., Broyde, S. & Geacintov, N. E. (2008). The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived lesions: Insights into the structural factors that favor dual incisions. Submitted.
Cai, Y., Patel, D. J., Geacintov, N. E. & Broyde, S. (2007). Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J. Mol. Biol. 374, 292-305.
References
- 1.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 2.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 3.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair: Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 6.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 7.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair. 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blagoev KB, Alexandrov BS, Goodwin EH, Bishop AR. Ultra-violet light induced changes in DNA dynamics may enhance TT-dimer recognition. DNA Repair (Amst) 2006;5:863–867. doi: 10.1016/j.dnarep.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst) 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Maillard O, Camenisch U, Clement FC, Blagoev KB, Naegeli H. DNA repair triggered by sensors of helical dynamics. Trends Biochem. Sci. 2007;32:494–499. doi: 10.1016/j.tibs.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J. Mol. Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geacintov NE, Naegeli H, Patel DJ, Broyde S. In: Structural aspects of polycyclic aromatic carcinogen-damaged DNA and its recognition by NER proteins. Siede W, Kow Y, Doetsch P, editors. New York: Talyor and Francis; 2005. pp. 263–296. [Google Scholar]
- 16.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 17.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N(6)-adenine adducts: unusual thermal stabilization of modified DNA duplexes. Chem. Res. Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 18.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 19.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 20.Koreeda M, Moore PD, Wislocki PG, Levin W, Yagi H, Jerina DM. Binding of benzo[a]pyrene 7,8-diol-9,10-epoxides to DNA, RNA, and protein of mouse skin occurs with high stereoselectivity. Science. 1978;199:778–781. doi: 10.1126/science.622566. [DOI] [PubMed] [Google Scholar]
- 21.Straub KM, Meehan T, Burlingame AL, Calvin M. Identification of the major adducts formed by reaction of benzo[a]pyrene diol epoxide with DNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5285–5289. doi: 10.1073/pnas.74.12.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstein IB, Jeffrey AM, Jennette KW, Blobstein SH, Harvey RG, Harris C, Autrup H, Kasai H, Nakanishi K. Benzo[a]pyrene diol epoxides as intermediates in nucleic acid binding in vitro and in vivo. Science. 1976;193:592–595. doi: 10.1126/science.959820. [DOI] [PubMed] [Google Scholar]
- 23.Kropachev K, Kolbanovskii M, Cai Y, Rodríguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived lesions: Insights into the structural factors that favor dual incisions. 2008 doi: 10.1016/j.jmb.2008.12.082. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vousden KH, Bos JL, Marshall CJ, Phillips DH. Mutations activating human c-Ha-ras1 protooncogene (HRAS1) induced by chemical carcinogens and depurination. Proc. Natl. Acad. Sci. USA. 1986;83:1222–1226. doi: 10.1073/pnas.83.5.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei SJC, Chang RL, Hennig E, Cui XX, Merkler KA, Wong CQ, Yagi H, Jerina DM, Conney AH. Mutagenic selectivity at the Hprt Locus in V-79 cells -Comparison of mutations caused by bay-region benzo[a]pyrene 7,8-diol-9,10-epoxide enantiomers with high and low carcinogenic activity. Carcinogenesis. 1994;15:1729–1735. doi: 10.1093/carcin/15.8.1729. [DOI] [PubMed] [Google Scholar]
- 26.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem Res Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez H, Loechler EL. Mutational specificity of the (+)-anti-diol epoxide of benzo[a]pyrene in a supF gene of an Escherichia coli plasmid: DNA sequence context influences hotspots, mutagenic specificity and the extent of SOS enhancement of mutagenesis. Carcinogenesis. 1993;14:373–383. doi: 10.1093/carcin/14.3.373. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez FA, Cai Y, Lin C, Tang Y, Kolbanovskiy A, Amin S, Patel DJ, Broyde S, Geacintov NE. Exocyclic amino groups of flanking guanines govern sequence-dependent adduct conformations and local structural distortions for minor groove-aligned benzo[a]pyrenyl-guanine lesions in a GG mutation hotspot context. Nucleic Acids Res. 2007;35:1555–1568. doi: 10.1093/nar/gkm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R, Mao B, Amin S, Geacintov NE. Bending and circularization of site-specific and stereoisomeric carcinogen-DNA adducts. Biochemistry. 1998;37:769–778. doi: 10.1021/bi971785x. [DOI] [PubMed] [Google Scholar]
- 31.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 32.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Kollman PA. AMBER 8. San Francisco, CA: University of California; 2004. [Google Scholar]
- 33.Simmerling C, Elber R, Zhang J. In: MOIL-View -A program for visualization of structure and dynamics of biomolecules and STO- a program for computing stochastic paths, in modeling of biomolecular structure and mechanisms. Pullman, editor. Netherlands: Kluwer; 1995. [Google Scholar]
- 34.Calladine CR. Mechanics of sequence-dependent stacking of bases in B-DNA. J. Mol. Biol. 1982;161:343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- 35.Gueron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 36.Patel DJ, Pardi A, Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982;216:581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- 37.Dickerson RE. DNA bending: The prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 39.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 41.Liu T, Xu J, Tsao H, Li B, Xu R, Yang C, Amin S, Moriya M, Geacintov NE. Base sequence-dependent bends in site-specific benzo[a]pyrene diol epoxide-modified oligonucleotide duplexes. Chem. Res. Toxicol. 1996;9:255–261. doi: 10.1021/tx9501086. [DOI] [PubMed] [Google Scholar]
- 42.Xu J. Ph.D. Thesis. New York University; 1999. Sequence dependence of carcinogen-induced DNA bending. [Google Scholar]
- 43.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 44.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, Van Houten B, Geacintov NE. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 46.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 47.Xu R, Mao B, Xu J, Li B, Birke S, Swenberg CE, Geacintov NE. Stereochemistry-dependent bending in oligonucleotide duplexes induced by site-specific covalent benzo[a]pyrene diol epoxide-guanine lesions. Nucleic Acids Res. 1995;23:2314–2319. doi: 10.1093/nar/23.12.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 49.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 50.Yan S, Wu M, Patel DJ, Geacintov NE, Broyde S. Simulating structural and thermodynamic properties of carcinogen-damaged DNA. Biophys. J. 2003;84:2137–2148. doi: 10.1016/S0006-3495(03)75020-6. Erratum in: Biophys J. 92, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER 7. San Francisco, CA: University of California; 2002. [Google Scholar]
- 52.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves”, dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 53.Ravishanker G, Beveridge DL. MD Toolchest 2.0. Middletown, CT: Wesleyan University; 1993. 06459. [Google Scholar]
- 54.Fratini AV, Kopka ML, Drew HR, Dickerson RE. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 55.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 56.Hingerty BE, Figueroa S, Hayden TL, Broyde S. Prediction of DNA structure from sequence: a build-up technique. Biopolymers. 1989;28:1195–1222. doi: 10.1002/bip.360280703. [DOI] [PubMed] [Google Scholar]
- 57.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.