Abstract
Transcription factor IIIA (TFIIIA) is required for eukaryotic synthesis of 5S ribosomal RNA by RNA polymerase III. Here we report the discovery of a structured RNA element with striking resemblance to 5S rRNA that is conserved within TFIIIA precursor mRNAs (pre-mRNAs) from diverse plant lineages. TFIIIA protein expression is controlled by alternative splicing of the exon containing the plant 5S rRNA mimic (P5SM). P5SM triggers exon skipping upon binding of ribosomal protein L5, a natural partner of 5S rRNA, which demonstrates the functional adaptation of its structural mimicry. Since the exon-skipped splice product encodes full-length TFIIIA protein, these results reveal a ribosomal protein-mRNA interaction that is involved in 5S rRNA synthesis and has implications for cross-coordination of ribosomal components. This study also provides insight into the origin and function of a newfound class of structured RNA that regulates alternative splicing.
A large portion of transcripts from diverse eukaryotes exhibit alternative splicing. Between 40-60% of expressed human genes are conservatively estimated to give rise to multiple splice products1. Alternative splicing events have recently been shown to be prevalent in plants as well, with an estimated ∼20% of expressed genes in Arabidopsis thaliana (thale cress) and Oryza sativa (rice) generating alternate splice products2. Many questions still remain about how alternative splicing is regulated, and what the biological significance of particular alternative splicing events is.
It is becoming apparent that RNA structures can influence splicing3,4. Structural elements present within pre-mRNAs can alter accessibility of the splicing sites and branch site that define the sequence to be excised. Also, binding of protein splicing factors to regulatory RNA motifs that influence recognition of the surrounding region as an exon or intron can be affected by the structural context of the motifs.
While many computationally predicted pre-mRNA structures have been associated with splicing regulation of specific transcripts, few examples have contained detailed experimental evidence for the RNA structure3 or for its interacting partner. Exceptions include some descriptions of splice factor binding to regulatory motifs5,6, ribosomal proteins binding to rRNA mimics7-10, and metabolite binding to structured RNA elements called riboswitches11,12. These studies demonstrate how detailed analyses of pre-mRNA-ligand interactions can provide insight into molecular mechanisms for splicing regulation.
For example, direct binding of thiamin pyrophosphate (TPP) to TPP riboswitches found in fungi and plants13,14 alters pre-mRNA splicing and leads to regulation of gene expression in response to the metabolite11,12,15,16. Sequences within the metabolite-binding domain of the riboswitch base pair with nucleotides at or adjacent to the proximal 5′ splice site and inhibit its use11,12. Under elevated TPP conditions, these sequences instead are engaged in binding TPP, resulting in alternative splicing utilizing the revealed splice site.
We have discovered a class of structured RNAs broadly conserved in plants that controls alternative splicing of TFIIIA, the specific transcription factor essential for synthesis of 5S rRNA. The newfound RNA resembles 5S rRNA to a similar degree as several retrotransposons derived from 5S rRNA 17-19, which supports its origination from an ancient 5S pseudogene. While there are thousands of copies of 5S genes and pseudogenes in eukaryotic genomes20-22, plant 5S rRNA mimic (P5SM) serves a specific function as a structured cis-regulatory RNA element.
Our findings reveal that A. thaliana P5SM binds to ribosomal protein L5 in a directly analogous fashion to the eukaryotic 5S-L5 interaction. This interaction within the context of the TFIIIA pre-mRNA promotes exon skipping and leads to generation of the splice product encoding full-length TFIIIA protein. Since an increase in TFIIIA protein upregulates synthesis of 5S rRNA23, the discovery of P5SM shows that ribosomal protein L5 can directly control the production of its partner, 5S rRNA. P5SM regulation of TFIIIA pre-mRNA splicing also provides an example of alternative splicing control in plants by a structured RNA distinct from metabolite-binding riboswitches.
RESULTS
Discovery of a plant 5S rRNA mimic (P5SM)
We performed a bioinformatics search for structured, cis-regulatory RNA elements conserved in plant species by comparative analysis. The search strategy was based on computational prediction of both nucleotide sequence and secondary structure conservation between non-coding regions of A. thaliana and O. sativa genes related by protein sequence homology (M.C.H. and R.R.B., unpublished data). Several candidate RNA classes were detected, including the previously characterized plant TPP riboswitch12,15. One structured RNA element identified is associated with the TFIIIA gene in both plant species. Over 30 additional unique representatives of this candidate across species were detected in available genomic and EST/cDNA databases (Supplementary Fig. 1). The RNA element is well-represented in angiosperms, and a more diverged version also has been identified in the moss, Physcomitrella patens (Supplementary Fig. 2), indicating conservation in both vascular and non-vascular land plants.
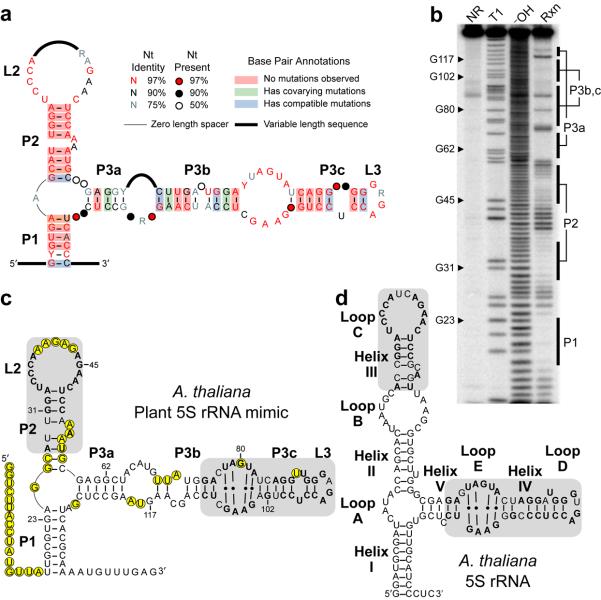
A consensus sequence and secondary structure model for the TFIIIA-associated RNA element was developed using the examples collected as inputs for the CMfinder program24. The RNA secondary structure predicted by the algorithm consists of two hairpin arms which are interrupted by internal loops and bulges (Fig. 1a). An additional pairing element (P1) forms a closing stem between sequences at the 5′ and 3′ termini.
Figure 1.
A conserved structured RNA element in plants resembles 5S rRNA. (a) Consensus sequence and secondary structure model for the angiosperm representatives of an RNA element identified by comparative bioinformatics. Sequences of all representatives are shown in Supplementary Fig. 1. Calculations for nucleotide conservation and base pair annotations were performed as previously described49. (b) In-line probing analysis of the 5′ 32P-labeled RNA shown in c, which encompasses the A. thaliana representative of the RNA element (nucleotides 649-793 of AtTFIIIA gene, NCBI gi 42592260). NR, T1, and −OH designate lanes containing nonreacted RNA, or RNA subjected to partial digest with RNase T1 (G-specific) or alkali, respectively. Rxn identifies RNA subjected to in-line probing. Labeled bars identify regions corresponding to pairing elements shown in c. (c) Sequence and secondary structure model for the A. thaliana plant 5S rRNA mimic. Positions with substantial backbone cleavage 3′ to the nucleotide upon in-line probing are circled. Shaded regions have sequence and structural similarity to corresponding shaded regions in A. thaliana 5S rRNA shown in d, with identical nucleotides in bold. Numbered nucleotides match labeled G nucleotides in b. (d) Sequence and secondary structure for the A. thaliana representative of 5S rRNA. Structural elements (helices and loops) are labeled using the 5S rRNA naming convention30. Shaded regions and bolded nucleotides are defined in c.
In-line probing analysis25 of the A. thaliana and O. sativa representatives support the secondary structure model (Fig. 1b, Supplementary Fig. 3). Typically, nucleotides that are base paired or otherwise structurally constrained are less subject to spontaneous phosphoester cleavage than more conformationally flexible internucleotide linkages. Accordingly, most predicted base paired regions exhibit reduced spontaneous cleavage (Fig. 1c). While some predicted bulges and loops are cleaved as expected for conformationally flexible regions, others (notably the P3b/P3c bulge) are not, which may indicate participation in non-canonical base pairings or in tertiary structures.
The sequence and structure of the candidate RNA do not match any RNA class models annotated in the Rfam database26. However, its unvarying association with the TFIIIA gene, which encodes a transcription factor required for the synthesis of 5S rRNA, suggests a connection between the newly identified RNA element and 5S rRNA. Comparison of sequences and secondary structures of 5S rRNA and the RNA element reveals two domains with striking homology between the two RNAs (Fig. 1c,d). Based on the structural similarity between these two RNAs, we named the newly discovered RNA element “plant 5S rRNA mimic” or “P5SM”.
Alternative splicing of P5SM controls TFIIIA expression
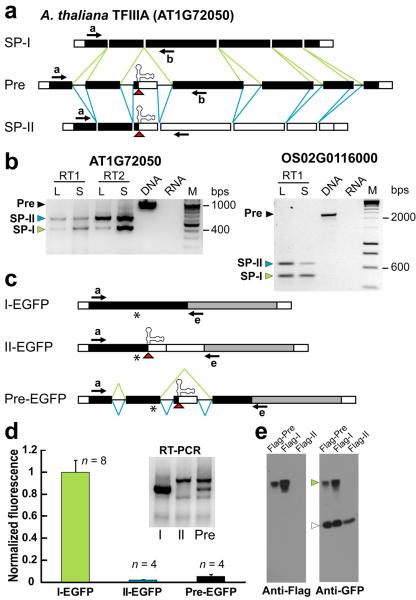
P5SM resides exclusively within an optional or “cassette” exon that is alternatively spliced in TFIIIA pre-mRNAs (Fig. 2a). In A. thaliana, two splice variants (NM_105863 and NM_202399) are annotated for the TFIIIA gene, which differ only in the presence or absence of the cassette exon. The existence of both splice products was confirmed by RT-PCR analysis (Fig. 2b). Similar results were obtained for the rice TFIIIA genes (Fig. 2b, Supplementary Fig. 4). Available EST/cDNA data also support the conservation of these two splice products for other plant TFIIIA genes. We have not identified any 5S rRNA-like element or alternate splice forms for annotated TFIIIA genes from non-plant species.
Figure 2.
The cassette exon containing the plant 5S rRNA mimic exhibits regulated alternative splicing and yields two splice products giving different gene expression. (a) Splicing model for TFIIIA pre-mRNAs from A. thaliana. Exon/intron organization (exons are rectangles, introns are lines) and exon features (untranslated regions are white, coding regions are black) are shown. Splicing reactions that lead to splice product I (SP-I, green lines) and splice product II (SP-II, blue lines) are shown. The cassette exon contains P5SM and its retention introduces a premature termination codon in SP-II (red triangle). Arrows indicate annealing sites of PCR primers used in b. (b) RT-PCR detection of two splice types for A. thaliana and O. sativa TFIIIA. cDNA templates were generated by reverse transcription reactions performed using poly-T primers (RT1) or random hexamers (RT2), then TFIIIA transcripts were detected by PCR. To assess qualitative developmental stage effects, the ratio of PCR products corresponding to SP-I and SP-II were analyzed in mature plant leaves (L) versus seedlings (S). Also shown are PCR reactions with genomic DNA, yielding a product the size of unspliced precursor (Pre), and with RNA as a control (no reverse transcription). DNA markers (M) are in increments of 100 base pairs (bps) from 200 to 1000 (top gel) or 200 bps from 400 to 1000 (bottom gel). Sequences of precursor and spliced mRNAs detected for A. thaliana and O. sativa TFIIIAs are shown in Supplementary Fig. 4. (c) Reporter constructs used in transient gene expression assays in Nicotiana benthamiana, depicted as described in a. The 5′ region of A. thaliana TFIIIA mRNAs (SP-I, SP-II, or precursor) that extends to the coding region for zinc finger 4 was fused in-frame to the EGFP reporter gene (grey). The resulting constructs are under control of the constitutive CaMV promoter. Arrows indicate annealing sites of PCR primers used in d. Asterisks indicate location of Flag peptide sequence introduced to constructs used for immunoblotting in e. (d) In vivo expression analysis for TFIIIA reporter constructs. Fluorescence was measured by laser-scanning of leaves. EGFP fluorescence was normalized to the value measured for I-EGFP. Numbers of independent leaf samples (n) measured are indicated. Error bars represent standard deviation (SD). Inset shows the corresponding RT-PCR data for the reporter constructs. (e) Western blot analysis of protein expressed in N. benthamiana from Flag-tagged versions of Pre-EGFP, I-EGFP, and II-EGFP reporters. A Flag peptide sequence was inserted into the coding region prior to the premature termination codon (asterisk, c) and does not affect fluorescence activity of the constructs (data not shown). The same immunoblot was probed with anti-Flag to detect N-terminal product and anti-GFP to detect C-terminal product following a wash step to allow reprobing with a second primary antibody. A green arrowhead labels the protein band detected for the reporter. Because I-EGFP gives very high protein expression (see d), this construct was transformed at one-tenth of the concentration of Pre-EGFP or II-EGFP constructs to enable parallel detection of weaker product signals. However, for each sample, the same concentration of an EGFP-only construct was transformed as an internal standard and detected with anti-GFP (open arrowhead) to normalize for transformation efficiency and equal sample loading.
All known examples of TFIIIA contain nine zinc fingers, except for Schizosaccharomyces pombe TFIIIA, which has ten27. The cassette exon carrying P5SM is always located between exons coding for zinc fingers 2 and 3 (Supplementary Fig. 4). Cassette retention introduces a premature termination codon in all splice product II (SP-II) transcripts identified, while skipping of the exon produces splice product I (SP-I) transcripts that encode the full-length protein. Truncation of TFIIIA protein before the third zinc finger is expected to eliminate 5S gene transcription initiation and 5S rRNA binding activities28.
We measured the effect of fusing the 5′ region of the different AtTFIIIA splice products on the activity of enhanced green fluorescent protein (EGFP) (Fig. 2c). The SP-I fusion (I-EGFP) results in robust fluorescence of the EGFP reporter. In contrast, the SP-II fusion (II-EGFP) shows very low activity even though the transcript is present (Fig. 2d), which is consistent with translational termination prior to the EGFP open reading frame.
Furthermore, analysis of an A. thaliana UPF1 mutant had identified SP-II as a target of nonsense-mediated decay29. Protein expression was analyzed for all reporter constructs modified with a Flag tag upstream of the premature termination codon. Western blot analysis with Flag and GFP antibodies to detect N- and C-terminal products, respectively, show expressed protein for Flag-I-EGFP but not Flag-II-EGFP (Fig. 2e). This result is consistent with the latter transcript being subject to nonsense-mediated decay, as no truncated N-terminal product was detected. The absence of C-terminal product for Flag-II-EGFP indicates that there is no alteration of start codon use. Full-length protein is detected by both antibodies for Flag-I-EGFP and Flag-Pre-EGFP constructs, as some of the latter is spliced to Flag-I-EGFP (Fig. 2d, inset).
Thus, SP-I gives expression of full-length protein, while SP-II gives no expression as premature translation termination targets the transcript for nonsense-mediated decay. Furthermore, in both A. thaliana and O. sativa, analogous changes in native TFIIIA splice product ratios were observed related to plant developmental stage (Fig. 2b), which suggests specific regulation of cassette splicing. The reporter data support the cassette exon as the major determinant of gene expression, and the conserved presence of P5SM within this exon implicates the RNA element as playing a key role in regulation.
L5 promotes skipping of the P5SM-containing cassette exon
Given its gene context and structure, we hypothesized that P5SM may exploit its structural mimicry of 5S rRNA to recruit a natural 5S rRNA binding partner to TFIIIA pre-mRNAs, and that this RNA-protein interaction influences cassette exon splicing. In eukaryotes, several proteins or protein complexes have been found in association with 5S rRNA that play roles in its maturation, transport, and storage outside the context of the ribosome30. In particular, mature 5S rRNA is known to associate with TFIIIA or ribosomal protein L5 to form 1:1 RNP complexes, and possible rationales exist for the coupling of TFIIIA expression to the levels of either protein.
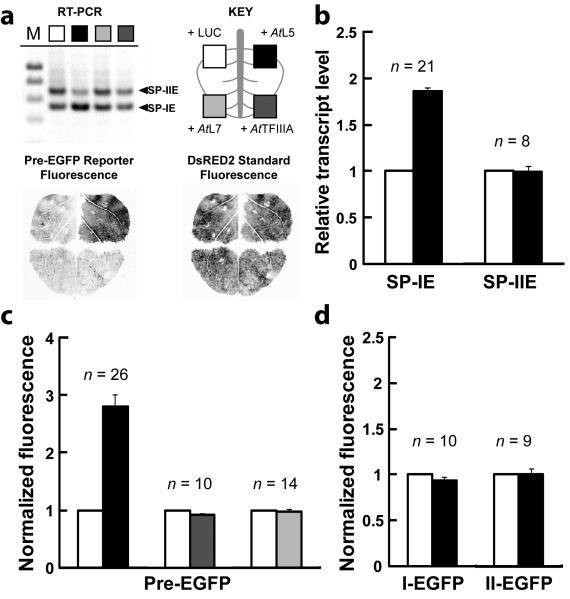
To monitor effects on cassette exon splicing and protein expression, a reporter was constructed with the 5′ region of unspliced TFIIIA precursor fused to EGFP (Pre-EGFP; Fig. 2c). This reporter construct enables mutational analysis to be performed on the RNA, but importantly maintains P5SM in its native context from the 5′ UTR through to the constitutive exon immediately following. The reporter transcript is expressed under a strong constitutive promoter (CaMV) to exclude promoter effects and fused in-frame to the EGFP coding region to allow facile quantitation of protein expression. Pre-EGFP is properly spliced in the in vivo plant model system, yielding transcripts that correspond to SP-I and SP-II, and expresses protein accordingly (Fig. 2d,e). These transcripts are identical in sequence to reporter constructs I-EGFP and II-EGFP but are products of splicing of the Pre-EGFP construct, and so will be referred to as SP-IE and SP-IIE.
We tested whether expression of A. thaliana L5 or TFIIIA protein (AtL5 or AtTFIIIA) affects splicing of the cassette exon. RT-PCR analysis shows increased splicing to SP-IE with AtL5 expression, relative to the transcript splice ratio observed for the same leaf with expression of an unrelated protein, luciferase (LUC) (Fig. 3a). Quantitative RT-PCR (qRT-PCR) measurement of many independent leaf samples shows an average 1.7-fold increase of SP-IE levels upon AtL5 expression (Fig. 3b). In contrast, expression of AtTFIIIA does not change the transcript splice ratio, nor does expression of another ribosomal protein that associates with 5S rRNA in the ribosome31, A. thaliana L7 (AtL7) (Fig. 3a). Similar expression levels for N-terminal Flag-tagged versions of L5, TFIIIA, and L7 proteins have been confirmed by western blot detection (Supplementary Fig. 5).
Figure 3.
Ribosomal protein L5 specifically increases TFIIIA gene expression by promoting splice product I formation. (a) RT-PCR analysis of transcript types SP-IE and SP-IIE resulting from splicing of a reporter containing the 5′ region of A. thaliana TFIIIA precursor mRNA fused to EGFP (Pre-EGFP, Fig. 2c) upon co-expression of luciferase (LUC), AtL5, AtTFIIIA, or AtL7 proteins in N. benthamiana. The splice products were detected using primers a and e. The key for tested proteins applies to all panels of this figure. Each RNA sample was extracted from the corresponding quadrant of a single leaf (cartoon), in which DNA constructs were introduced that express the Pre-EGFP reporter, the DsRED2 fluorescent protein as a normalization standard, a viral suppressor of RNA interference (p19), and the indicated test protein. Laser-scanned fluorescence of a representative leaf shows the delineation of protein expression by quadrants and the effect of AtL5 co-expression on Pre-EGFP fluorescence. (b) Quantitative RT-PCR analysis of relative amount of transcript types SP-IE and SP-IIE resulting from splicing of the Pre-EGFP reporter upon expression of AtL5 versus LUC. Numbers of independent RNA samples (n) analyzed are indicated. Error bars represent SEM. (c) In vivo expression analysis of the Pre-EGFP reporter upon co-expression of AtL5, AtTFIIIA, or AtL7 proteins in N. benthamiana. For all constructs and conditions, EGFP fluorescence was normalized to the value measured with expression of LUC, which was set to a value of 1. Fluorescence was measured by protein extraction. Numbers of independent leaf samples (n) measured are indicated. Error bars represent standard error of the mean (SEM). (d) Reporter fusion constructs corresponding to spliced products (I-EGFP, II-EGFP) also were analyzed with expression of AtL5, similar to as described in c.
The expected functional consequence of a change in TFIIIA exon splicing is a change in protein expression. Fluorescence from the Pre-EGFP reporter was measured upon expression of AtL5, AtTFIIIA, or AtL7, compared under each condition to the value measured upon expression of LUC. Consistent with the observed increase in SP-IE transcript, which encodes full-length reporter protein, AtL5 expression results in an approximately three-fold increase of Pre-EGFP reporter activity, whereas AtTFIIIA or AtL7 expression do not change reporter activity (Fig. 3a,c).
AtL5 expression does not affect I-EGFP or II-EGFP reporter constructs, which simulate the splice products (Fig. 3d). Thus, the change in SP-IE abundance is unlikely to be due to an effect on its stability. Rather, the increase of SP-IE is consistent with its enhanced production from altered splicing of the pre-mRNA in response to AtL5.
Assuming that splicing of TFIIIA pre-mRNA involves a choice between SP-I or SP-II, production of more SP-I should come at the expense of generating SP-II. Although we observe an increase in SP-IE levels when AtL5 is constitutively expressed, there is no corresponding decrease in SP-IIE levels (Fig. 3b). The levels of native TFIIIA SP-II transcript are increased 17-fold in an A. thaliana mutant deficient in the nonsense-mediated decay pathway29. Thus, the stability and steady state levels of SP-II type transcripts appear to be dominated by nonsense-mediated decay, which could obscure modest effects achieved by L5 expression. A small but sustained conversion in pre-mRNA splicing from SP-II to SP-I would lead to an accumulation of the more stable TFIIIA-encoding transcript that is disproportionate to the decrease in SP-II transcript. Overall, the in vivo reporter assays and RT-PCR analyses indicate that splicing of TFIIIA pre-mRNA is selectively influenced by L5 toward the exon-skipped product.
P5SM binds L5 via structural mimicry of 5S rRNA helix III
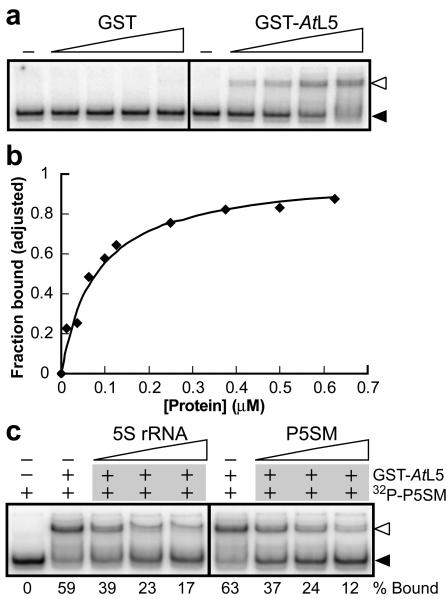
To determine whether splicing control involves a direct interaction between P5SM and L5, the binding of P5SM to the ribosomal protein was assessed in vitro. Recombinant AtL5 was expressed as a C-terminal fusion to glutathione-S-transferase (GST), similar to the protein construct previously demonstrated to bind 5S rRNA in vitro27. A 5′-radiolabeled RNA was transcribed in vitro corresponding to the region of AtTFIIIA pre-mRNA that contains P5SM and proximal splice sites (termed P5SM RNA). In a non-denaturing gel shift assay, GST-AtL5 binds P5SM RNA in vitro with an apparent dissociation constant (KD) of ∼75 nM, whereas the purification tag alone (GST) exhibits no appreciable binding at concentrations up to 1 μM (Fig. 4a,b). The KD for the A. thaliana 5S-L5 interaction is ∼10 nM (Supplementary Fig. 6), which is similar to that previously measured for the Xenopus laevis 5S-L5 interaction (2 nM)32.
Figure 4.
GST-AtL5 fusion protein binds to P5SM RNA in vitro. (a) In vitro binding analysis for the P5SM RNA (transcribed from nucleotides 603-810 of the AtTFIIIA gene, NCBI gi 42592260) with either GST or GST-AtL5. Radiolabeled RNA was incubated at 25°C in binding buffer in the absence or presence of protein (0-1 μM). RNA-protein complex formation was analyzed by non-denaturing PAGE. Unbound RNA (filled arrowhead) and RNA-protein complex (open arrowhead) are indicated. (b) Representative plot used to determine the apparent KD for the interaction between P5SM RNA and GST-AtL5 protein. Maximal binding observed at 1.25 μM protein was normalized to 1. Graphed line corresponds to the best-fit curve for a two-state binding model with 1:1 stoichiometry and KD of 75 nM. (c) In vitro competition binding analysis. Radiolabeled P5SM RNA mixed with buffer alone or unlabeled RNA (5S rRNA or P5SM, 50-200 nM) was incubated with GST-AtL5 protein (0.6 μM) at 25°C and analyzed by non-denaturing PAGE. The percent bound for each lane is shown below the gel. Other details are as described in a.
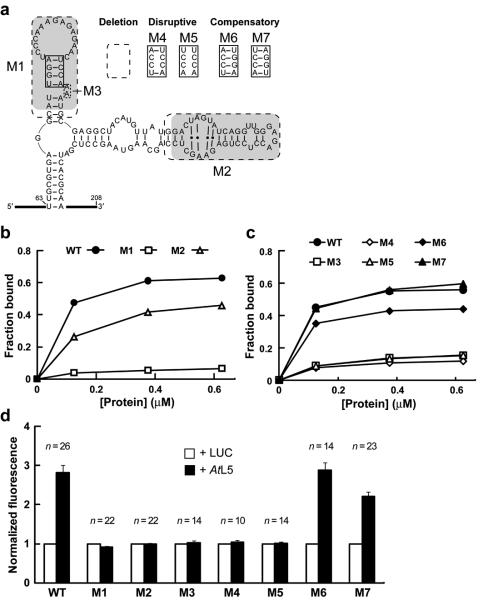
A similar binding mode for both RNAs was first supported by the observation that unlabeled 5S rRNA competes with radiolabeled P5SM for binding to L5 (Fig. 4c), indicating the RNAs form mutually exclusive complexes with the ribosomal protein. Furthermore, P5SM has two domains that are highly homologous to regions of 5S rRNA. Two deletion constructs (Fig. 5a) were made to test the individual contribution of each 5S-like domain to L5 binding. Removal of the P2/L2 hairpin (P5SM mutant M1) eliminates most binding by GST-AtL5, whereas deleting half of the P3/L3 hairpin (M2) only has a modest effect on protein binding (Fig. 5b). Structural evaluation of M1 and M2 constructs by in-line probing confirmed that the effects of deleting these regions are localized (Supplementary Fig. 7).
Figure 5.
The P2 stem of P5SM RNA is critical for binding to GST-AtL5 in vitro and disruption of AtL5 binding abolishes regulation of splicing in vivo. (a) Sequences of wild type (WT), deletion mutants, and P2 stem mutants of P5SM RNA used to assess domain contributions to protein binding. Shaded regions have sequence and structural similarity to corresponding shaded regions in 5S rRNA (Fig. 1d). Dashed boxes indicate nucleotides removed in deletion constructs (M1 through M3). The solid boxed region in the P2 stem was subjected to the disruptive (M4, M5) and compensatory (M6, M7) mutations shown. (b, c) Representative graphs of in vitro binding analyses for WT and mutant P5SM RNAs with different concentrations (0-0.625 μM) of GST-AtL5 protein. In c, the same symbol designates a disrupted mutant (open symbols) and its corresponding compensation mutant (filled symbols). The data lines for M3 and M5 are overlapping. (d) In vivo expression analysis for Pre-EGFP reporter constructs with co-expression of AtL5 in N. benthamiana. Pre-EGFP variants incorporate mutations M1 through M7 in the P5SM element. Data for the Pre-EGFP WT construct from Fig. 3c is shown for reference. For each construct, the EGFP fluorescence measured with expression of luciferase (LUC) was set to a value of 1 to enable comparison of relative change in response to AtL5 overexpression. This normalization procedure does not allow comparison of relative expression levels between the P5SM constructs, which instead were measured as shown in Supplementary Fig 9. Numbers of independent leaf samples (n) measured are shown. Error bars represent SEM.
The P2/L2 hairpin of P5SM, which corresponds to helix III/loop C of 5S rRNA, appears to be a major contributor to L5 binding. Accordingly, more subtle mutations to the P2 stem were analyzed (Fig. 5c). Substitutions of two or four nucleotides that disrupt the P2 stem (M4, M5) are almost as detrimental to protein binding as removal of the entire hairpin, and a similar loss of binding is observed upon deletion of the conserved dinucleotide bulge (M3). Compensatory mutations which restore base pairing interactions but have different nucleotide identities in the stem are able to partially (M6) or fully (M7) rescue binding. The in-line probing patterns for these mutant constructs also are consistent with the predicted structural effects on the P2 stem (Supplementary Fig. 7).
The combined in vitro binding and structural probing data for the mutant RNAs provide additional validation of our structural model of P5SM and support a binding mode in which the P2 stem interacts with L5. These results parallel binding data for helix III in the X. laevis 5S-L5 interaction32, which is also consistent with the archaeal large ribosomal subunit structure31 (Supplementary Fig. 8). Thus, binding of P5SM to L5 appears to mimic the 5S-L5 interaction.
P5SM recruitment of L5 is responsible for splicing control
With direct binding between P5SM and L5 established, we sought to determine whether splicing control in vivo requires this interaction. Mutations corresponding to M1 through M5, which disrupt the RNA structure, were introduced to the P5SM element within the Pre-EGFP reporter construct. The wild-type Pre-EGFP reporter (WT) responds to expression of AtL5 protein by increased splicing to generate SP-IE, which leads to higher reporter protein expression. In contrast, reporter constructs M1 through M5 are unaffected by AtL5 expression (Fig. 5d). Full deletion of either of the two 5S-like domains (M1, M2) leads to deregulation, consistent with their strong conservation in P5SM. More revealing is that subtle mutations of a few nucleotides distal to splice sites but within P5SM (M3 through M5) are able to completely abolish splicing regulation by AtL5 in parallel to their effect of disrupting L5 binding in vitro. Furthermore, the restoration of base pairing in the P2 stem through compensatory mutations M6 and M7 rescues splicing regulation by AtL5 (Fig. 5d) in parallel to their effect of rescuing L5 binding in vitro. Overall, the results of these in vivo reporter assays are consistent with splicing regulation involving the recruitment of L5 to TFIIIA pre-mRNA by P5SM.
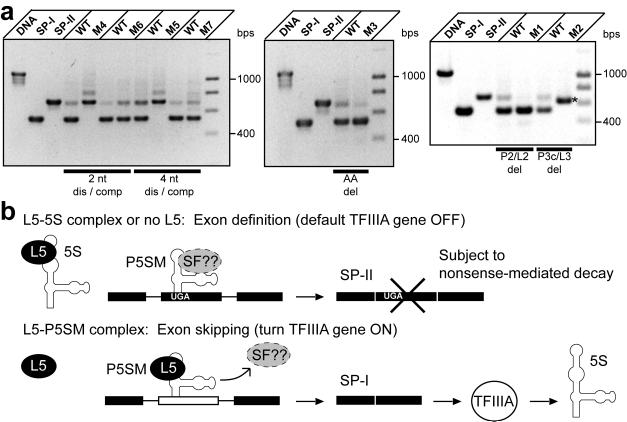
Also, reporter splicing for the P5SM mutants was compared to WT by RT-PCR analyses and reporter fluorescence (Fig. 6a, Supplementary Fig. 9). To minimize effects from varying endogenous L5 levels between individual samples, these experiments were performed with constitutive co-expression of AtL5. Whereas splicing to SP-IE is promoted by L5 binding for WT P5SM, both P2 stem pairing mutations (M4, M5) result in constitutive splicing to SP-IIE and low fluorescence activity relative to WT, as expected for loss of L5 binding. However, the bulge deletion (M3) unexpectedly favors constitutive splicing to SP-IE and gives high fluorescence activity. This result reveals that the bulge deletion may affect another aspect of P5SM function in addition to disrupting L5 binding. The more extensive deletion in M1 includes the bulge deletion and yields the same splicing outcome as M3, which suggests that both of these deletions are disrupting exon definition. This effect apparently dominates over the simple loss of L5 binding.
Figure 6.
The constitutive splicing patterns of deregulated mutants reveal that P5SM is involved in both exon definition and skipping, leading to a proposed model for regulation of TFIIIA pre-mRNA splicing. (a) RT-PCR detection of splice products arising from splicing of WT versus mutant Pre-EGFP reporter constructs. Each set consists of WT and one mutant reporter construct transformed on half of the same leaf to ensure near identical conditions for comparison. Effects from varying endogenous L5 levels were minimized by analyzing splice ratios upon constitutive AtL5 expression. Reporter fluorescence for these constructs measured under the same conditions for many individual leaves are shown in Supplementary Fig. 9. Total RNA was isolated from the same tissue amount for each leaf half, and roughly 1 μg was used in the RT reactions. Bars define disruptive (dis) and compensatory (comp) mutant pairs or deletion (del) mutants. Also shown are PCR products corresponding to unspliced precursor, SP-IE, and SP-IIE derived from DNA templates. The major splice product for the M2 mutant (labeled with an asterisk) is an SP-II type product which is shorter than the normal SP-II because of deleted sequence in the construct. DNA markers (M) from 400 to 1000 are in 200 bps increments. (b) The P5SM-L5 complex activates exon skipping, leading to the splice product (SP-I) that encodes full-length TFIIIA, whereas the default splice product (SP-II) is a nonsense-mediated decay substrate. Evidence suggests that P5SM is important in both exon recognition and skipping events. In the proposed mechanism, L5 displaces a putative exon-defining splice factor (SF) from P5SM. Thus, the discovery and analysis of this structured RNA element has elucidated a role for ribosomal protein L5 in the regulation of 5S rRNA synthesis in plants.
Importantly, all disruptive mutants including M1 and M3 are unresponsive to AtL5 (Fig. 5d), so these findings are consistent with loss of L5 binding causing splicing deregulation. Both compensatory mutations (M6, M7) display behavior similar to WT in splice product ratios and reporter activity (Fig. 6a), which suggests that normal regulation is restored. The character of the deletion mutant M2 indicates that additional aspects of P5SM function remain to be deciphered. However, the cumulative mutagenesis data reveal that P5SM may contribute to both definition and skipping of the exon in which it resides, and provide evidence for a regulatory model of TFIIIA splicing controlled by the RNA element.
DISCUSSION
In bacteria, ribosomal protein expression is commonly controlled by negative feedback from binding of ribosomal proteins to their own transcripts, resulting in inhibition of transcription or translation33. Structural studies have confirmed rRNA mimicry for some ribosomal protein mRNAs7,8 although it is not as general as originally proposed34. In eukaryotes, a few ribosomal proteins have been shown to auto-inhibit splicing and/or translation of their own transcripts35. Direct correspondence of eukaryotic rRNA mimics to rRNA binding sites has only been established for the yeast L30 pre-mRNA9.
The plant 5S rRNA mimic does not autoregulate ribosomal protein synthesis, but instead regulates the synthesis of 5S rRNA via control of TFIIIA expression. Also, while other rRNA mimics are similar to only the region of the rRNA that binds the ribosomal protein, P5SM exhibits much more extensive similarity in both sequence and structure to 5S rRNA. The resemblance between the two RNAs is even more striking in the lineage of non-vascular plants, as represented by the P. patens P5SM, which shares 75% nucleotide identity with an entire moss 5S rRNA (Supplementary Fig. 2).
One intriguing rationale for this apparent homology is that P5SM may have evolved from a 5S pseudogene. Its preservation within both vascular and non-vascular plants suggests that the element was acquired within the TFIIIA gene before the divergence of these two lineages. RNA polymerase III transcribed RNAs are prevalent originators of retrotransposed pseudogenes, and abundant retrotransposons derived from 5S rRNA have been described recently in both plants and animals17-19.
It has been proposed that conserved 5S-related retrotransposon sequences in mammals are functional17. Sequences derived from the Alu element in humans36 and a tRNA-related retrotransposon in animals37 also have been implicated in alternative splicing. P5SM illustrates how an ancient 5S pseudogene in plants has been adapted to serve this cis-regulatory function at the RNA level.
Our data reveal that P5SM exploits the homology of its P2 stem to helix III of 5S rRNA to bind ribosomal protein L5 (Fig. 5). In contrast, deletion of the conserved portion of the P3 stem in P5SM does not substantively affect L5 binding in vitro. These data are consistent with the 5S-L5 interaction in the archaea large ribosomal subunit structure. The function of the P3 domain in P5SM remains to be determined. Additional experiments are required to explain why the deletion mutant M2 disrupts splicing regulation and gives constitutive splicing to an SP-II type product. Initial results show that cassette exon splicing and reporter expression are unaffected by L7, the ribosomal protein that contacts loop E of 5S rRNA in the ribosome (Supplementary Fig. 8).
Unlike a plant retrotransposon that utilizes the embedded 5S gene as an internal RNA polymerase III promoter19, the P5SM-encoding sequences of TFIIIA genes have lost or mutated the A-box promoter element, which also corresponds to part of loop B in 5S rRNA38. In addition, P5SM lacks regions critical for TFIIIA protein binding to 5S rRNA, comprising the core of the rRNA that is not mimicked (loop A/helix II/loop B)39. The P. patens P5SM does have homology extending to helix II, but deviates from 5S rRNA in the relevant parts of loops A and B. Thus, P5SM appears to be an ancient copy of 5S rRNA whose structure was adapted for specific regulation of TFIIIA splicing in plants.
Our data support a model whereby alternative splicing of TFIIIA pre-mRNA is controlled by the binding of ribosomal protein L5 to P5SM (Fig. 6b). When L5 is depleted due to low production or complex formation with 5S rRNA, splicing of TFIIIA pre-mRNA retains the cassette exon. This splicing pathway generates SP-II, which is a transcript that is subject to nonsense-mediated decay because it carries a premature termination codon. In reporter assays, WT Pre-EGFP is not exclusively spliced to SP-IIE in the absence of AtL5 co-expression, most likely due to the presence of some endogenous L5. However, constructs M4 and M5 that carry mutations disrupting L5 binding exclusively yield SP-IIE (Fig. 6a), which implies that splicing defaults to SP-II when free L5 concentrations are low.
Alternatively, if P5SM is bound by L5, such as when there is an insufficient amount of 5S rRNA to occupy the ribosomal protein, the splicing pathway switches to favor generation of SP-I. This transcript type encodes TFIIIA protein, which in turn should activate synthesis of 5S rRNA. The regulatory logic in this system is converse to that of autoregulatory circuits, in which protein binding results in splicing to nonsense-mediated decay targets40,41.
Raising the level of AtL5 in vivo by constitutive over-expression results in an approximately two-fold increase in SP-IE levels (Fig. 3b). This change may not reflect the true dynamic range of the splicing switch, as changes to endogenous L5 levels under native conditions may be different than those tested. Comparison between P5SM mutants that are constitutively spliced to SP-IE or SP-IIE does reveal that the full dynamic range for change in protein expression as regulated by P5SM cassette splicing is up to ∼40 fold (Supplementary Fig. 9).
Increase of TFIIIA mRNA or TFIIIA protein has been shown to promote 5S rRNA transcription23,27. The expected consequence of P5SM-mediated regulation is that ribosomal protein L5 promotes the synthesis of its partner, 5S rRNA. In eukaryotes, 5S rRNA must be bound by L5 prior to incorporation into the large ribosomal subunit 30. Thus there is a strong rationale for the cell to balance synthesis of these two essential ribosomal components. Previous work in X. laevis oocytes demonstrated that a network of ribonucleoprotein complexes connecting L5 to TFIIIA could be responsible for homeostatic maintenance of 5S rRNA relative to L542. In plants, P5SM apparently functions to coordinate the transcription of 5S rRNA by RNA polymerase III directly to the level of free ribosomal protein L5, which is a translated product of RNA polymerase II.
Unlike other structured RNA elements that influence splicing, such as the yeast L30 mRNA element43 and eukaryotic TPP riboswitches11,12,16, P5SM does not directly overlap or have clear nucleotide complementarity with regulated splice sites. However, our data indicate that P5SM plays opposing roles in both exon definition and skipping, suggesting a possible mechanism for regulation. L5 binding to P5SM may displace an exonic splice factor from the pre-mRNA, resulting in regulated exon skipping (Fig. 6b). Competitive displacement of splice factors in other systems is precedented44. The displacement mechanism implies that mutations to the RNA may directly perturb splice factor binding and exon definition without need of L5. The M1 and M3 mutations to P5SM cause constitutive exon skipping even though binding to L5 is disrupted (Fig. 6a). These observations suggest that another factor may be required for exon definition.
Purine-rich motifs in A. thaliana have been shown to enhance recognition of surrounding sequences as exonic45, and these exonic splicing enhancers (ESE) are often recognized by splice factors46. Thus, the variable purine-rich insertions in L2 of angiosperm P5SMs (Supplementary Fig. 1) are attractive as potential ESEs that may place a splice factor in proximity to the P2 stem and within the influence of L5 binding. Replacement of the five purine nucleotides in L2 of P5SM with the UC sequence from Loop C of 5S rRNA (M8) does not affect L5 binding in vitro (Supplementary Fig. 10). But unlike other P5SM reporter constructs that bind L5, M8 is insensitive to L5 and constitutively splices to the exon-skipped product. This result is consistent with disruption of its proposed role as an ESE and is similar to the observations for M1 and M3. The data for these P5SM mutants reveal that some sequences in L2 contribute to exon definition, potentially through recruitment of an exonic splice factor. Further experiments will be needed to fully test this hypothesis, and to analyze the contribution of the complete P5SM structure to splicing regulation. Nevertheless, the discovery of P5SM and detailed analysis of its interaction with ribosomal protein L5 has led to a model for the molecular logic behind splicing regulation of TFIIIA pre-mRNAs.
Intriguingly, in several grasses, we have identified two separate expressed TFIIIA genes which are distinguished by the presence of a single copy or tandem arrangement of P5SM cassette exons. Conservation of the single and tandem P5SM arrangements suggests distinctive regulation of the two TFIIIA genes in response to L5 levels in these plants. Preliminary experiments on rice TFIIIA containing tandem P5SM cassettes have identified transcripts retaining none, one, or both cassette exons (Supplementary Fig. 11). The tandem P5SM architecture may function like some metabolite-binding riboswitches that use tandem arrangements to increase responsiveness to changing ligand concentrations47. P5SM elements, either in their single or tandem configurations, show that the original protein-binding function of the universally conserved 5S rRNA has been co-opted to serve in cis-regulation.
METHODS
Bioinformatics
We identified the plant 5S rRNA mimic by comparative analysis of non-coding sequences from annotated genes in the genomes of A. thaliana and O. sativa (M.C.H. and R.R.B., unpublished data). Additional candidate sequences were detected in available genomic and EST databases either by sequence similarity or by contextual searching via similarity to flanking translated regions. We used the CMfinder program24 to computationally predict the RNA secondary structure from the unaligned sequences.
Oligonucleotides and DNA constructs
Sequences of synthetic primers used and details of plasmid construct cloning are described in Supplemental Table 1 [AU: Correct?]. We isolated genomic DNA using the Plant DNAzol reagent standard protocol (Gibco BRL). DNA constructs were generated by PCR from cDNA or genomic templates, and confirmed by sequencing after cloning. We introduced nucleotide changes to DNA templates for RNA transcription by two-piece PCR ligation using overlapping primers that contain the desired mutation. Reporter constructs are based on the pBinAR plasmid48 modified by the addition of different fluorescent protein coding sequences as previously described12. We introduced nucleotide changes to reporter constructs by Quikchange (Stratagene) using complementary primers that contain the desired mutation.
In-line probing
RNAs were transcribed in vitro using T7 RNA polymerase from DNA templates, dephosphorylated, and radiolabeled with γ-32P ATP, then in-line probing of the 5′ 32P-labeled RNA was performed as previously described47.
RNA isolation and RT-PCR analyses
Total RNA was isolated, and RT-PCR and qRT-PCR were performed as previously described12. For the developmental study, we collected tissue from A. thaliana seedlings (14 days old) and rosette plants (6-8 weeks old), and from O. sativa seedlings (20 days old) and mature plants (2 months old). For reporter RT-PCR studies, we collected leaf tissue from N. benthamiana plants (3-5 weeks old) 48 h after infiltration with constitutively expressed reporter and tested protein constructs. cDNAs were generated using either poly-T or random hexamer primers and used as PCR templates for detection of TFIIIA transcripts or TFIIIA-derived reporter transcripts. qRT-PCR analysis was performed using the ABI 7500 Real-time PCR system (Applied Biosystems).
Protein expression
The C-terminal fusion of AtL5 protein to a GST tag was constructed in the pGEX-4T-2 expression vector in a similar manner to that previously described27. We expressed the recombinant protein in E. coliBL21-CodonPlus(DE3)-RIPL with 1 mM IPTG induction for 3 h at 28°C. Protein purification was performed as previously described27. Protein concentration was quantitated using the Quick Start Bradford Protein Assay (Bio-Rad).
Non-denaturing gel shift assays
To perform the gel shift assay, we incubated in vitro transcribed 5′ 32P-labeled RNA for 3 min at 55°C in renaturation buffer (50 mM Tris-HCl [pH 8.0 at 25°C], 50 mM KCl, 5 mM MgCl2), then allowed it to cool at 25°C. RNA (∼0.2-2 nM) was added to binding buffer (20 mM Tris-HCl [pH 7.5 at 25°C], 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% (v/v) glycerol, 100 μg ml−1 BSA, 3.75 μg ml−1 tRNA) and incubated for 10 min at 25°C in the absence or presence of different concentrations of protein (GST or GST-AtL5), which had been pre-incubated with RNase inhibitor (SUPERasin, Ambion). The reaction was quenched with addition of 50% glycerol and placed on ice. Samples were run in a 10% polyacrylamide gel (25 mM Tris-HCl [pH 7.5 at 25°C], 200 mM glycine, 5% (v/v) glycerol) at 4°C for 3 h at 300 V.
In vivo reporter fluorescence assays
We performed in vivo reporter studies using the Agrobacterium-mediated leaf infiltration assay in N. benthamiana as described previously12. All reporters and tested proteins were constitutively expressed under the cauliflower mosaic virus 35S promoter, and expression was analyzed 72 h after infiltration. The fluorescent proteins EGFP and DsRED2 served as reporter proteins and their fluorescence was measured in vivo with the Typhoon laser-based scanning system (Molecular Dynamics). Alternatively, we extracted 100 mg of tissue per sample with 300 μL buffer (50 mM Tris-HCl [pH 7.5 at 25°C], 150 mM NaCl, 0.1% (v/v) Tween 20, 0.1% (v/v) beta-mercaptoethanol). After centrifugation, fluorescence activity of the supernatant was measured in a 96-well plate fluorometer. EGFP was excited at 480 nm (10 nm bandpass) and detected at 510 nm (10 nm bandpass). Constitutive DsRED2 expression was monitored to normalize for transformational efficiency and to verify protein expression. DsRED2 was excited at 544 nm and detected at 590 nm (10 nm bandpass).
Immunoblots
We introduced Flag-tagged reporter constructs along with untagged EGFP, DsRED2, and p19 (RNAi suppressor) by leaf infiltration as described previously12, and crude protein was extracted from plant tissue as described above. SDS-PAGE and western blots were performed according to standard procedures. Anti-Flag from mouse (Sigma-Aldrich, F3165) or anti-GFP from rabbit (Invitrogen, 460092) were used as primary antibodies and detected by chemiluminescence detection (Super Signal West Dura, Pierce) of secondary antibodies conjugated to horse radish peroxidase.
Statistical analysis
For experiments in which 8 or more independent biological replicates (n) were analyzed, standard error of the mean (SEM) is calculated to show the variability of the mean. Otherwise, standard deviation (SD) is calculated.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Keith Corbino, Kathrin Leppek, and Alexander Westermann for technical assistance, Zasha Weinberg for advice on bioinformatics, Elena Puerta-Fernandez for advice on gel-shift assays, and Adam Roth for critical reading of the manuscript. M. C. H. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, and A. W. was supported by the German Research Foundation while in the Breaker laboratory. The research was supported by a grant from the NIH (GM068819). RNA science in the Breaker laboratory also is supported by the Howard Hughes Medical Institute.
Footnotes
Author Contributions:
M. C. H. conceived of and led project, performed bioinformatics analysis, designed and carried out in vitro structural probing and binding experiments, and wrote the manuscript. A. W. designed and carried out RT-PCR and qRT-PCR analyses, in vivo reporter assays, and edited the manuscript. R. R. B. advised on the project and edited the manuscript. All authors contributed to discussions regarding the data and their interpretation.
References
- 1.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 2.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA. 2006;103:7175–80. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–14. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLOS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–19. [PMC free article] [PubMed] [Google Scholar]
- 7.Ehresmann C, et al. Molecular mimicry in translational regulation: the case of ribosomal protein S15. RNA Biol. 2004;1:66–73. [PubMed] [Google Scholar]
- 8.Merianos HJ, Wang J, Moore PB. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–64. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao JA, Williamson JR. Joint x-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. RNA. 2000;6:1773–80. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 12.Wachter A, et al. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–50. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubodera T, et al. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–20. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 14.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–7. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bocobza S, et al. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–5. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–74. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- 19.Kalendar R, et al. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA. 2008;105:5833–8. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolffe A. The role of transcription factors, chromatin structure and DNA replication in 5S RNA gene regulation. J Cell Sci. 1994;107:2055–2063. doi: 10.1242/jcs.107.8.2055. [DOI] [PubMed] [Google Scholar]
- 21.Rogers SO, Bendich AJ. Ribosomal RNA genes in plants - variability in copy number and in the intergenic spacer. Plant Mol Biol. 1987;9:509–520. doi: 10.1007/BF00015882. [DOI] [PubMed] [Google Scholar]
- 22.Douet J, Tourmente S. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity. 2007;99:5–13. doi: 10.1038/sj.hdy.6800964. [DOI] [PubMed] [Google Scholar]
- 23.Andrews MT, Brown DD. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell. 1987;51:445–53. doi: 10.1016/0092-8674(87)90640-4. [DOI] [PubMed] [Google Scholar]
- 24.Yao Z, Weinberg Z, Ruzzo WL. CMfinder--a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–52. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 25.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–25. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:121–4. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathieu O, et al. Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res. 2003;31:2424–2433. doi: 10.1093/nar/gkg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pieler T, Theunissen O. TFIIIA: nine fingers - three hands? Trends Biochem Sci. 1993;18:226–230. doi: 10.1016/0968-0004(93)90194-r. [DOI] [PubMed] [Google Scholar]
- 29.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 30.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S rRNA: structure and interactions. Biochem J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 32.Scripture JB, Huber PW. Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5S ribosomal RNA - the major determinants of recognition are located in helix III-loop C. J Biol Chem. 1995;270:27358–27365. doi: 10.1074/jbc.270.45.27358. [DOI] [PubMed] [Google Scholar]
- 33.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–70. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 34.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc Natl Acad Sci U S A. 1980;77:7084–8. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIntosh KB, Bonham-Smith PC. Ribosomal protein gene regulation: what about plants? Can J Botany. 2006;84:342–362. [Google Scholar]
- 36.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–7. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 38.Cloix C, Yukawa Y, Tutois S, Sugiura M, Tourmente S. In vitro analysis of the sequences required for transcription of the Arabidopsis thaliana 5S rRNA genes. Plant J. 2003;35:251–261. doi: 10.1046/j.1365-313x.2003.01793.x. [DOI] [PubMed] [Google Scholar]
- 39.McBryant SJ, et al. Interaction of the RNA binding fingers of Xenopus transcription factor IIIA with specific regions of 5S ribosomal RNA. J Mol Biol. 1995;248:44–57. doi: 10.1006/jmbi.1995.0201. [DOI] [PubMed] [Google Scholar]
- 40.Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–71. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 41.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. Embo J. 2001;20:1785–96. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pittman RH, Andrews MT, Setzer DR. A feedback loop coupling 5 S rRNA synthesis to accumulation of a ribosomal protein. J Biol Chem. 1999;274:33198–33201. doi: 10.1074/jbc.274.47.33198. [DOI] [PubMed] [Google Scholar]
- 43.Li B, Vilardell J, Warner JR. An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc Natl Acad Sci USA. 1996;93:1596–600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283:1217–21. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 45.Egoavil C, Marton HA, Baynton CE, McCullough AJ, Schuler MA. Structural analysis of elements contributing to 5′ splice site selection in plant pre-mRNA transcripts. Plant J. 1997;12:971–80. doi: 10.1046/j.1365-313x.1997.12050971.x. [DOI] [PubMed] [Google Scholar]
- 46.Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol. 2007;58:267–94. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 47.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–4. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 48.Hofgen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum) Plant Sci. 1990;66:221–230. [Google Scholar]
- 49.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–19. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.