Abstract
microRNAs are functional, 22 nt, noncoding RNAs that negatively regulate gene expression. Disturbance of microRNA expression may play a role in the initiation and progression of certain diseases. A microRNA expression signature has been identified that is associated with pancreatic cancer. This has been accomplished with the application of real-time PCR profiling of over 200 microRNA precursors on specimens of human pancreatic adenocarcinoma, paired benign tissue, normal pancreas, chronic pancreatitis and nine pancreatic cancer cell lines. Hierarchical clustering was able to distinguish tumor from normal pancreas, pancreatitis and cell lines. The PAM algorithm correctly classified 28 of 28 tumors, 6 of 6 normal pancreas and 11 of 15 adjacent benign tissues. One hundred micro-RNA precursors were aberrantly expressed in pancreatic cancer or desmoplasia (p < 0.01), including microRNAs previously reported as differentially expressed in other human cancers (miR-155, miR-21, miR-221 and miR-222) as well as those not previously reported in cancer (miR-376a and miR-301). Most of the top aberrantly expressed miRNAs displayed increased expression in the tumor. Expression of the active, mature microRNA was validated using a real-time PCR assay to quantify the mature microRNA and Northern blotting. Reverse transcription in situ PCR showed that three of the top differentially expressed miRNAs (miR-221, -376a and -301) were localized to tumor cells and not to stroma or normal acini or ducts. Aberrant microRNA expression may offer new clues to pancreatic tumorigenesis and may provide diagnostic biomarkers for pancreatic adenocarcinoma.
Keywords: cancer, noncoding RNA, gene expression, real-time PCR
Pancreatic cancer is the fourth leading cause of cancer-related death in the United States.1 The annual death rate over the last five years has been ~30,000 with a similar number of new cases diagnosed each year. The prognosis for pancreatic cancer is the worst of all cancers with a mortality/incidence ratio of 0.99.2 The incidence of pancreatic cancer in the United States is ~9 per 100,000.3 These discouraging numbers, reflecting the increasing rates of incidence and death, are due to the lack of improvement in detection and diagnosis strategies and the paucity of breakthroughs in treatment regimens.
miRNAs were first discovered in c. elegans in 19934 and have subsequently been discovered in all multicellular organisms.5–7 miRNAs are negative regulators of gene expression and are believed to function primarily through imperfect base pair interactions to sequences within the 3′ untranslated region of protein coding mRNAs. Presently 326 miRNAs have been discovered in humans.8 While the role for each of these miRNAs is unknown, specific miRNAs have been implicated in the regulation of a diverse number of cellular processes, including differentiation of adipocytes,9 maturation of oocytes,10 maintenance of the pluripotent cell state11 and regulation of insulin secretion.12
A growing number of direct and indirect evidence suggests a relationship between altered miRNA expression and cancer. These include miR-15a and miR-16-1 in chronic lymphocytic leukemia, 13,14 miR-143 and miR-145 in colorectal cancer,15 let-7 in lung cancer16,17 and miR-155 in diffuse large B cell lymphoma.18 Expression profiling has identified other cancers with differential expression of several miRNAs, including breast cancer,19 glioblastoma 20,21 and papillary thyroid cancer.22 A polycistron encoding five miRNAs is amplified in human B-cell lymphomas and forced expression expression of the polycistron along with c-myc was tumorigenic, suggesting that this group of miRNAs may function as oncogenes.23,24
The purpose of this study was to profile the miRNA expression in clinical specimens of pancreatic adenocarcinoma. A real-time, quantitative PCR assay25,26 was used to profile the expression of over 200 miRNA precursors in clinical specimens of pancreatic cancer and pancreatic cancer cell lines. A unique miRNA signature was identified that distinguished pancreatic cancer from normal and benign pancreas.
Material and methods
Tissue procurement
The tissue samples analyzed in this study were derived from patients undergoing a surgical procedure to remove a portion of the pancreas at the University of Oklahoma Health Sciences Center and The Ohio State University. The collection of samples conformed to the policies and practices of the facility’s Institutional Review Board. Upon removal of the surgical specimen, research personnel immediately transported the tissue to the surgical pathology lab. Pathology faculty performed a gross analysis of the specimen and selected cancerous appearing pancreatic tissue and normal appearing pancreatic tissue for research. Each sample was placed in a cryovial and flash-frozen in liquid nitrogen and stored at −150°C until analysis. Subsequent pathologic analysis by the institutes providing the surgical specimens confirmed the histopathology of the samples taken for research. A second level of quality control was performed on the adjacent benign tissues by the laboratory who performed the RNA analysis. Histological slides were prepared from the section of the frozen tissue directly adjacent to tissue from which RNA was isolated. These slides were examined by one of us (W.L.F.) to determine if the benign tissues contained any pancreatic tumor cells. Benign tissue that contained residual tumor was not included in the study. The clinical data on the specimens are listed in Table I.
TABLE I.
Clinical data and tumor pathology
| Normal pancreas tissues | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample No. | Supplier | Age (years)/gender | Cause of death | RIN1 | ||||
| N1 | Ambion | 82/Female | Emphysema | 9.6 | ||||
| N2 | Ambion | 20/Male | Head trauma | 7.8 | ||||
| N3 | Ambion | 71/Male | Cerebral vascular accident | 9.2 | ||||
| N4 | Ambion | 71/Male | Failure to thrive | 9.1 | ||||
| BD | Clontech | 35/Male | Sudden death | 7.5 | ||||
| ST | Stratagene | 72/Male | Chronic obstructive pulmonary disease | 6.8 | ||||
| Pancreatic adenocarcinoma tissues | ||||||||
| Sample No. | Tumor pathology | Differentiation | Age (years)/gender | RIN |
||||
| Tumor | Benign | |||||||
| OUP2 | T3NxMx | Moderately | 85/Male | 8.2 | 7.0 | |||
| OUP14 | T3N0Mx invasive | Poor-moderate | 71/Male | 7.8 | 3.8 | |||
| OUP16 | T3N1bMx | Well | 50/Male | 5.6 | 5.6 | |||
| OUP20 | T3N1aMx | Poorly | 70/Male | 8.2 | 3.4 | |||
| OUP27 | T3N0Mx invasive | Moderate | 86/Male | 3.3 | 5.8 | |||
| OUP29 | T3N0Mx invasive | Poorly | 63/Male | 8.3 | 7.0 | |||
| OUP28 | T3N1bMx invasive | Poorly | 48/Female | 7.9 | 6.9 | |||
| OUP31 | T3N0Mx | Poorly | 38/Male | 7.7 | 7.0 | |||
| OUP33 | T3N1Mx | Poor-moderate | 42/Female | 5.1 | 8.3 | |||
| OUP36 | T3N1bMx | Moderate | 56/Female | 7.7 | 7.822 | |||
| OUP37 | T3N1bMx invasive | Mod-poor | 79/Male | 7.1 | No tissue | |||
| OUP42 | T3N0Mx invasive | Moderate | 78/Female | 6.6 | 7.6 | |||
| OUP48 | T3N1bMx invasive | Moderate | 61/Male | 8.1 | 4.3 | |||
| OUP49 | T3N1aMx invasive | Poor-moderate | 62/Female | 7.6 | 8.6 | |||
| OUP91 | T3N1bMx | Moderate | 65/Male | 5.9 | 6.1 | |||
| OUP97 | T3N1bMx | Moderate | 56/Male | 8.0 | 2.6 | |||
| OUP101 | T2N0Mx invasive | Moderate | 52/Female | 8.5 | 6.6 | |||
| OUP105 | T2N1aMx | Moderate | 55/Male | 8.3 | 8.9 | |||
| OUP122 | T3N1aMx | Moderate | 64/Male | 9.1 | 8.4 | |||
| OUP123 | T3N1bMx | Moderate-well | 52/Male | 6.4 | 7.62 | |||
| OUP135 | T3N0Mx | Moderate | 82/Male | 7.8 | 8.22 | |||
| OUP139 | T2N0Mx | Moderate | 50/Male | 6.6 | 6.7 | |||
| 0206C077B(T) | T3N0Mx | Moderately | 49/Male | 7.2 | No tissue | |||
| 0206C138B(T) | T3N0M1 | Intermediate to high grade | 44/Female | 6.1 | No tissue | |||
| 1050005A2(T) | T3N1Mx | Poorly | 73/Female | 7.0 | 7.3 | |||
| 3030800A3(T) | T3N1Mx | Moderately | 46/Male | 7.5 | No tissue | |||
| 3030396A1(T) | Intraductal papillary mucinous tumor | – | 59/Male | 7.3 | No tissue | |||
| 3030401A1(T) | Intraductal papillary mucinous tumor | – | 72/Male | 8.9 | No tissue | |||
RNA integrity number from Agilent bioanalysis.
Bengin tissue contained residual tumor and was not included in the study.
Cell lines
The following pancreatic tumor cell lines were purchased from American Type Tissue Collection (Manassas, VA). Panc-1, HS766T, MIA PaCa-2, HPAF-II, BxPC-3, Mpanc-96, PL45, Panc03.27 and Panc10.05. Cell lines were cultured in RPMI 1640 medium with 10% FBS or other optimized complete medium using standard conditions.
miRNA precursor expression profiling
Total RNA was isolated from the cell lines or tissues in 1 ml of Trizol (Invitrogen, Carslbad, CA). Frozen tissue (~10 mg) were first pulverized in a stainless steel mortar and pestle. Total RNA from normal pancreases were purchased from Ambion (Austin, TX), BD Biosciences (Mountain View, CA) and Stratagene (La Jolla, CA). All donors of the normal tissue died from complications other than pancreatic diseases (Table I). RNA concentration was determined by analyzing 1 µl of solution using the ND-1000 micro-spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA integrity number (RIN) was determined using the RIN algorithm of the Agilent 2100 expert software27 (Table I). RNA with a RIN ≥ 4 was included in the study. RNA was briefly treated with RNase-free DNAase I and cDNA was synthesized from 1 µg of total RNA using gene specific primers to 222 miRNA precursors plus 18S rRNA as described.25,26 The expression of 222 miRNA precursors was profiled using a real-time quantitative PCR assay.25,26 Duplicate PCRs were performed for each miRNA precursor gene in each sample of cDNA. The mean CT was determined from the duplicate PCRs. Relative gene expression was calculated as 2−(CTmiRNA−CT18S rRNA). Relative gene expression was multiplied by 106 to simplify the presentation of the data. The raw relative miRNA precursor expression data are provided in Supplemental Table 1.
Real-time PCR of mature miRNA
Assays to quantify the mature miRNA (i.e. TaqMan® microRNA Assays, Applied Biosystems Foster City, CA) were conducted as described28 with one modification. A 5× cocktail containing 28 different antisense looped RT primers was prepared by concentrating the 2× stock solutions in a Speed Vac. One hundred nanograms of total RNA was heated for 5 min at 80°C and then incubated for 5 min at 60°C with 10 µM of the 18S rRNA antisense primer followed by cooling to room temperature. Three microliters of the 5× looped primer mix was then added and the cDNA was made as described.28 This allowed for the creation of a library of 28 miRNA cDNAs plus the 18S rRNA internal control. Real-time PCR (10 µl total reaction) was performed as described28 using 1 µl of a 1:50 dilution of cDNA. Duplicate PCRs were performed for each mature miRNA gene in each sample of cDNA. The mean CT was determined from the duplicate PCRs. Gene expression was calculated relative to 18S rRNA as described earlier and multiplied by 106 to simplify data presentation.
Northern blotting
Northern blotting was performed as previously described.6,26 DNA oligonucleotides of the reverse compliment to the mature miRNA were used as probes. Blots were successfully stripped and reprobed up to 3 times.
RT in situ PCR
The RT in situ PCR protocol was performed as previously described.29 Briefly, optimal protease digestion time was determined using nonspecific incorporation of the reporter nucleotide digoxigenin dUTP. Optimal protease digestion was followed by overnight incubation in RNase-free DNase (10 U per sample, Boehringer Mannheim, Indianapolis, IN) and one step RT/PCR using the rTth system and digoxigenin dUTP. The chromogen is nitro-blue tetrazolium and bromochloroindolyl phosphate (NBT/BCIP) with nuclear fast red as the counterstain. The primer sequences to the precursors of miR-221, miR-301 and miR-376a were the same as those used for the profiling.25 The negative controls included omission of the primers and substitution with irrelevant (human papillomavirus specific) primers, as this virus does not infect pancreatic tissue. RT in situ PCR was performed on the archived, formalin-fixed paraffin-embedded sample 1050005A2(T) (Table I).
Statistics
The ΔCT data for 201 gene expression values (relative to control gene expression) were mean-centered and analyzed using the following strategy. The expression patterns of unfiltered data were assessed using unsupervised hierarchical clustering of samples and unsupervised hierarchical clustering of genes based on average linkage and Euclidian distance.30,31 To determine genes that are differentially expressed between groups of samples, the data were filtered on significance of differences using multi-group permutations-based ANOVA test (Welch approximation) with p < 0.01 (10,000 random permutations) and multiple testing correction (Westfall-Young step-down correction with maxT). To compare the expression patterns of differentially expressed genes, the filtered data were analyzed using hierarchical clustering of samples and hierarchical clustering of genes based on average linkage and Euclidian distance.30,31
Additional cluster analysis of filtered data was done using an expression terrain map.32 Terrain maps provide a three-dimensional overview of the major clusters inherent in the data. Samples were first mapped into a two-dimensional grid in which the placement of each element is influenced by a number of nearest neighbors based on Euclidian distance that was calculated using miRNA precursor expression data. The third dimension is determined by the density of points over the two-dimensional grid and its’ value is projected as a surface; higher peaks indicate larger numbers of very similar elements. The average correlation between each pair of samples was also calculated. Each pair of samples with an average correlation above the threshold (0.8) is indicated on the expression terrain map by a line connecting the two samples. This allows visualizing subsets of samples with a highly correlated pattern of miRNA expression. Peaks representing samples from a particular cluster are labeled with color coded spheres on top of each peak.
A supervised machine learning algorithm was applied for classification of samples based on unfiltered miRNA expression data. The predictive scores for each miRNA were calculated based on 2 class comparison (normal vs. tumor) of expression data using the prediction analysis of micro-arrays algorithm (PAM)33 based on training, take-one-out cross validation and testing procedures. The division of the samples into training and test sets is done using the commonly accepted approach of randomly splitting data into training and test sets (usually 2/3 for train, 1/3 for test).33 For this study, tumor samples were randomly split into ~75% for training and 25% for testing. Since the number of normal cases was small (N = 6), normal cases were included in the training set only. The small number of chronic pancreatitis samples (N = 4) were not included in the PAM analysis. While, two levels of quality control were performed to eliminate adjacent benign tissue that contained any tumor cells, the RNA was extracted from whole tissue rather than microdissected tissue. Thus, we could not completely rule out the possibility that some tumor cells contaminated the adjacent benign, nor do we know if early premalignant changes have occurred in these benign samples that are obtained from tissue adjacent to tumor. Therefore benign samples were excluded from the training set in order to train the classifier on true normal and tumor cases. Benign cases were used in the test set.
Results
Validation
Pancreatic tissue is rich in ribonuclease and care must be taken during RNA isolation to reduce the possibility of autolysis. To validate the integrity of the RNA isolated from the pancreatic tissue, ~100 ng of each RNA sample was assayed using the Agilent 2100 Bioanalyzer. Fifty-two tissues had a RIN ≥ 4 (median 7.6, range 4.3–9.6, Table I).
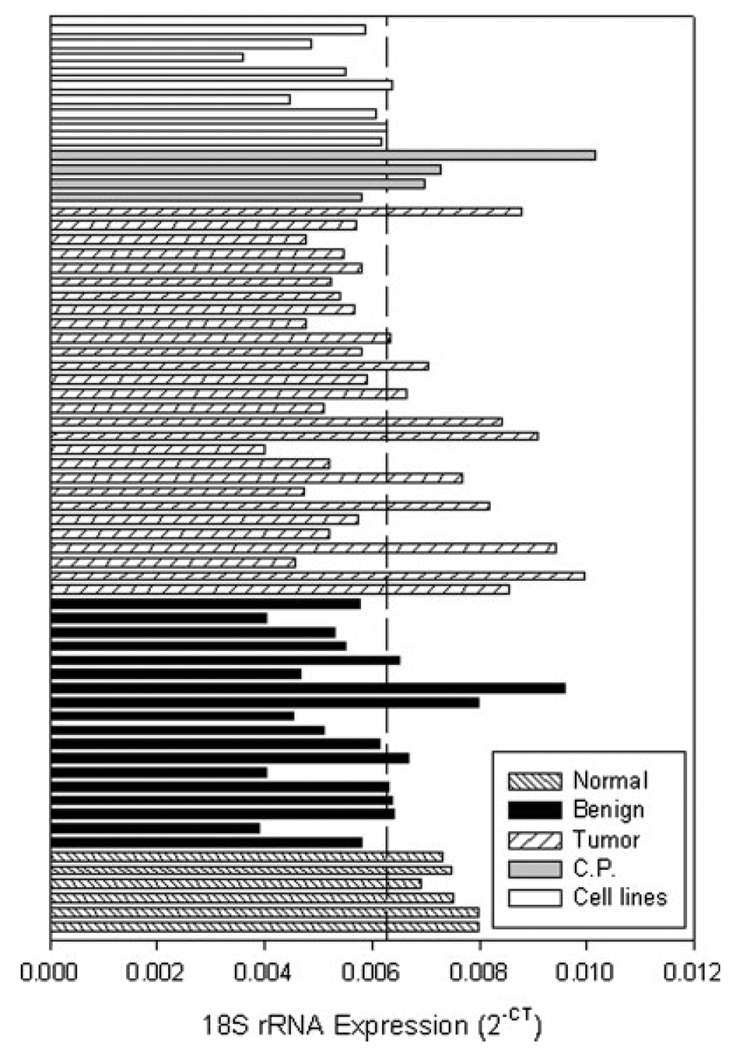
miRNA precursor expression in each of the samples was determined using real-time PCR and normalized to an internal control gene. Since equivalent amounts of total RNA was added to each RT reaction, 18S rRNA was validated as the internal control by comparing the mean expression among the various samples (i.e. tumor, normal and benign). There was no statistically significant difference in the mean 18S rRNA expression between the tumor samples or the normal pancreas (p = 0.116, Fig. 1). The 18S rRNA expression in the tumor and normal tissue determined here are in agreement with those previously reported in pancreatic tissue. 34 Since there was no significant difference in the 18S rRNA expression between the tumor, normal and benign groups, 18S rRNA was selected as the internal control gene in the study.
FIGURE 1.
18S rRNA expression in pancreatic tissue. The expression of the 18S rRNA internal control is shown in pancreatic tumors, adjacent benign tissue, normal pancreas, chronic pancreatitis and pancreatic cancer cell lines. 18S rRNA expression, determined using real-time PCR as described in Material and Methods, is presented as 2−CT. Dashed line, mean value.
miRNA precursor profiling in pancreatic tissues
The expression of 201 miRNA precursors, representing the 222 miRNAs discovered as of April, 2005 was profiled in 28 tumors, 15 adjacent benign tissues, 4 chronic pancreatitis specimens, 6 normal pancreas tissues and 9 pancreatic cancer cell lines. Unsupervised hierarchical clustering of samples was performed on the entire set of unfiltered data. The heatmap demonstrates that unfiltered expression data of only 201 miRNA precursors sufficiently sorts the samples into clusters of normal pancreas, tumor and cell lines (Supplemental Fig. 1). While most of the benign samples clustered together with the normal pancreas samples, several benign samples clustered with the tumor samples or pancreatitis.
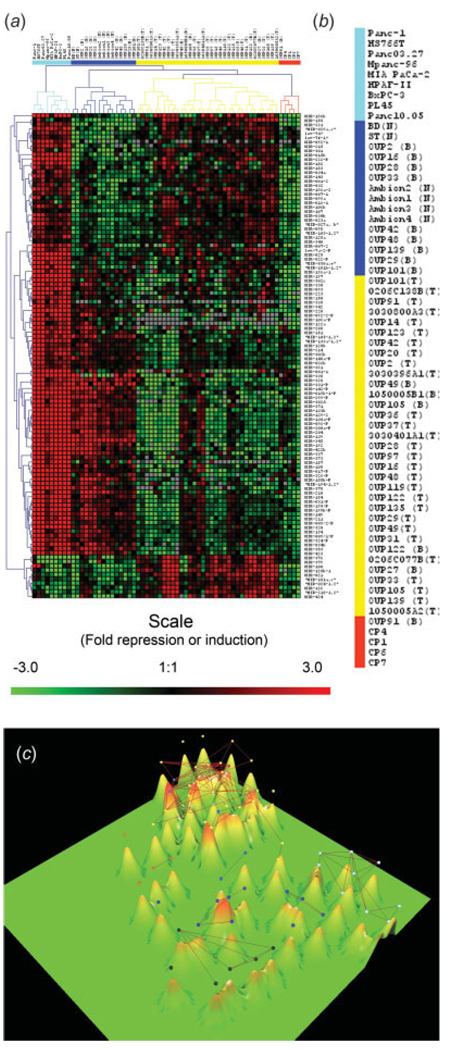
The miRNA precursor expression data was filtered using multigroup ANOVA. Statistical filtering of data is conventionally considered to be a required step of the expression data preprocessing because gene expression data are inherently noisy. Hierarchical clustering of samples and genes was performed on the resulting 112 miRNAs. Hierarchical clustering of filtered data allowed us to identify major groups of miRNAs that have different patterns of expression in the resulting four clusters of samples (Fig. 2a,b).One cluster contained only cell lines. Another cluster contained all 6 normal pancreases and 9 of 15 benign tissues. A third cluster contained the 4 chronic pancreatitis specimens and 1 benign tissue. Finally, a large cluster contained 28 of 28 tumors and 5 benign tissues (Fig. 2a,b).
FIGURE 2.
miRNA precursor expression in pancreatic samples. (a) The relative expression of each miRNA precursor was determined by real-time PCR; data are presented as ΔCT. Hierarchical clustering was performed on a subset of 112 genes that are differentially expressed (p < 0.001) among groups (tumor, chronic pancreatitis, cell lines and normal tissue) as determined by ANOVA multi-group comparison test. A median expression value equal to 1 was designated black; red, increased expression; green, reduced expression; grey, undetectable expression. (b) Dendrogram representing the results of hierarchical clustering analysis of the miRNA precursor expression pattern in 62 samples. Samples include primary pancreatic tumors (N = 28), normal pancreatic tissues (N = 6), adjacent benign pancreas (N = 15), chronic pancreatitis (N = 4) and pancreatic cancer cell lines (N = 9). (c) Three-dimensional expression terrain map was created from the filtered miRNA precursor expression data presented in (a). Each mountain represents an individual sample (tumor, adjacent benign, chronic pancreatitis, normal pancreas or pancreatic cancer cell line). The individual mountains sort into small groups based upon their similarities or differences to each other. Colored dots represent the same clusters as in (b). Black dots represent the 6 normal pancreases. The lines connecting pairs of samples indicate those samples which have very similar patterns of miRNA expression with average correlation above the threshold (>0.8).
The filtered data were also analyzed using a different clustering technique known as expression terrain maps.32 The terrain map separated the expression data into 5 main groups of samples (Fig. 2c). Like the hierarchical clustering, the terrain map separates groups of samples from the normal pancreas, the cell lines, pancreatic adenocarcinomas and chronic pancreatitis. This analysis showed that each of the clusters of samples occupies distinct regions on the expression terrain map thus providing additional evidence that each of the 4 groups of samples has distinct patterns of miRNA expression and suggests the possibility of finding subsets of microRNAs that discriminate normal and tumor samples. Most of the adjacent benign tissue grouped in between the normal and tumor, while those benign samples that clustered with the tumor also grouped with the tumor on the terrain map (Fig. 2). As an additional feature of terrain map, the average correlations of all miRNA expression values were calculated between each pair of samples. Those correlations above the threshold of 0.8 are shown as lines connecting pairs of samples with similar patterns of miRNA expression. This allows visualizing subsets of samples with a highly correlated pattern of miRNA expression. The samples within each of the 4 main groups are connected (i.e. average correlation >0.8) but there were no between the groups connections, except for two pancreatitis samples that correlated with some of the tumor samples (Fig. 2c).
Comparing the gene expression profile among different pancreas tissues has been used to eliminate the stroma and cell proliferation (e.g. cell line) contributions and identify genes expressed in pancreatic tumors.35–37 An attempt to perform such analysis on the miRNA expression data in Figure 2a was unsuccessful due to the uniformly high expression in the cancer cell lines and the uniformly low expression in the pancreatitis samples. However, a number of possible tumor-related miRNAs were identified that were increased in the cell lines or tumors but not in the normal or pancreatitis (Fig. 2a).
Data analysis by prediction analysis of micro-arrays algorithm (PAM)
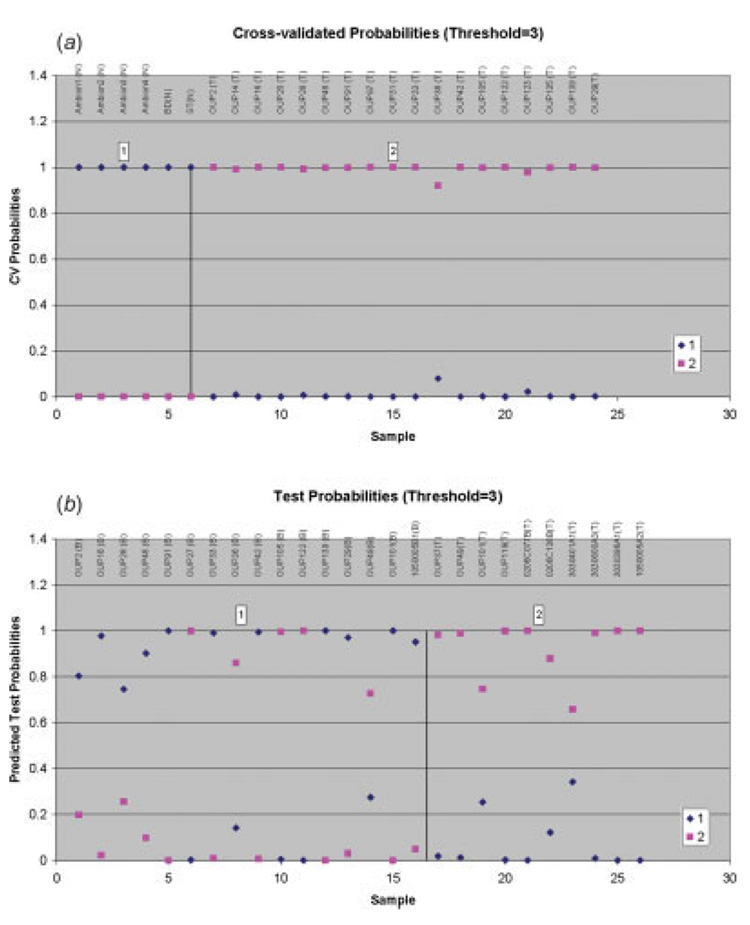
The PAM classification algorithm was used to determine if the miRNA expression data could predict which class the samples fit (tumor or normal) and to determine the most important, differentially expressed miRNAs related to pancreatic adenocarcinoma. The unfiltered data on 201 pre-miRNAs were analyzed by the PAM algorithm.33 The 3 genes with more than 75% of missing data were eliminated from the analysis in order to prevent possible artifacts of the imputation algorithm. PAM training and cross-validation were conducted using the 6 normal pancreas and 18 pancreatic tumors. PAM has correctly classified 100% of the normal and tumor samples (Fig. 3a). PAM testing was conducted on 10 tumor samples and 15 adjacent benign samples. PAM has correctly classified 100% of the tested tumor samples and 11 of 15 of the tested benign samples (Fig. 3b). The 20 top ranked differentially expressed miRNAs in pancreatic adenocarcinoma as selected by PAM are listed in Table II.
FIGURE 3.
Estimated probabilities for the training and test data. All training data including 6 normal pancreas samples and 18 of the samples known to be pancreatic tumors are correctly classified (a). Eleven out of 15 adjacent benign samples and 10 samples known to be pancreatic tumors are correctly classified in the testing group (b). Samples are partitioned by the true class (a) and the predicted class (b).
TABLE II.
Top 20 aberrantly expressed miRNA precursors in pancreatic adenocarcinoma
| Rank | Name | p-value (t-test) | Fold change | Chromosome location |
|---|---|---|---|---|
| 1 | miR-221 | 5.66E–05 | 26.2 | Xp11.3 |
| 2 | miR-424 | 3.62E–08 | 56.3 | Xq26.2 |
| 3 | miR-301 | 1.11E–05 | 34.2 | 17q23.2 |
| 4 | miR-100 | 4.40E–06 | 36.9 | 11q24.1 |
| 5 | miR-376a | 7.00E–04 | 7.79 | 14q32.31 |
| 6 | miR-125b-1 | 1.00E–04 | 23.2 | 11q24.1 |
| 7 | miR-021 | 2.00E–04 | 15.7 | 17q23.2 |
| 8 | miR-345 | 1.44E–15 | −14.5 | 14q32.2 |
| 9 | miR-016-1 | 3.73E–04 | 14.3 | 13q14.2 |
| 10 | miR-181a,c | 8.31E–04 | 18.6 | 9q33.3, 19p13.13 |
| 11 | miR-092-1 | 3.40E–03 | 19.6 | 13q31.3 |
| 12 | miR-015b | 4.00E–04 | 8.55 | 3q25.33 |
| 13 | miR-142-P | 3.63E–07 | −15.4 | 17q23.2 |
| 14 | miR-155 | 1.51E–03 | 14.0 | 21q21 |
| 15 | let-7f-1 | 4.00E–04 | 10.9 | 9q22.32 |
| 16 | miR-212 | 2.00E–04 | 22.2 | 17p13.3 |
| 17 | miR-107 | 3.86E–05 | 8.20 | 10q23.31 |
| 18 | miR-024-1,2 | 9.12E–08 | 8.17 | 9q22.32, 19p13 |
| 19 | let-7d | 7.06E–04 | 8.38 | 9q22.32 |
| 20 | miR-139 | 6.79E–11 | −7.91 | 11q13.4 |
Validation of miRNA expression
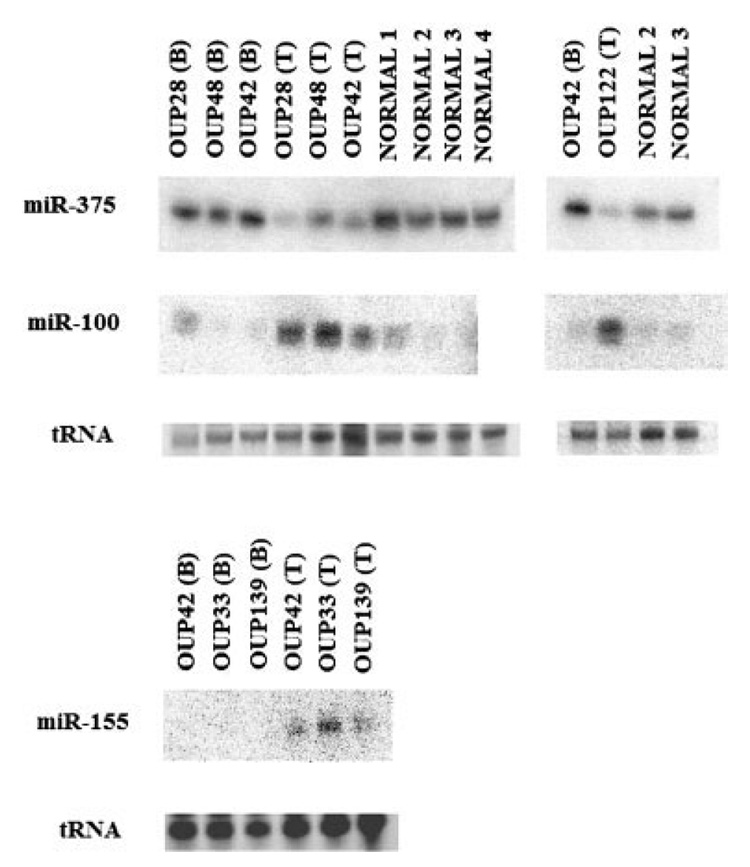
The real-time PCR assay used in this study quantifies the miRNA precursors and not the active, mature miRNA.25,26 Northern blotting was performed on the identical RNA used in the realtime PCR analysis to validate if the mature miRNA correlated with the precursor. Expression levels of mature miR-100, mir-375 and miR-155 from 4 normal pancreases and 5 pairs of tumor/adjacent benign tissue paralleled the miRNA precursor levels by PCR (Fig. 4). miR-100 and miR-155 were among the top 20 differentially expressed miRNAs (Table II). miR-375 was validated by Northern blotting because it was one of the few miRNAs with decreased expression in pancreas cancer. While miR-375 was not among the top 20 miRNAs (Table II), the expression of both precursor and mature miR-375 was significantly decreased in pancreas cancer by real-time PCR (p < 1 × 10−5). miRNA expression in the paired benign and tumor tissue was consistently increased or decreased in all cases, demonstrating that the differences in miRNA expression between tumor and benign are due to differences in individual patient’s tissues and not due to differences in the mean expression of the group.
FIGURE 4.
miRNA expression by Northern blotting. The expression of miR-100, miR-375 and miR-155 was determined in tissue specimens of pancreatic cancer (T), adjacent benign tissue (B) or normal pancreas (N). Blots were stripped and reprobed. tRNA, visualized by ethidium bromide staining, was used as a loading control.
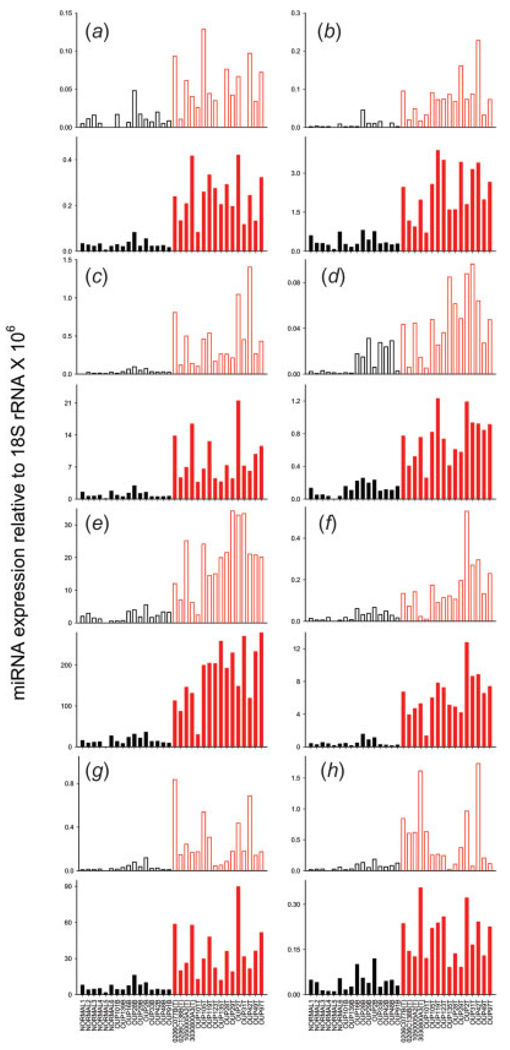
The real-time PCR data was further validated using a commercially available real-time PCR assay to amplify and quantify the mature miRNA. Mature miRNA expression was validated on 8 of the top aberrantly expressed miRNAs from PAM. cDNA from the following tissues were assayed: 6 normal pancreases, 16 pancreatic adenocarcinomas and 10 adjacent benign tissues that were predicted as normal from PAM. The mature miRNA expression highly correlated with the miRNA precursor (Fig. 5). To our knowledge, this is the initial presentation of both precursor and mature miRNA expression determined by sensitive, real-time PCR assays. A direct comparison between the precursor and mature PCR data presented in Fig. 5 is not possible. Among other reasons, different amounts of RNA were added to each RT, 1 µg for the precursor assay and 100 ng for the mature. Despite these differences, the relative amount of mature miRNA is greater than the precursors in all cases except for panel H. This demonstrates that the steady state levels of mature miRNA are more predominant than the precursors, a fact that is documented on nearly all published Northern blots that show the band intensity of the mature is more intense than the precursor. We have no explanation as to why this situation is apparently reversed in the case of panel H (miR-212). Of interest is that while the relative expression values for these mature miRNAs spanned 3-logs (from 0.1 to 100), the trend in differential expression between the tumor and normal tissues remained (Fig. 5). This suggests that miRNAs function in tumor and normal pancreas at different expression levels, yet the differential expression is maintained between cancer and normal tissue.
FIGURE 5.
Validation of precursor and mature miRNA levels. The expression of 8 miRNAs was validated in 6 normal pancreas specimens, 10 adjacent benign tissues and 16 pancreatic adenocarcinomas. The relative expression of the miRNA precursors (open bars) was determined using a real-time PCR assay to the miRNA precursors while the relative expression of the mature miRNA (closed bars) was determined using a real-time PCR assay to the mature miRNAs. The mean differences in miRNA expression between the normal pancreas black) and tumors (red) was significant p < 0.01 (student’s t-test). (a) let-7i, (b) miR-221, (c) miR-100, (d) miR-301, (e) miR-21, (f) miR-181a,c (precursor) and miR-181a (mature), (g) miR-125b-1 (precursor) and miR-125b (mature), (h) miR-212.
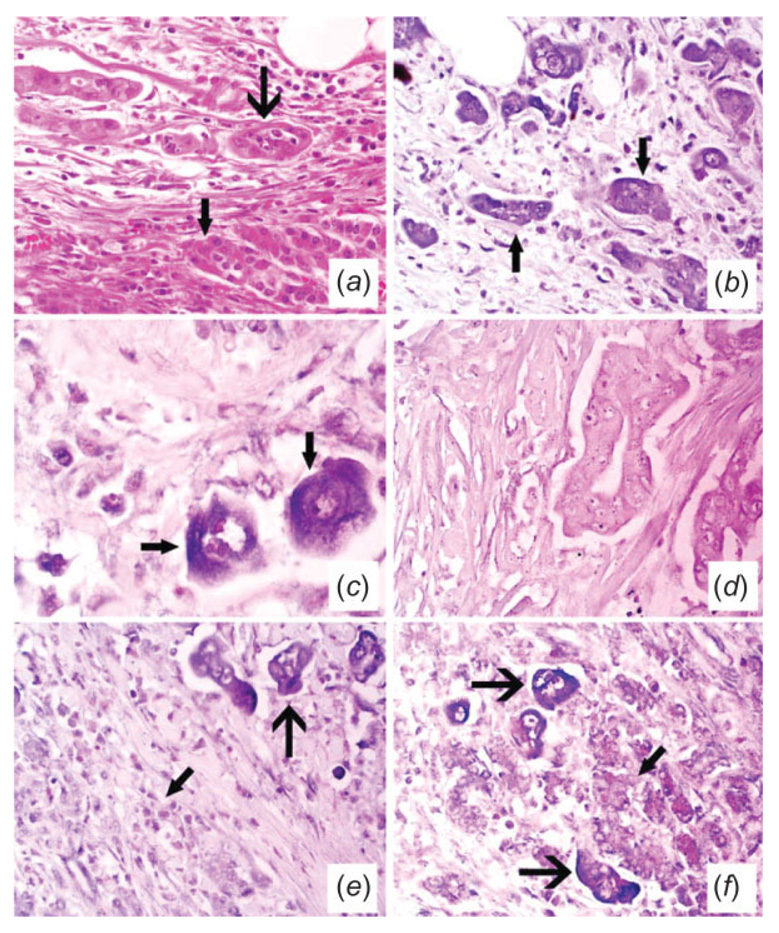
Finally, the cell-type miRNA expression was studied using RT in situ PCR. miR-221, miR-376a and miR-301 were selected for the in situ PCR since they were among the top differentially expressed miRNAs (Table II) and had increased expression in the tumor and cell lines compared to the normal pancreas and pancreatitis (Fig. 2). We were particularly interested in the cell type expression of miR-376a since it was cloned from pancreas cells.12 miR-221 and miR-376a are localized to the tumor cells and not to the benign pancreatic acini or stromal cells (Fig. 6) or benign ducts (not shown). miR-301 was also localized to tumor (data not shown).
FIGURE 6.
Histologic and molecular analyses of pancreatic cancer for microRNA expression. Panel (a) (×400) depicts the hematoxylin and eosin analysis of a pancreatic adenocarcinoma. The normal pancreatic glands (small arrow) are being invaded by the poorly formed glands of the carcinoma (large arrow). Serial section analysis of miR-221 after in situ amplification of the corresponding cDNA showed that many of the tumor cells contained the target sequence; note the cytoplasmic localization (arrows, panel (b) – ×400 and at higher magnification, panel (c) – ×1,000; the signal is blue due to NBT/BCIP with negative cells counterstained with fast red). The signal was lost with either omission of the primers or substitution with HPV-specific primers (panel (d), ×400). The adjacent serial section also showed many of the tumor cells expressed miR-376a after in situ amplification of the cDNA (e, f). Panel (e) (×400) shows the positive tumor cells (large arrow) and the negative stromal cells in the areas of desmoplasia (small arrow) while panel (f) (×400) depicts the positive tumor cells (large arrow) adjacent to the negative benign pancreatic gland acini (small arrow).
Discussion
Reported here are the results of the first detailed miRNA expression profiling study in pancreatic ductal adenocarcinoma. Expression profiling identified a large number of miRNAs that are aberrantly expressed in pancreatic ductal adenocarcinoma. A bead-based flow cytometric assay was used previously to profile the expression of 217 miRNAs in nine samples of pancreatic adenocarcinoma. 38 A micro chip assay was recently used to profile the expression of miRNAs in 39 samples of endocrine pancreas cancer.39 Our results in pancreatic ductal adenocarcinoma more closely approximate those of Volinia, et al.,39 in endocrine pancreas tumors and show that the majority of miRNAs are increased in the tumor compared to normal pancreas. Many of the miRNAs that are increased in both pancreas adenocarcinoma and endocrine pancreas cancer are similar, including miR-221, -100, -125b and -21. On the other hand, Lu, et al.,38 report an almost universal decrease in miRNA expression in the pancreatic adenocarcinomas compared to normal pancreas.
miRNAs are believed to function primarily as negative regulators of gene expression following binding to conserved sequences within the 3′ untranslated region of target mRNAs. While the biological roles of miRNA are under intense investigation, they are believed to define and maintain cellular fate in a manner similar to transcription factors40 by regulating developmental timing and differentiation. 41 Since alterations in developmental pathways play a critical role in pancreatic cancer development,42,43 alterations in miRNA expression may be an important contributor to the development of pancreatic adenocarcinoma.
Our study is unique in that a sensitive, real-time PCR assay was used to profile a relatively small number of nonconding RNAs. Real-time PCR is the gold standard of RNA quantification and has much less technical noise and greater reproducibility than traditional cDNA micro-arrays. miRNA expression profiling correctly identified 28 of 28 tissues as tumor (Fig. 3). All 6 normal pancreases were correctly predicted and 11 of 15 adjacent benign tissues were classified as normal tissue (Fig. 3). The data presented here reinforces the argument that miRNA expression profiling may generate a unique molecular signature for a given cancer. This concept is supported by several recent studies. Profiling of various cancers revealed that the pattern of miRNA expression varies markedly across different tumors and that a small number of miRNAs define the cancer better than expression data from 16,000 mRNAs.38 A unique expression signature of only 13 miRNAs differentiated cases of the more aggressive form of chronic lymphocytic leukemia from the more indolent form and was associated with the presence or absence of disease progression.44
The 3 factors that are likely driving the differences in gene expression are the normal acini, stroma and tumor cells. We cannot conclude that each of the aberrantly expressed miRNAs (Table II) reflect a difference in expression between normal ductal epithelium and tumor and may reflect differences in expression among the different cell types (stoma, acini and tumor). However, we confirmed by RT in situ PCR that 3 of the top differentially expressed miRNAs miRNAs that were identified in the screen are localized to the tumor cells (Fig. 6). We cannot explain why some of the benign tissues failed to cluster with the normal pancreas (Fig. 2). While 2 levels of quality control were used to reduce the possibility of contaminating tumor cells, it is possible that some tumor cells were present in the benign tissue since the RNA was isolated from whole tissue and not microdissected tissue. Another possibility is that premalignant changes have already occurred in some of the benign tissues as those samples are obtained from tissue adjacent to tumor. It is interesting that most of the benign samples (and chronic pancreatitis as well) lie in between the normal pancreas and tumor on the expression terrain map (Fig. 2c). This observation may indeed describe the premalignant alterations that have occurred in these benign tissues. Future studies using RT in situ PCR and perhaps laser microdissection will be able to address these issues in more detail.
Some of the differentially expressed miRNAs in pancreatic cancer were aberrantly expressed in other cancers. These include miR-155, which was increased in the present study and in diffuse large B-cell lymphoma18; miR-21 was increased here and in glioblastomas, 20,21 breast cancer19 and papillary thyroid cancer22; miR-221 was increased in pancreatic cancer, in glioblastoma21 and in thyroid cancer.22 miR-221 is located ~700 bp from miR-222 on the X chromosome; both miR-221 and miR-222 are predicted to bind to and regulate kit.22,45 miR-222 precursor was not among the top 20 differentially expressed miRNAs (Table II); however, subsequent analysis of mature miR-222 by PCR showed that miR-222 was increased in pancreas cancer at levels that were similar to miR-221 (data not shown). Thus, deregulation of the miRNAs mentioned earlier may be unique to cancer in general. miRNAs differentially expressed in other cancers were not deregulated to the same degree in pancreatic cancer. The let-7 family, decreased in lung cancer, 16,17 was increased here. Expression of the miR-17-92 polycistron (encoding miR-17, -18, -19a, -19b-1 and -92-1) was increased in lymphoma and colorectal cancer24 but was not significantly altered in pancreatic cancer. We report deregulation of a number of miRNAs in pancreatic cancer such as miR-376a and miR-301 that have not been reported in any other cancers to our knowledge. Also of interest is the fact that most of the deregulated miRNAs reported here show increased expression in the tumors compared to the normal pancreas. A few miRNAs had reduced expression in pancreatic cancer including miR-375 (Fig. 4). miR-375 was cloned from pancreas and is believed to be islet cell specific.12
We report significant changes in miRNA expression between pancreatic adenocarcinoma and normal pancreas. Since each miRNA may regulate scores of mRNAs,46 the impact on gene expression in pancreatic cancer may be profound. The miRNA field is currently hampered by a lack of methods to sort through the many hundreds of predicted miRNA target genes. As more sophisticated approaches become available to identify and validate miRNA targets, the role of aberrant miRNA expression in pancreas cancer will become better understood.
Acknowledgements
Supported by an NIH grant CA107435 to T.D.S. The Tissue Procurement Shared Resource at The Ohio State University funded by the National Cancer Institute, grant P30 CA16058. We thank Dr. Caifu Chen at Applied Biosystems for his assistance with the TaqMan looped primer assays.
Footnotes
This article contains supplementary material available via the Internet at http://www.interscience.wiley.com/jpages/0020-7136/suppmat/
References
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Niederhuber JE, Brennan MF, Menck HR. The National Cancer Data Base report on pancreatic cancer. Cancer. 1995;76:1671–1677. doi: 10.1002/1097-0142(19951101)76:9<1671::aid-cncr2820760926>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 3.Warshaw AL, Fernandez-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, et al. Micro-RNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 10.Nakahara K, Kim K, Sciulli C, Dowd SR, Minden JS, Carthew RW. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci USA. 2005;102:12023–12028. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 12.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 16.Johnson S, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert K, Brown D, Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 18.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 20.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 21.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 22.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 24.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:E43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 32.Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 33.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes. 2005;19:101–109. doi: 10.1016/j.mcp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 37.Binkley CE, Zhang L, Greenson JK, Giordano TJ, Kuick R, Misek D, Hanash S, Logsdon CD, Simeone DM. The molecular basis of pancreatic fibrosis: common stromal gene expression in chronic pancreatitis and pancreatic adenocarcinoma. Pancreas. 2004;29:254–263. doi: 10.1097/00006676-200411000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 39.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 42.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, Antoniu B, McMahon M, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, et al. Notch mediates TGF α-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 44.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 45.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, et al. Micro-RNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]