Abstract
Actin and tubulin cytoskeletons are conserved and widespread in bacteria. A strikingly intermediate filament (IF)-like cytoskeleton, composed of crescentin, is also present in Caulobacter crescentus and determines its specific cell shape. However, the broader significance of this finding remained obscure, because crescentin appeared to be unique to Caulobacter. Here we demonstrate that IF-like function is probably a more widespread phenomenon in bacteria. First, we show that 21 genomes of 26 phylogenetically diverse species encoded uncharacterized proteins with a central segmented coiled coil rod domain, which we regarded as a key structural feature of IF proteins and crescentin. Experimental studies of three in silico predicted candidates from Mycobacterium and other actinomycetes revealed a common IF-like property to spontaneously assemble into filaments in vitro. Furthermore, the IF-like protein FilP formed cytoskeletal structures in the model actinomycete Streptomyces coelicolor and was needed for normal growth and morphogenesis. Atomic force microscopy of living cells revealed that the FilP cytoskeleton contributed to mechanical fitness of the hyphae, thus closely resembling the function of metazoan IF. Together, the bioinformatic and experimental data suggest that an IF-like protein architecture is a versatile design that is generally present in bacteria and utilized to perform diverse cytoskeletal tasks.
Introduction
Research of the two past decades has revealed that bacterial cells are architecturally surprisingly complex. Bacteria, like eukaryotes, use protein filaments for spatial organization of their cells, and a large variety of cytoskeletal elements has already been identified in bacteria (Graumann, 2007). Best studied of these are the FtsZ-and MreB-family proteins, which are structurally and evolutionarily related to tubulin and actin of eukaryotic organisms. FtsZ and MreB are widespread and conserved in bacteria and fulfil important functions, including spatial orchestration of cell division, growth and morphogenesis [recently reviewed in Michie and Lowe (2006),Cabeen and Jacobs-Wagner (2007),Graumann (2007),Harold (2007),Pichoff and Lutkenhaus (2007),Thanbichler and Shapiro (2008)]. The morphogenetic function of the bacterial cytoskeleton appears to depend greatly on its ability to recruit and spatially organize proteins involved in the synthesis of the cell wall peptidoglycan (PG). PG consists of long glycan strands cross-linked by short peptide side-chains into one huge molecule encasing the cell and functions as an exoskeleton to maintain cell shape and to withstand turgor pressure. At the same time, turgor pressure may act as a driving force in cell wall expansion during growth (Koch, 1985; Harold, 2002). Thus, according to this model, growth and morphogenesis are intimately coupled in bacteria. By allowing cell wall expansion only at certain positions dictated by cytoskeletal structures and driven by turgor pressure (and\or other forces), different shapes can be generated (Cabeen and Jacobs-Wagner, 2007; Harold, 2007). The helical cytoskeletal structure formed in vivo by the bacterial actin MreB defines a cylinder and thus determines the common rod shape of many bacteria (Jones et al., 2001; Kruse et al., 2003; Figge et al., 2004; Dye et al., 2005; Carballido-Lopez, 2006; Carballido-Lopez et al., 2006; Divakaruni et al., 2007; Mohammadi et al., 2007). Ends of the cell cylinder are capped by hemispherical poles, generated by a cell division process guided by the action of bacterial tubulin FtsZ, which forms a constricting ring structure at the cell division site (Bi and Lutkenhaus, 1991; Daniel and Errington, 2003; Varma and Young, 2004; Margolin, 2005; Aaron et al., 2007). Cocci generally do not contain MreB and are made up of two poles created by FtsZ-directed cell division (Jones et al., 2001; Pinho and Errington, 2003). However, the cell shape arsenal of bacteria contains much more than just spheres and straight rods. Obviously, other spatial organizers besides MreB and FtsZ must exist in bacteria to determine more complicated forms. A step further in understanding bacterial morphogenesis was the identification of the intermediate filament (IF)-like protein crescentin in an aquatic bacterium with a characteristic crescent-like cell shape, Caulobacter crescentus (Ausmees et al., 2003). The laterally localized crescentin cytoskeleton converts the FtsZ-and MreB-dependent simple straight rod into a curved or helical rod, depending on the length of the cell (Ausmees et al., 2003). Surprisingly, despite the remarkable architectural and biochemical relatedness of crescentin to IF proteins, a sequence similarity search failed to reveal crescentin homologues in other bacteria. Thus, the additional morphogenetic factors in other MreB\FtsZ-bacteria with complex shapes remain to be uncovered. How general is the above outlined morphogenetic model based on the actions of MreB, FtsZ and additional cytoskeletal elements, such as crescentin? Remarkably, at least one large and important group of bacteria, the actinomycetes, seem to use a different modus operandi for cell growth and morphogenesis that is independent of MreB and FtsZ. These bacteria grow in a polarized fashion: rod-shaped species, such as Mycobacterium and Corynebacterium assemble new cell wall at both cell poles, while filamentous ones, such as Streptomyces, grow by hyphal tip extension analogously to how filamentous fungi grow (Daniel and Errington, 2003; Flärdh, 2003a,Flb; Ramos et al., 2003; Nguyen et al., 2007; Letek et al., 2008). The actinomycetes exhibit extraordinary diversity, regarding their morphology (from simple coccoid to complex branching and multicellular filaments), lifestyle (e.g. pathogenic like Mycobacterium or symbiotic like Frankia), physiology, metabolism (e.g. production of a multitude of secondary metabolites by Streptomyces) and colonization of various environmental niches (Ventura et al., 2007). The coiled coil protein DivIVA seems to have an important role in generation of polarity in these bacteria (Flärdh, 2003a; Letek et al., 2008), but other than that, the molecular mechanisms underlying the diverse morphologies of actinomycetes are relatively poorly understood.
One of the main objectives of the present study was to investigate the incidence of the IF-like function in bacteria, prompted by the intriguing observation that crescentin, the bacterial counterpart of metazoan IF, did not appear to be conserved in bacteria. Was acquisition of crescentin by C. crescentus just an isolated event or might a similar function, perhaps carried by structurally similar proteins albeit with diverging primary sequences, be more common? Here we demonstrate that proteins with a basically IF-or crescentin-like architecture are in fact present and widespread in bacteria. Experimental analysis of a conserved group of such proteins from actinobacteria demonstrated that IF-like biochemical properties accompanied the basic IF-like architecture of these proteins and also revealed a novel cytoskeletal function in actinobacterial growth and morphogenesis. Together our data suggest that an IF-like cytoskeleton, based on a versatile structural element of a segmented coiled coil rod, is more widespread in bacteria than previously thought.
Results
Proteins with a potential segmented coiled coil rod domain are widespread in bacteria
Despite poor sequence conservation all eukaryotic IF proteins share a similar building plan, consisting of a central rod domain of alternating coiled coil segments and linkers, flanked by more globular head and tail domains (Fig. 1, top) (Herrmann and Aebi, 2004). The rod domain is absolutely essential for filament formation (Parry et al., 2007). The bacterial IF-like protein crescentin also possesses an apparent rod domain with a different arrangement of coiled coil segments and linkers (Fig. 1) yet exhibits remarkably IF-like biochemical properties and has a cytoskeletal function (Ausmees et al., 2003). We chose arbitrarily 26 bacterial genomes to survey for the presence of rod-domain proteins among the encoded proteomes (Table 1). A rod domain was defined as a sequence of more than 80 amino acids in coiled coil conformation, positioned either as one continuous block or interrupted by short non-coiled coil sequences. An additional condition was that the candidates should contain no known functional domains, such as a signal sequence, transmembrane segments, enzymatic activity, etc. Out of 26 genomes 21 encoded at least one candidate protein with above mentioned properties, and 16 genomes encoded several (Table 1). One putative crescentin-like rod-domain protein from each genome is schematically shown in Fig. 1. The analysed genomes represent distant phylogenetic groups, suggesting that proteins with a rod-like architecture are widespread among bacteria. Further database searches revealed a conserved rod-domain protein family with members from at least 14 actinomycete species. The hallmark of this family was a pair of clearly conserved sequence motifs at the N-terminal borders of the two first coiled coil segments, regardless of the different lengths of the respective segments and intervening linkers in individual proteins (Fig. 2). These conserved motifs are not discernible in crescentin. However, the basically crescentin-like architecture of the actinomycete proteins and their lack of other known domains suggested that they might similarly have a structural or cytoskeletal function in vivo, so we subjected this family of proteins to further analysis.
Fig. 1.
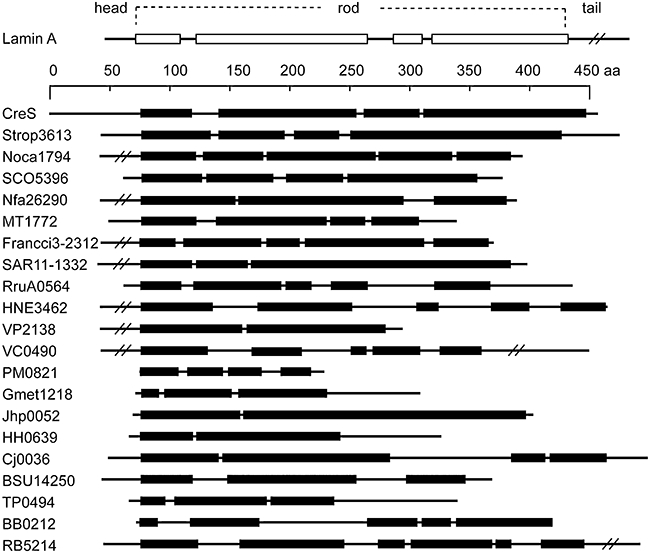
Architectures of bacterial rod-domain proteins. The tripartite building plan of a human IF protein nuclear lamin A is depicted at the top of the figure. The scale bar refers to amino acid residues of crescentin (CreS), the IF-like protein from C. crescentus. Other proteins are drawn in scale, and aligned in respect to their first coiled coil domains. Boxes represent domains in coiled coil conformation and lines non-coiled coil sequences. Long head or tail domains are in some occasions truncated. Designations refer to locus tags of respective proteins from the following species (in order of occurrence): S. tropica, Nocardioides sp. JS614, S. coelicolor, N. farcinica, M. tuberculosis, Frankia sp. Ccl3, P. ubique, R. rubrum, H. neptunium, V. parahaemolyticus, V. cholerae, P. multocida, G. metallireducens, H. pylori, H. hepaticus, C. jejuni, B. subtilis, T. pallidum, B. burgdorferi, R. baltica.
Table 1.
Summary of COILS prediction of 26 bacterial genomes.
| Species | Class | > 100a | > 200a | Rodb |
|---|---|---|---|---|
| S. coelicolor | Actinobacteria | 15 | 6 | 3 |
| S. tropica | Actinobacteria | 11 | 4 | 4 |
| N. farcinica | Actinobacteria | 10 | 5 | 3 |
| Frankia sp. CcI3 | Actinobacteria | 5 | 0 | 2 |
| Nocardioides sp. JS614 | Actinobacteria | 4 | 2 | 2 |
| M. tuberculosis | Actinobacteria | 2 | 1 | 2 |
| R. rubrum | Alphaproteobacteria | 5 | 1 | 2 |
| H. neptunium | Alphaproteobacteria | 5 | 1 | 2 |
| C. crescentus | Alphaproteobacteria | 4 | 2 | 1 |
| Maricaulis maris | Alphaproteobacteria | 3 | 1 | 0 |
| Agrobacterium tumefaciens | Alphaproteobacteria | 3 | 1 | 0 |
| P. ubique | Alphaproteobacteria | 3 | 2 | 2 |
| G. metallireducens | Deltaproteobacteria | 12 | 2 | 1 |
| Desulfovibrio desulphuricans | Deltaproteobacteria | 7 | 0 | 0 |
| H. pylori | Epsilonproteobacteria | 18 | 3 | 4 |
| C. jejuni | Epsilonproteobacteria | 5 | 0 | 5 |
| H. hepaticus | Epsilonproteobacteria | 1 | 0 | 2 |
| V. parahaemolyticus | Gammaproteobacteria | 11 | 1 | 2 |
| P. multocida | Gammaproteobacteria | 10 | 1 | 2 |
| E. coli 536 | Gammaproteobacteria | 8 | 2 | 0 |
| V. cholerae | Gammaproteobacteria | 10 | 2 | 1 |
| B. subtilis str. 168 | Bacillales | 13 | 5 | 1 |
| R. baltica | Planctomycetacia | 22 | 7 | 2 |
| L. interrogans | Spirochaetes | 9 | 2 | 0 |
| B. burgdorferi | Spirochaetes | 7 | 1 | 2 |
| T. pallidum | Spirochaetes | 5 | 2 | 1 |
Number of proteins with more than 100 and 200 amino acid residues in coiled coil conformation encoded in each genome.
Number of proteins with a putative rod domain and no known function encoded in the respective genome.
Fig. 2.
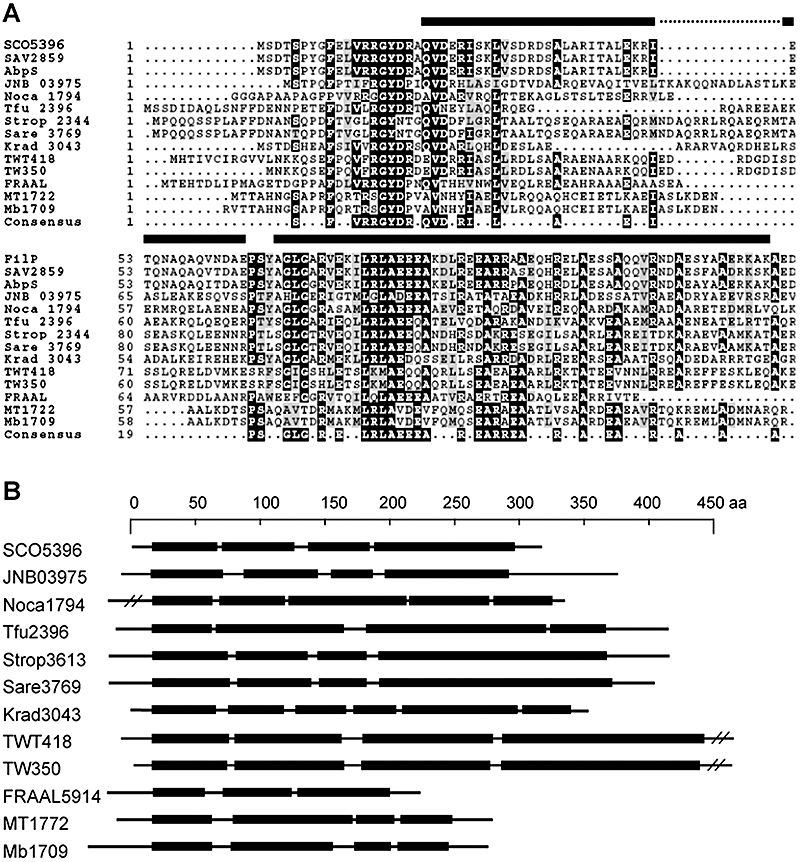
Conserved sequence motifs define a protein family in actinomycetes. A. MULTALIGN alignment (Corpet, 1988) of the N-termini of the FilP-family proteins. AbpS designates the Avicel-binding proteins from S. reticuli. Other proteins are identified by their locus tags shown to the left of the alignment. SCO –S. coelicolor, SAV –S. avermitilis, Krad –Kineococcus radiotolerans, Noca –Nocardioides sp., Tfu –Thermobifida fusca, FRAAL –Frankia alni, JNB –Janibacter sp., TWT – Tropheryma whipplei strain Twist, TW –Tropheryma whipplei strain TW, MT –M. tuberculosis, Mb –M. bovis, Strop –S. tropica, Sare –Salinispora arenicola. Black bars represent the positions of the two first coiled coil domains of SCO5396 (FilP). Multiple alignment was performed with default settings and the output order reflects the relatedness of the input sequences. B. Schematic representation of the various coiled coil architectures of the proteins belonging to the conserved family in (A). Black bars represent coiled coil domains, lines represent non-coiled coil sequences. Long head and tail domains are truncated in some cases.
Rod-domain proteins from actinomycetes form filaments in vitro
A characteristic property of cytoskeletal proteins in general is to form filamentous structures in vitro. Differently from actin and tubulin, IF proteins and crescentin do not require binding of nucleotide or other cofactors but polymerize spontaneously into regular filaments in vitro upon denaturation and subsequent renaturation in physiological buffers (Steinert et al., 1976; Domingo et al., 1992; Ausmees et al., 2003; Parry et al., 2007). Because members of the actinobacterial rod-domain family displayed considerably different arrangements of coiled coil segments and linkers despite conserved sequence motifs (Figs 2B and 3E), we chose three proteins with distinct domain architectures for biochemical analysis. Proteins corresponding to locus tags SCO5396 from Streptomyces coelicolor A3(2), Mb 1709 from Mycobacterium bovis and JNB03975 from Janibacter sp. were purified as recombinant N-terminally polyhistidine-tagged proteins in denatured form and subjected to renaturation conditions. Scanning and transmission electron microscopy revealed that, indeed, all three proteins assembled into filaments, albeit with different morphologies, in buffers with neutral pH and physiological salt concentrations (Fig. 3). It is also known that filament morphology can vary depending on the individual IF protein and buffer conditions (Goulielmos et al., 1996; Geisler et al., 1998; Ausmees et al., 2003). The 35 kDa rod-domain protein from S. coelicolor produced two distinct types of filaments in a 50 mM TrisHCl buffer at pH 7.0. A long smooth and rope-like filament with a diameter of ˜60 nm (Fig. 3A) constituted the less frequent type. The even width and lack of branches indicate that this is a stable polymer, unable to add subunits\filaments laterally. The more abundant type consisted of branching striated filaments of varying diameter (Fig. 3B). The latter filaments visually resembled those formed by nuclear lamins under similar conditions (Stuurman et al., 1998). The relative abundance of smooth and striated filaments did not change significantly in buffers containing 150 mM NaCl or 20 mM MgCl2. The Mb 1709 protein from M. bovis formed a network of laterally aggregating smooth filaments (Fig. 3C), whereas JNB03975 from Janibacter sp. formed smooth filaments with a constant diameter of approximately 40 nm, which did not aggregate laterally (Fig. 3D). Formation of in vitro filaments by three members with distinctly different architectures of the rod domains and belonging to phylogenetically divergent species of Actinobacteria suggests that assembly into regular structures in vitro is a common property of this conserved protein family (Figs 2 and 3). Because S. coelicolor is a genetically tractable model organism for actinomycetes, we chose SCO5396 (hereafter called filP for filament-forming protein) for further in vivo studies.
Fig. 3.
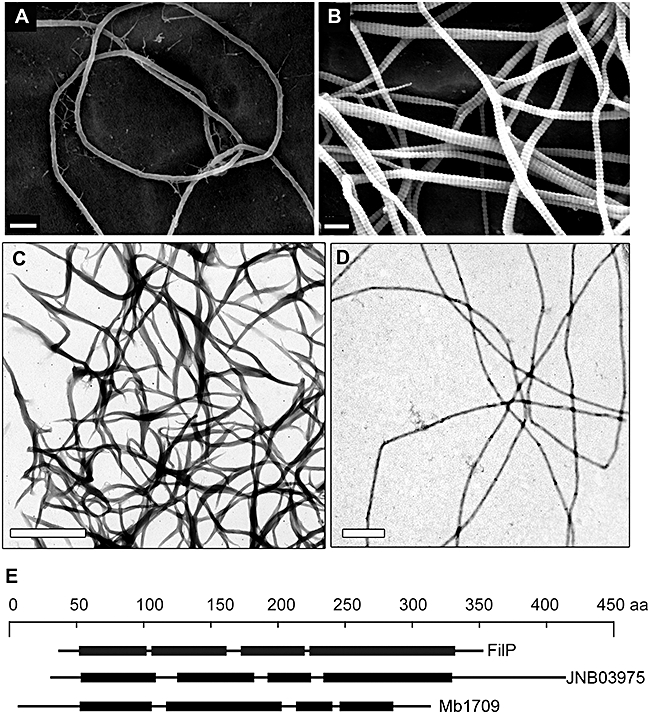
In vitro filaments formed by rod-domain proteins of actinomycetes. A and B. Scanning electron micrographs of filaments formed by S. coelicolor protein SCO5396 (FilP) in 50 mM TrisHCl at pH 7.0. A smooth non-branching filament is shown in A and the striated branching filaments are shown in B. C and D. Transmission electron micrographs of negatively stained filaments formed by Janibacter sp. protein JNB03975 and by M. bovis protein Mb 1709 respectively. E. Schematic representation of the various rod-domain structures of the proteins in A-C. Size bars represent 200 nm for A and B, and 1 μm for C and D.
FilP forms filamentous structures in vivo
Streptomyces coelicolor is a soil-dwelling filamentous and truly multicellular bacterium with a complex lifestyle [reviewed in Chater and Losick (1997),Chater (2000),Flärdh (2003a),Elliot et al. (2007)]. First, a network of apically growing and branching vegetative hyphae invade the growth substrate. In response to exhaustion of the nutrients and after complex signalling, submerged vegetative hyphae give rise to aerial hyphae which break the water–air interface. Aerial hyphae then develop into spore chains and ultimately produce unicellular heat-and desiccation-resistant spores. We fused the enhanced green fluorescent protein (EGFP) to the C-terminus of FilP for its visualization in vivo. Samples for live-cell fluorescence microscopy were withdrawn from developing cultures containing filP-egfp grown for 12, 24, 36 and 48 h to visualize all different cell types: germinating spores, vegetative mycelium, aerial hyphae and spore chains. A filP deletion strain in which filP-egfp was expressed from the native promoter at an ectopic locus in the chromosome (NA446 in Table S1) exhibited prominent fluorescent filamentous structures already in vegetatively growing hyphae (Fig. 4A). We also detected FilP-EGFP filaments in immature and still growing aerial hyphae, but not in mature spore chains in which growth had stopped (data not shown), suggesting that FilP might be involved in normal hyphal growth or germination. However, the filP-egfp strain had a slight morphological defect similar to that of the †filP mutant (see below), suggesting that the FilP-EGFP filaments were not fully functional. A merodiploid filP+\filP-egfp strain (NA282 in Table S1), on the other hand, was morphologically indistinguishable from the wild-type and displayed different spatial organization of FilP-EGFP fluorescence compared with the filP-egfp strain. Redistribution of the FilP-EGFP fluorescence in the presence of the wild-type FilP protein indicates that the tagged and untagged FilP proteins form hybrid structures. Figure 4C shows young vegetative hyphae of the merodiploid strain grown in liquid culture and containing various FilP\FilP-EGFP structures. First, the tip regions of nearly all young hyphae exhibited strong FilP-EGFP fluorescence as a distinct filament and\or as a more diffuse spot (arrowheads in Fig. 4C). This consistent localization to apical areas was not observed in the filP-egfp strain, which produces no wild-type FilP (Fig. 4A). FilP\FilP-EGFP filaments were also present in tip-distal regions. Similar localization was observed in hyphae grown on solid medium (Fig. 4B). Hybrid FilP\FilP-EGFP filaments were often longer (compare Fig. 4A with B) and displayed weaker fluorescence intensity than those of FilP-EGFP. Second, condensed spots or foci were also seen in most hyphae (Fig. 3C). Unfortunately, we were unable to localize the native FilP in the wild-type situation. An attempt to produce a polyclonal anti-FilP antiserum was unsuccessful, and the fusion of a short antigenic FLAG tag to the C-terminus of FilP rendered it partially non-functional. However, the wild-type morphology and growth rate of the merodiploid filP+\filP-egfp strain suggest that the hybrid FilP\FilP-EGFP structures are functional. This closely resembles the in vivo behaviour of tagged and untagged crescentin derivatives. Filaments formed entirely by crescentin-EGFP are short, brightly fluorescent and non-functional. Addition of the wild-type crescentin causes redistribution of crescentin-EGFP into dimmer and slightly discontinuous hybrid crescentin\crescentin-EGFP structures, which stretch from pole to pole along the concave side of the cells and are able to cause cell curvature (Ausmees et al., 2003). Thus, the resemblances in the domain architecture and in vitro properties of FilP and crescentin extend also to their behaviour in vivo.
Fig. 4.
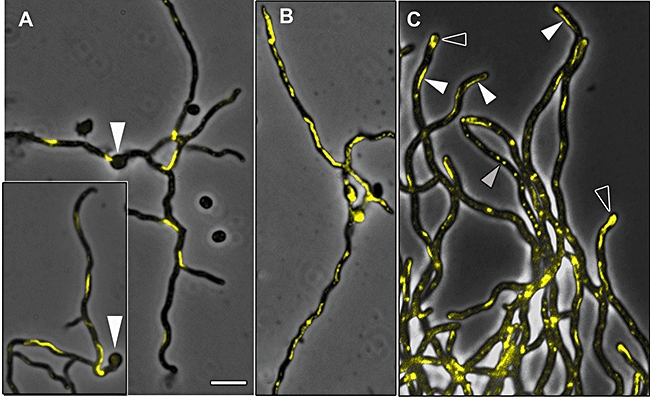
FilP-EGFP forms filamentous structures in growing S. coelicolor hyphae. A–C. Overlays of fluorescence and phase contrast micrographs, FilP-EGFP fluorescence is false coloured yellow. Young vegetative hyphae (12 h) of the filP-egfp strain grown in solid MS agar medium show pronounced filamentous structures of FilP-EGFP (A). White arrowheads mark germinated spores. Young vegetative hyphae (12 h) of the merodiploid filP+\filP-egfp strain are shown in B. The hybrid FilP\FilP-EGFP filaments are longer and display weaker fluorescence than those of FilP-EGFP. Hyphae of the merodiploid filP+\filP-egfp strain grown in liquid YEME medium for 14 h are slightly larger than those grown on solid MS medium and contain several types of fluorescent structures (C). White arrowheads indicate FilP\FilP-EGFP filaments, open arrowheads indicate diffuse apical spots, and grey arrowheads indicate condensed foci. Size bar corresponds to 4 μm in all panels.
We also considered the possibility that FilP might form hybrid structures with other rod-domain proteins SCO3114 and SCO5397 of S. coelicolor in vivo (Table 1). SCO3114 displayed a weak sequence similarity to FilP but lacked the characteristic N-terminal sequence motifs. SCO5397 (a very large coiled coil protein of more than 1300 aa) was not conserved in other bacteria. SCO5397-egfp strain and a merodiploid SCO3114+\SCO3114-mcherry strain (NA360 and NA399 respectively, in Table S1) exhibited wild-type morphology and displayed fluorescence patterns very different from those of the filP-egfp strains. SCO5397-EGFP localized in a weak, indistinct punctate pattern, and SCO3114-mCherry (mCherry is a red fluorescent protein) displayed a relatively strong diffuse signal (Fig. S1). As FilP did not colocalize with other rod-domain proteins of S. coelicolor we assume that a straightforward functional redundancy of these proteins is unlikely, although we cannot exclude some overlap in their respective cellular roles.
filP deletion causes defects in growth and morphology
To address the biological function of the FilP filamentous structures, we studied a strain in which filP was replaced by an apramycin resistance cassette (†filP, NA335 in Table S1). First, the †filP strain had a pronounced growth defect. For example, growth tests in various liquid and solid media showed that after inoculation of an equal number of viable spores, the mutant accumulated only 49–78% of the biomass of the wild-type strain in 30–36 h (data not shown). Total growth is affected by germination efficiency of the spores, the linear growth rate and the branching frequency of the hyphae. Growth curves (Fig. S2) indicated that biomass accumulation started with a delay and proceeded more slowly in the †filP strain compared with wild-type in a liquid medium. As we did not observe overt differences in the frequency of branching between the †filP and the wild-type strains, we assumed that a combination of delayed germination and slower linear growth was responsible for the total growth defect of the †filP strain. Slower growth of the mutant on solid media was also evident, although normal amounts of dark grey spores were finally produced (approximately a day later than the wild-type), indicating that sporulation was not significantly affected. Furthermore, vegetative hyphae of the mutant showed a characteristic distorted morphology when grown in the angle between an inserted coverslip and the surface of the solid agar medium for microscopic observation (Fig. 5B). Wild-type hyphae grew in a straight fashion under the same conditions (Fig. 5A, see also Fig. S3). A similar difference in morphology between the strains was observed also in liquid cultures (data not shown), thus ruling out the possibility that the apparent morphological differences were caused by different surface adhesion properties of the two strains. Aerial hyphae of the †filP strain, however, did not display any striking morphological defects. Both the morphological and growth defects of the †filP strain were complemented by addition of wild-type filP in trans, confirming that the mutant phenotypes were caused by the lack of filP (data not shown).
Fig. 5.
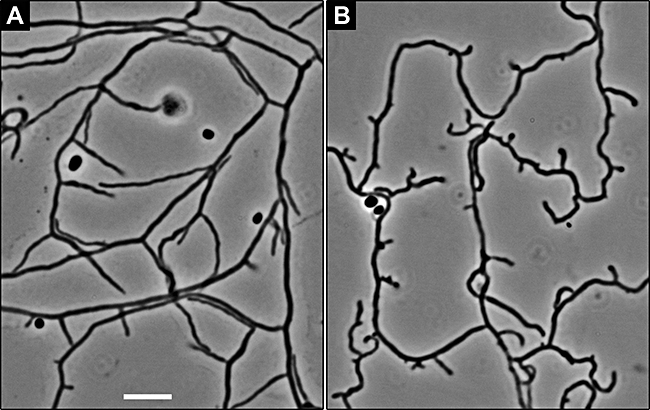
Deletion of filP causes a morphological defect. A and B. Live hyphae of the wild-type strain M145 (A) and the †filP mutant strain (B) grown for 20 h in the angle between an inserted coverslip and the MS agar surface and visualized by phase contrast microscopy.
Atomic force microscopy shows that filP is needed for normal rigidity of the hyphae
The PG sacculus of bacteria is the most important cellular component to maintain cell shape and integrity, and many mutations affecting assembly of PG are known to cause altered cell morphology. Thus, we speculated that the morphological defect of the †filP hyphae is perhaps caused by a weakened PG exoskeleton, which renders the hyphae less rigid and is unable to elongate in a straight fashion typical to those of the wild-type. To test this, we probed living vegetative hyphae of the wild-type and the †filP strains by atomic force microscopy (AFM). First, topological images were collected of both strains in a completely hydrated state by using the contact imaging mode (Fig. 6A and B and Fig. S3). Interestingly, †filP hyphae seemed to yield more under the tip pressure and shear forces, as compared with wild-type hyphae. Height profiles in Fig. 6A show significant distortion of the †filP hyphae, especially in the parts lying perpendicular to the fast scan direction (see Supporting information text for additional information). In contrast, the wild-type hyphae remained undistorted and exhibited uniform height and width (Fig. 6B). This indicates that †filP hyphae are more deformable and ‘softer’ than those of the wild-type. Further, to measure the elastic properties of the hyphae, we recorded force curves as the tip of the cantilever approached, indented the hyphal surface for about 100 nm and then retracted. Force volume (an array of force curves) was recorded over several areas containing apical parts of wild-type or †filP hyphae. Figure 6C displays an image reconstructed using compliance values calculated from an 80 × 80 array of force curves over a tip of a wild-type hypha. Compliance is the inverse of stiffness and the latter is defined as dFdI-1 with F being the force and I being the indentation. Because of the pyramidal shape of the tip the edges of a cell experience deformation simultaneously in downwards and sidewise direction, which results in high apparent compliance. To avoid edge effects 50 force curves along a line running parallel to the long axis of the hyphae in the highest part of the hyphal cylinder were used for analysis, resulting in average compliance values of 34.4 m N-1 and 44.1 m N-1 for wild-type and †filP hyphae respectively. Single typical force-separation curves of †filP, and wild-type hyphae are shown in Fig. 6D. The steeper slope of the wild-type force curve implies lower compliance of the wild-type hypha compared with the †filP hypha. As both hyphae were indented by the same cantilever, this indicates that the hyphae of the wild-type strain are stiffer than those of the †filP strain. Another significant observation is that the ‘load’ and ‘unload’ curves match reproducibly over the entire surface of the wild-type hyphae (Fig. 6D). This indicates that the wild-type hyphae undergo a fully reversible elastic deformation and revert to their original shape as the tip retracts. On the other hand, the †filP hyphae seemed to suffer a more persistent deformation, indicated by the significant hysteresis effect consistently present in all force curves (manifested as a loop between the load and unload curves as the cells demonstrate a delayed elastic recovery after deformation, Fig. 6D). This is in agreement with other AFM data and can be explained by a softer cell surface and\or by lower turgor pressure of the †filP hyphae. Thus, our data indicate that the FilP cytoskeleton affects the mechanical properties of the cells. This is consistent with the current model of bacterial morphogenesis, according to which cytoskeletal elements control mechanical properties of the cell by spatially orchestrating the deposition of the stress-bearing exoskeleton. Nevertheless, this to our knowledge the first time the impact of bacterial cytoskeleton has been demonstrated by direct probing of the physical properties of the cells.
Fig. 6.
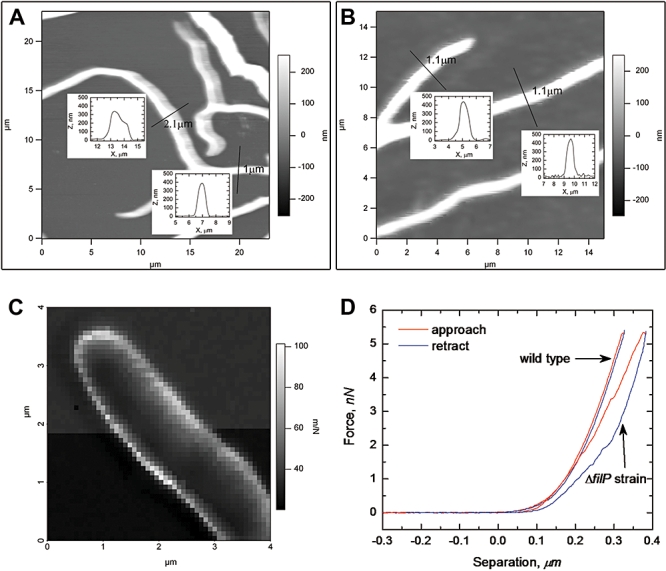
Deletion of filP causes changes in the visco-elastic properties of S. coelicolor cells. A and B. Topological images of live hyphae of the wild-type M145 (B) and the †filP strain (A) grown as in Fig. 4. Insets show height profiles of the hyphae at positions marked with perpendicular lines, indicating that mutant hyphae undergo deformation in the imaging process, whereas wild-type hyphae maintain their form. C. An image of a tip of a wild-type hypha reconstructed using data from 80 × 80 force curves. Each pixel corresponds to a compliance value calculated from an individual force curve and depicted in greyscale, as shown by a scale bar to the right. Thus, areas of the sample characterized by dark pixels are relatively stiff (low compliance), and areas with lighter pixels are softer (high compliance). D. Single representative force curves of wild-type and †filP hyphae showing that wild-type hyphae are less compliant than those of the mutant. The hysteresis effect (manifested as a loop between the load and unload curves) was consistently present in all force curves of the mutant hyphae, and absent in those of the wild-type.
Discussion
Diversity of putative IF-like proteins and cytoskeletal functions in bacteria
One of the objectives of this study was to address whether the IF-like cytoskeleton is confined to only one bacterial species or if it might be a more widespread phenomenon. The latter notion was evoked by the high degree of diversity displayed by the sequences of IF proteins. For example, there is only approximately 30% sequence identity between the rod domains of human and fruit fly lamins, and 27% between two human IF proteins lamin A and cytokeratin 18. Also, individual IF perform quite different functions and are present in different cell types. This is in strong contrast with the strict conservation of the sequences and functions of the actin and tubulin proteins. Apparently, the basic structure of IF proteins, consisting of a segmented coiled coil rod (supports assembly into cytoskeletal filaments) flanked by the head and tail domains (accommodate additional functional motifs), is a versatile design and can be adapted to various cellular functions. This could be a reason why the evolution of IF is less constrained than that of actin and tubulin. Thus, to address our objective, we first estimated the occurrence of the basic IF architecture among bacterial proteins. Our analysis suggests that proteins with a segmented coiled coil rod domain are present in many diverse species (Figs 1 and 2). However, a cytoskeletal function has to be shown experimentally in each case. We chose a protein (FilP) from a conserved actinobacterial rod-domain family and demonstrated that it indeed performed a cytoskeletal function in S. coelicolor. Interestingly, members of that sequence-wise conserved actinomycete protein family display different architectures of the segmented rod domain (Fig. 2). Nevertheless, three tested members, representing different architectures, were able to form regular filaments in vitro. This supports the idea of a versatile coiled coil rod, which does not have to conform to a strictly defined sequence and architecture to support filament formation and a cytoskeletal function. Rather, the specific architecture is probably tailored for the specific function of each protein. The common occurrence of rod domains in bacteria together with our experimental data suggest that diverse bacteria may possess a filament system composed of proteins sharing the basic design of IF elements. However, it is not clear whether these proteins originate from a common ancestor or have been evolved independently.
Our analysis was restricted to previously uncharacterized rod-domain proteins without known functional domains in order to identify crescentin-like cytoskeletal elements. However, there are also examples of proteins in which rod domains are coupled to a known function. For example, rod-domain proteins TlpA from a Salmonella enterica virulence plasmid (Hurme et al., 1994) and Scc from Leptospira interrogans (Mazouni et al., 2006) were both shown to form filaments in vitro and in vivo in Escherichia coli cells. A common feature of these otherwise very different proteins is that both also bind DNA. Another example is the AglZ protein from Myxococcus xanthus which carries a filament-forming coiled coil module coupled to a response regulator receiver domain (Yang et al., 2004). Very recently, a short (only 81 aa long) coiled coil protein ZapB was also shown to form striated filaments in vitro and have a role in FtsZ-ring assembly and cell division in E. coli (Ebersbach et al., 2008). Thus, filament formation via coiled coil rod domains might be relevant in various functional contexts in a bacterial cell.
FilP cytoskeleton in cell growth and morphology
The second goal was to characterize the novel cytoskeletal function identified in this study, performed by an S. coelicolor rod-domain protein FilP. We have shown that a filP mutant was affected in growth and morphology, indicating that FilP has a role in both processes. The observation that filP mutant hyphae were more compliant under direct mechanical probing with AFM suggests that the softness of the cells might lie behind both defects – the distorted morphology and the slower growth. One can envision that mechanical forces of the environment, such as turbulence in liquid or roughness of the solid substrate, would distort the softer †filP hyphae to a greater extent than the more rigid wild-type hyphae. Thus, frequent change of growth direction might cause the undulating morphology of the mutant hyphae, while the wild-type hyphae are able to grow in a straight fashion in similar conditions (Fig. 5). It has previously been shown that externally applied mechanical constraints induce long-persisting changes in bacterial cell shape. For example, E. coli cells forced to grow as cell filaments in circular microchambers retained a bent cell shape upon release into a liquid medium even after growth and cell division (Takeuchi et al., 2005). Reduced rigidity might also cause less efficient tip elongation of the †filP strain and explain its slower accumulation of biomass.
Rigidity of the bacterial cells is mainly determined by the cell wall and by turgor pressure. At present we cannot convincingly determine whether the increased compliance of the †filP strain is due to changes in the cell envelope structure or lower turgor pressure. As discussed above, the strategic site of new cell wall synthesis in S. coelicolor is the tip of a growing or emerging hypha, where also a key protein in this process, DivIVA, localizes (Flärdh, 2003a,b). Interestingly, the tip regions of growing hyphae of the merodiploid filP+\filP-egfp strain consistently contained a strong fluorescence signal of FilP-EGFP evident as a filamentous structure, an apical spot or both (Fig. 4B and C). Together, the presence of FilP within the zone of active growth and the abnormal softness of the cells in the absence of FilP suggest that it might have a role, although nonessential, in cell wall synthesis or maturation. Another possibility is that FilP somehow changes the composition or permeability of the cell membrane and by that affects turgor pressure and thus the rigidity. Furthermore, it cannot be excluded that FilP structures might act as a mechanical support in resemblance with the IF network in mammalian cells. Cell wall expansion in bacteria involves lytic activity in order to break already existing cross-links and incorporate new PG precursors. Thus, in the growing region additional mechanical support might be needed to compensate for the relative weakness of the exoskeleton. A similar function has very recently been proposed for a cellulose synthase-like protein CslA of S. coelicolor (Xu et al., 2008). CslA localized to the hyphal tips and was responsible for the accumulation of a β-glycan polysaccharide. The authors proposed that this polysaccharide, most likely cellulose, acts as a bandage to reinforce the growing tips of the hyphae. Interestingly, a FilP orthologue from S. reticuli has previously been shown to bind Avicel, a derivative of cellulose and was therefore named AbpS (Avicel-binding protein) (Walter et al., 1998). The authors also showed that AbpS oligomerizes (Walter and Schrempf, 2003), which agrees with our data showing that FilP forms filaments in vitro and in vivo. We assume that FilP and other proteins of the family might share the cellulose-binding ability of AbpS. As FilP is also present at the hyphal tips and in the subapical regions, it must at least partially colocalize with CslA. Future studies might clarify the significance of the cellulose-binding ability of FilP and the interaction network between the proteins populating the tip regions of elongating hyphae.
The apparent involvement of FilP in determining the mechanical properties of S. coelicolor hyphae relates to the functions of metazoan IF. Although IF have become more and more implicated in signalling, protein targeting and other dynamic cellular processes, one important aspect of their functions is mechanical, providing structural support to the cells and dealing with mechanical stress (Chou et al., 2007; Kim and Coulombe, 2007; Pekny and Lane, 2007). Indeed, several disease-causing mutations in genes encoding IF-proteins have been shown to, directly or indirectly, lead to increased fragility of affected cells and tissues (Omary et al., 2004; Herrmann et al., 2007). Similarly, the compromised rigidity of the †filP strain might render it more susceptible to the mechanical stresses inflicted by the environment and affect survival and fitness in nature. The presence of FilP-like proteins in other actinomycetes and in vitro filament formation by FilP-family proteins from Janibacter sp. and M. bovis suggest that a similar IF-like cytoskeletal function is conserved. Janibacter and Mycobacterium species display slightly irregular rod shape and are also known to adopt different morphologies under certain conditions (Martin et al., 1997; Ojha et al., 2000). It would be interesting to study the cellular roles of FilP-family proteins in non-filamentous bacteria. Further research is also needed to understand the details of FilP localization, the relevance of its different cellular structures and whether FilP shares the property of other cytoskeletal elements, such as MreB and FtsZ, to act as a spatial organizer of cell wall construction. To this end, we believe that our novel application of AFM offers a useful tool to address the roles of various cytoskeletal elements in the important task of building up a bacterial cell.
Experimental procedures
Bacterial strains and media
The S. coelicolor A3(2) and E. coli strains used in this work are listed in Table S1. Cultivation of E. coli strains was performed as described in (Sambrook et al., 1989). S. coelicolor strains were grown on mannitol soy flour agar plates (MS agar), in yeast extract-malt extract medium (YEME), in tryptone soy broth or on R2YE agar (Kieser et al., 2000).
Construction of plasmids and recombinant S. coelicolor strains
The plasmids used are listed in Table S1. DNA manipulation and cloning were carried out according to standard protocols (Sambrook et al., 1989). DNA fragments to be cloned into plasmid vectors were created by PCR, and final constructs were verified by DNA sequencing. Sequences of primers used for cloning and construction of mutants and exact cloning strategies are available upon request. The PCR-targeting procedure was used for generation of filP gene knock-out mutant (NA335) essentially as described in Gust et al. (2003) (see also Table S1). To generate the merodiploid filP+\filP-egfp strain (NA282), the entire filP gene with 526 bp upstream region was first fused in frame to egfp resulting in plasmid pNA859. S. coelicolor protoplasts were then transformed with pNA859, followed by selection of transformants where homologous recombination via a single crossing-over event had created a full-length recombinant fusion allele in the wild-type locus under control of the native promoter, as well as a second full-length copy of the gene. A similar strategy was used to create the merodiploid SCO3114+\SCO3114-mcherry strain NA399. A derivative of the cloning vector pBluescript, containing an apramycin resistance cassette and full-length SCO3114 fused to mcherry (Shaner et al., 2004), was used for transformation of S. coelicolor protoplasts. SCO5397-egfp strain (NA360) was constructed by insertion of pEGFP-N2 encoding a C-terminal part of SCO5397 fused to EGFP into the chromosome of M145 by homologous recombination. For generation of the filP-egfp strain (NA446) the insert from pNA859 encoding FilP-EGFP was subcloned into the vector pIJ82 and by conjugation introduced into strain NA335 (†filP) where it integrated in the chromosome at the phage ΦC31 attachment site. Mutant strains were verified by diagnostic PCR.
Database searches
Genome sequences of C. crescentus CB15 (AE005673.1), Salinispora tropica CNB-440 (NC009380), Nocardioides sp. JS614 (NC008699), S. coelicolor A3(2) (NC003888), Nocardia farcinica IFM 10152 (NC006361), Mycobacterium tuberculosis CDC1551 (NC002755), Frankia sp. Ccl3 (NC007777), Pelagibacter ubique HTCC1062 (NC007205), Rhodospirillum rubrum (NZAAAG00000000), Hyphomonas neptunium ATCC 15444 (NC008358), Vibrio parahaemolyticus RIMD 2210633 (NC004603), Vibrio cholerae (NC002505, NC002506), Pasteurella multocida (NC002663), Geobacter metallireducens (NZAAAS00000000), Helicobacter pylori J99 (NC000921), Helicobacter hepaticus ATCC51449 (NC004917), Campylobacter jejuni (NC002163), Bacillus subtilis strain 168 (NC000964), Treponema pallidum (NC000919), Borrelia burgdorferi (NC001318), Rhodopirellula baltica SH1 (NC005027) were analysed with the COILS algorithm (Lupas, 1997) as described in Ausmees et al. (2003). The coiled coil domain architecture was then determined individually for all proteins containing more than 80 amino acid residues in coiled coil conformation using different settings of the COILS prediction. Proteins exhibiting a potential rod-domain architecture were further analysed with various publicly available bioinformatic tools to detect conserved domains and putative functions.
In vitro filament formation and electron microscopy
N-terminally polyhistidine-tagged proteins (FilP, JNB03975 from Janibacter sp. and Mb 1709 from M. bovis) were expressed from the pET-28a(+) vector in E. coli BL21(DE3) and purified using the Ni-NTA His-Bind® Resin (Novagen) under denaturing conditions (8 M urea) as described in the user manual. To induce filament formation, protein samples were dialysed against a buffer containing 10 mM Tris-HCl with 150 mM NaCl at pH 7.0. Dialysis was performed overnight at 4°C or for 2 h at room temperature with two bath changes. The dialysed samples were then prepared for scanning (SEM) or transmission (TEM) electron microscopy. For SEM the samples were prefixed with 2.5% glutaraldehyde, washed three times in phosphate-buffered saline and fixed in 1% osmium tetraoxide. After dehydration in ethanol the samples were injected through nucleopore filters with 0.2 μm pore size, critical-point dried, mounted on Cambridge alloy Stubbs, silver sputtered and examined in a Zeiss Supra 35-VP field emission SEM equipped with a STEM detector, EDAX Genesis 4000 EDS. For TEM, the dialysed protein samples were applied on carbon-coated and glow discharged grids, negatively stained with 1% uranyl acetate and observed in Hitachi H7100 transmission electron microscope.
Light microscopy
Samples for phase contrast and fluorescence microscopy were obtained by growing the strains in liquid medium or in the angle between an inserted coverslip and the agar surface (Kieser et al., 2000). EGFP fluorescence was observed directly after mounting the coverslips or a drop of liquid culture to a glass slide covered with 1% agarose in phosphate-buffered saline. All fluorescence and phase-contrast microscopy was performed using an Axioplan II imaging fluorescence microscope equipped with appropriate filter sets, an Axiocam charge-coupled device camera and Axiovision software (Carl Zeiss Light Microscopy). Digital images were processed using Axiovision and Adobe Photoshop CS version 8.0 software.
Atomic force microscopy
Atomic force microscopy of S. coelicolor cells was carried out with the MFP3D instrument (Asylum Research) in contact mode in fluid. Cells were hydrated in water during all imaging and force volume measurements. The water reached a maximum temperature of 30°C as monitored with an IR-thermometer. All imaging and force volume measurements were performed using gold coated silicon nitride probes (MLCT-AU) with measured spring constant of 0.05 N m-1. A variety of surfaces were tested for culturing cells, including glass, polylysin treated glass, Au-Pd-coated glass and MPS coated glass. The best cell-to-surface adherence was obtained using freshly cleaved muscovite mica (SPI supplies) glued to the glass coverslips. Mica is the surface used as support for all the AFM measurements reported in this paper.
Acknowledgments
We are grateful to Steven Giovannoni and Jaydip Ghosh for gifts of Janibacter sp. and M. bovis chromosomal DNA respectively. We also thank Klas Flärdh and Gerhart Wagner for critical reading and helpful comments on the manuscript, as well as Magnus Östberg and Olov Svartström for their contribution in recombinant protein purification. We are also deeply grateful to Stefan Vinzelberg (Atomic Force F and E GmbH) for providing the FV software code and his most valuable help and discussions. This work was supported by Carl Trygger Foundation (HT and NA) and the Swedish Research Council (NA). LB acknowledges financial support from Knut and Alice Wallenberg Foundation, Swedish Foundation for Strategic Research and Swedish Research Council.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol. 2007;179:381–387. doi: 10.1083/jcb.200708001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R. The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homologue MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Chater KF. Developmental decisions during sporulation in the aerial mycelium in Streptomyces. In: Brun YV, Shimkets LJ, editors. Prokaryotic Development. Washington, DC: American Society for Microbiology Press; 2000. pp. 33–48. [Google Scholar]
- Chater KF, Losick R. Multicellular lifestyle of Streptomyces coelicolor A3 (2) and its relatives. In: Shapiro JA, Dworkin M, editors. Bacteria as Multicellular Organisms. New York, NY: Oxford University Press; 1997. pp. 149–182. [Google Scholar]
- Chou YH, Flitney FW, Chang L, Mendez M, Grin B, Goldman RD. The motility and dynamic properties of intermediate filaments and their constituent proteins. Exp Cell Res. 2007;313:2236–2243. doi: 10.1016/j.yexcr.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Domingo A, Sarria AJ, Evans RM, Klymkowsky MW. Studying intermediate filaments. In: Carraway KL, Carraway CAC, editors. The Cytoskeleton; a Practical Approach. Oxford: IRL Press; 1992. pp. 223–255. [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Galli E, Moller-Jensen J, Lowe J, Gerdes K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol. 2008;68:720–735. doi: 10.1111/j.1365-2958.2008.06190.x. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Buttner MJ, Nodwell JR. Multicellular development in Streptomyces. In: Kaplan HB, Whitworth DE, editors. Multicellularity and Differentiation Among the Myxobacteria and Their Neighbors. Washington, DC: American Society for Microbiology Press; 2007. [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3 (2) Mol Microbiol. 2003a;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- Flärdh K. Growth polarity and cell division in Streptomyces. Curr Opin Microbiol. 2003b;6:564–571. doi: 10.1016/j.mib.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Geisler N, Schunemann J, Weber K, Haner M, Aebi U. Assembly and architecture of invertebrate cytoplasmic intermediate filaments reconcile features of vertebrate cytoplasmic and nuclear lamin-type intermediate filaments. J Mol Biol. 1998;282:601–617. doi: 10.1006/jmbi.1998.1995. [DOI] [PubMed] [Google Scholar]
- Goulielmos G, Gounari F, Remington S, Muller S, Haner M, Aebi U, Georgatos SD. Filensin and phakinin form a novel type of beaded intermediate filaments and coassemble de novo in cultured cells. J Cell Biol. 1996;132:643–655. doi: 10.1083/jcb.132.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold FM. Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet Biol. 2002;37:271–282. doi: 10.1016/s1087-1845(02)00528-5. [DOI] [PubMed] [Google Scholar]
- Harold FM. Bacterial morphogenesis: learning how cells make cells. Curr Opin Microbiol. 2007;10:591–595. doi: 10.1016/j.mib.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Hurme R, Namork E, Nurmiaho-Lassila EL, Rhen M. Intermediate filament-like network formed in vitro by a bacterial coiled coil protein. J Biol Chem. 1994;269:10675–10682. [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood D. Practical Streptomyces Genetics. Norwich: The John Innes Foundation.; 2000. [Google Scholar]
- Kim S, Coulombe PA. Intermediate filament scaffolds fulfill mechanical, organizational, and signalling functions in the cytoplasm. Genes Dev. 2007;21:1581–1597. doi: 10.1101/gad.1552107. [DOI] [PubMed] [Google Scholar]
- Koch AL. How bacteria grow and divide in spite of internal hydrostatic pressure. Can J Microbiol. 1985;31:1071–1084. doi: 10.1139/m85-204. [DOI] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letek M, Ordonez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol. 2008;190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Schumann P, Rainey FA, Schuetze B, Groth I. Janibacter limosus gen nov., sp. nov., a new actinomycete with meso-diaminopimelic acid in the cell wall. Int J Syst Bacteriol. 1997;47:529–534. doi: 10.1099/00207713-47-2-529. [DOI] [PubMed] [Google Scholar]
- Mazouni K, Pehau-Arnaudet G, England P, Bourhy P, Saint Girons I, Picardeau M. The scc spirochetal coiled-coil protein forms helix-like filaments and binds to nucleic acids generating nucleoprotein structures. J Bacteriol. 2006;188:469–476. doi: 10.1128/JB.188.2.469-476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol. 2007;189:7896–7910. doi: 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha AK, Mukherjee TK, Chatterji D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun. 2000;68:4084–4091. doi: 10.1128/iai.68.7.4084-4091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N Engl J Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Pekny M, Lane EB. Intermediate filaments and stress. Exp Cell Res. 2007;313:2244–2254. doi: 10.1016/j.yexcr.2007.04.023. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Overview of cell shape: cytoskeletons shape bacterial cells. Curr Opin Microbiol. 2007;10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol. 2003;50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- Ramos A, Honrubia MP, Valbuena N, Vaquera J, Mateos LM, Gil JA. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology. 2003;149:3531–3542. doi: 10.1099/mic.0.26653-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Idler WW, Zimmerman SB. Self-assembly of bovine epidermal keratin filaments in vitro. J Mol Biol. 1976;108:547–567. doi: 10.1016/s0022-2836(76)80136-2. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–1823. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. Getting organized – how bacterial cells move proteins and DNA. Nat Rev Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]
- Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Schrempf H. Oligomerization, membrane anchoring, and cellulose-binding characteristics of AbpS, a receptor-like Streptomyces protein. J Biol Chem. 2003;278:26639–26647. doi: 10.1074/jbc.M212792200. [DOI] [PubMed] [Google Scholar]
- Walter S, Wellmann E, Schrempf H. The Cell Wall-Anchored Streptomyces reticuli Avicel-Binding Protein (AbpS) and Its Gene. J Bacteriol. 1998;180:1647–1654. doi: 10.1128/jb.180.7.1647-1654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chater KF, Deng Z, Tao M. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J Bacteriol. 2008;190:4971–4978. doi: 10.1128/JB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Bartle S, Otto R, Stassinopoulos A, Rogers M, Plamann L, Hartzell P. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J Bacteriol. 2004;186:6168–6178. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


