Abstract
New DNA sequencing technologies can sequence up to one billion bases in a single day at low cost, putting large-scale sequencing within the reach of many scientists. Many researchers are forging ahead with projects to sequence a range of species using the new technologies. However, these new technologies produce read lengths as short as 35–40 nucleotides, posing challenges for genome assembly and annotation. Here we review the challenges and describe some of the bioinformatics systems that are being proposed to solve them. We specifically address issues arising from using these technologies in assembly projects, both de novo and for resequencing purposes, as well as efforts to improve genome annotation in the fragmented assemblies produced by short read lengths.
New technologies: more data and new types of data
The ongoing revolution in sequencing technology has led to the production of sequencing machines with dramatically lower costs and higher throughput than the technology of just 2 years ago. Sequencers from 454 Life Sciences/Roche, Solexa/Illumina and Applied Biosystems (SOLiD technology) are already in production, and a competing technology from Helicos should appear soon. However, the increase in the volume of raw sequence that can be produced from these sequencers is threatening to swamp our available data archives, because genomics centers are gearing up to produce much more data in the next several years. For example, major National Institutes of Health (NIH) sequencing centers are planning to sequence 100 complete human genomes in the next 2–3 years [1]. Furthermore, the increased throughput of the new sequencing machines makes it possible for biologists to sequence large numbers of bacterial strains and isolates, leading some microbiologists to suggest that we characterize the genomes of all organisms present in culture collections.
These technologies greatly increase sequencing throughput by laying out millions of DNA fragments on a single chip and sequencing all these fragments in parallel. The various technologies differ in the procedures used to array the DNA fragments: 454 and Applied Biosystems first attach the DNA to coated beads, whereas Solexa and Helicos attach the DNA directly to the chip. (For a more detailed description of these technologies, see the companion article by Mardis [2].)
As sequence production is increasing, however, a major difference between the new technologies and the old, ‘Sanger’ sequencing has not yet been addressed. Succinctly put, this difference is one of quality: DNA sequences (‘reads’) produced by the new technologies are much shorter than sequences produced by capillary sequencers such as the ABI 3730xl. Capillary sequencers produce reads up to 900 bp in length, whereas 454 sequencer reads are 250 bp and Illumina reads are 35 bp. Further difficulties arise because of the unavailability of paired-end reads, although limited forms of paired-end sequencing are just becoming available. These features of the new technologies present major bioinformatics challenges, particularly for genome assembly. The short read lengths and absence of paired ends make it difficult for assembly software to disambiguate repeat regions, therefore resulting in fragmented assemblies (Figure 1). Nevertheless, the sequences are coming, and the bioinformatics community needs to act quickly to keep pace with the expected flood of raw sequence data. In this review, we describe the challenges facing those who use genome assembly and annotation software and review the initial efforts to develop new bioinformatics software for short read sequencing (SRS) technology.
Figure 1.
Effect of repeats on long and short read assemblies. The middle section of the figure represents a set of repeats longer than 30 bp within a 50-kbp region of the Yersinia pestis CO92 genome. The blue (red) boxes represent repeats longer (shorter) than 800 bp. An assembly of this region using Sanger data would result in the set of contigs represented at the top, correctly resolving the short repeats and only breaking at the boundaries of long repeats. Furthermore, paired-ends (indicated by thin lines connecting the contigs) would provide long range connectivity across repeats. The assembly generated from short read sequencing data (bottom line) is considerably more fragmented, breaking at all repeat boundaries, and lacking long range connectivity caused by the absence of paired-end data.
Sequence assembly using SRS technology
The development of automated sequencing technologies has revolutionized biological research by allowing scientists to decode the genomes of many organisms. SRS technologies can accelerate the pace at which we explore the natural world, yet pose new challenges to the software tools used to reconstruct genetic information from the raw data produced by sequencing machines.
Genome resequencing
The near completion of a reference human genome has greatly accelerated research on genetic diversity within our species. Resequencing efforts have thus far targeted individual genes or other genomic regions of interest [3], but advances in SRS technologies have opened up the possibility of whole genome resequencing. The resequencing of multiple strains of several model organisms (e.g. Drosophila melanogaster and Caenorhabditis elegans) and the large-scale resequencing of human cancers are currently underway. Any resequencing effort requires that the reads are sufficiently long to be mapped accurately onto the genome. The mapping process must efficiently handle the millions of sequences generated while being robust in the presence of sequencing errors and polymorphisms. Although existing sequence alignment tools such as Blast [4] or Blat [5] can handle this mapping [6,7], several new programs have been developed that are designed specifically for SRS data, including Illumina’s Eland short-read aligner and the PET-Tool of Chiu et al. [8] for ditag transcriptome sequencing. The choice of alignment tool also depends on the types of polymorphisms being studied. For example, Dahl et al. [6] studied sequence variation within cancer genes, relying on Blast for the identification of single nucleotide polymorphisms (SNPs) and short indels, whereas a survey of structural variants in the human genome by Korbal et al. [9] required customized software. The latter study involved the use of paired 454 reads to identify large scale differences (large indels and inversions) between individual human genomes. The mapping process alone required 200 000 CPU hours, further underscoring the need for efficient mapping algorithms.
Resequencing is also widely used in microbial ecology studies, where population structure is inferred from the sequence of the short subunit of the rRNA (16S rRNA). Sogin et al. [10] used 454’s SRS technology in a survey of microbial diversity within the ocean, targeting the V6 hypervariable region of the 16S rRNA. To reduce the effect of sequencing errors, they removed reads that did not perfectly match the PCR primer, were too short or contained ambiguous base-calls (nucleotide N). In total, 24% of the sequences were removed before analysis, showing the need for a better understanding of the error characteristics of short read data.
Studies of pyrosequencing data generated by 454 Life Sciences machines [11,12] have characterized several types of errors commonly encountered in these data, including incorrect estimates of homopolymer lengths, ‘transposition-like’ insertions (a base identical to a nearby homopolymer is inserted in a nearby nonadjacent location) and errors caused by multiple templates attached to the same bead. Furthermore, whereas the quality values associated with base-calls generally correlate with error rates, they are overly pessimistic in homopolymers [11] and generally not as reliable as the quality values for Sanger sequencing data. Together these results suggest that SRS data cannot be analyzed with software developed for Sanger data (such as polyphred [13] or polyscan [14]) and that any polymorphisms identified through resequencing must be carefully analyzed to rule out technology-specific error patterns.
Assembly of closely related species – mind the gap
Genome scientists have sequenced and assembled the human genome, most model organisms, and almost all major human pathogens to high degrees of accuracy. Many of these genomes – particularly the bacterial and viral species – have been finished, meaning that all chromosomes are sequenced end-to-end with no gaps. Almost as soon as the first genome from each species was published, scientists started to make plans to sequence additional strains and isolates. The dramatically lower cost of sequencing using SRS technology has accelerated plans to sequence additional strains of already-sequenced genomes. To take just one example, scientists at the Broad Institute are sequencing multiple strains of Mycobacterium tuberculosis to better understand the mechanisms of antibiotic resistance in this organism [15]. Although few publications have yet appeared, projects are in progress or planned to sequence hundreds of bacterial species and several larger eukaryotic genomes.
If recent history is a guide, most of the resequencing projects will initially focus on bacteria, just as most of the genome projects of the late 1990s did. Because of the relatively small size of bacterial genomes, sequencers can compensate for short reads – to a limited extent –by increasing the depth of coverage. No amount of coverage, however, will truly compensate for the lack of linking information across repetitive sequences. Essentially, reads of length N cannot ‘get across’ a repeat of length N or longer; thus, if read length is short, the unavoidable consequence is that every repeat in the genome will cause a break in the assembly. Even relatively short genomes will be broken into hundreds of pieces (Figure 2). This problem can be remedied with paired-end sequences, a standard technique for Sanger sequencing, but the paired-end strategies of 454 and Illumina are not yet mature, so it remains to be seen how effective they will be.
Figure 2.
Repeats longer than 30 bp in the genomes of Bacillus anthracis Ames and Yersinia pestis CO92. Tic marks on the outer concentric circles correspond to direct (same strand) and palindromic (opposite strand) repeats; every tic mark represents a breakpoint (where a gap would occur) for an assembly based on reads of 30 bp or less. Contigs generated from short read sequencing data would cover 97% and 93% of these genomes, respectively, with N50 sizes of 30 387 and 25 894 bp. The fractions of these genomes covered by unique segments longer than 10 kbp (1 kbp) are 84% (96%) and 66% (91%), respectively.
An appealing alternative is to sequence a genome for which a closely related species has already been sequenced. For these projects, the related genome can be used as a substrate and the new genome can be assembled onto it. The resequencing of humans is really just a special case of this strategy, for which all the individuals are particularly closely related. Sequencing of multiple strains of pathogenic bacteria is already under way using 454 sequencing technology; for example, 20 strains of Francisella tularensis have been sequenced, 11 of them using 454 sequencers [data available from the National Center for Biotechnology Information (NCBI), http://www.ncbi.nih.gov]. NCBI’s Trace Archive contains >182 million reads representing 123 species, all from 454-based sequencing projects. A small selection of these projects is shown in Table 1. To handle the rapidly increasing volume and variety of SRS data, NCBI has recently established a Short Read Archive, which already contains entries for 47 species from multiple U.S. and international sequencing centers.
Table 1.
Small sample of the 170 genomes sequenced (as of December 2007) by short-read technology and deposited in NCBI’s Trace Archive
| Species | Sequencing center | Number of reads |
|---|---|---|
| Francisella tularensis | Baylor College of Medicine | 1 218 271 |
| Staphylocooccus aureus COL | Baylor College of Medicine | 575 197 |
| Mammuthus primigenius | Pennsylvania State University | 303 789 |
| Bacillus weihenstephanensis KBAB4 | Joint Genome Institute, U.S. Department of Energy | 1 034 406 |
| Delftia acidovorans SPH-1 | Joint Genome Institute, U.S. Department of Energy | 1 409 615 |
| Pseudomonas putida GB-1 | Joint Genome Institute, U.S. Department of Energy | 1 047 806 |
| Fossil metagenome | Max Planck Institute for Evolutionary Anthropology | 590 264 |
| Burkholderia thailandensis | Broad Institute | 1 100 515 |
| Listeria monocytogenes | Broad Institute | 6 620 471 |
| Pseudomonas aeruginosa | Broad Institute | 588 691 |
| Caenorhabditis remanei | Washington University Genome Sequencing Center | 419 542 |
| Clostridium difficile | Washington University Genome Sequencing Center | 417 941 |
| Drosophila mauritiana | Washington University Genome Sequencing Center | 2 569 374 |
Genome assembly technology has not yet produced a standard solution for assembly of closely related species, although a few options are available. This ‘assembly gap’ means that many new genome sequences are likely to appear before the software is available to assemble them. Whiteford et al. [16] presented an analysis that showed the limits of what a perfect assembler could do with such reads. A perfect assembler will detect all true overlaps in error-free data and will assemble all regions for which a unique solution exists. For bacterial genomes, this perfect assembler should produce excellent results even with reads of 30 bp, approximately the read length produced by Illumina sequencers. At this length, Escherichia coli can be assembled (in theory) so that 75% of the genome is covered by contigs >10 000 bp in length, and 96% of its genes are successfully assembled within single contigs (as opposed to being split across multiple contigs). However, with longer eukaryotic genomes such as C. elegans, short reads will leave significant amounts of the genome uncovered: even with 50-bp reads, considerably longer than current Illumina sequencers can produce, only 51% of the genome can be assembled into contigs >10 000 bp.
Although theoretical results such as those from White-ford et al. [16] are valuable at this stage of technology, it is crucial to have assemblers that can use SRS data. Currently there is one assembler that can handle short reads to assemble closely related species. It uses a comparative assembly algorithm, in which the new genome (the ‘target’) is assembled by mapping it onto a close relative (the ‘reference’). This system, AMOSCmp [17], has been available for several years and works with either traditional paired-end sequences, unpaired sequences or a mixture of the two. The comparative assembly strategy works best when the two species are >90% identical. AMOSCmp can tolerate substantial sequencing errors because of its use of a reference genome: as long as a read maps uniquely to the reference, the assembler can place it. It also handles exact repeats fairly robustly: reads that map to multiple copies of a repeat on the reference are scattered randomly among the repeats. This limits its ability to detect expansions and contractions of long tandem repeats, but isolated repeats do not normally cause breaks in the assembly as they do with a de novo strategy.
De novo assembly
Despite a dramatic increase in the number of complete genome sequences available in public databases, the vast majority of the biological diversity in our world remains unexplored. SRS technologies have the potential to significantly accelerate the sequencing of new organisms. De novo assembly of SRS data, however, will require the development of new software tools that can overcome the technical limitations of these technologies. An overview of genome assembly is provided in Box 1.
Box 1. De novo genome assembly
De novo genome assembly is often likened to solving a large jigsaw puzzle without knowing the picture we are trying to reconstruct. Repetitive DNA segments correspond to similarly colored pieces in this puzzle (e.g. sky) that further complicate the reconstruction.
Mathematically, the de novo assembly problem is difficult irrespective of the sequencing technology used, falling in the class of NP-hard problems [45], computational problems for which no efficient solution is known. Repeats are the primary source of this complexity, specifically repetitive segments longer than the length of a read. An assembler must either ‘guess’ (often incorrectly) the correct genome from among a large number of alternatives (a number that grows exponentially with the number of repeats in the genome) or restrict itself to assembling only the nonrepetitive segments of the genome, thereby producing a fragmented assembly.
The complexity of the assembly problem has partly been overcome in Sanger projects because of the long reads produced by this technology, as well as through the use of mate-pairs (pairs of reads whose approximate distance within the genome is known). Paired reads are particularly useful as they allow the assembler to correctly resolve repeats and to provide an ordering of the contigs along the genome.
Studies by Chaisson et al. [18] and Whiteford et al. [16] showed a rapid deterioration in assembly quality as the read length decreases. Chaisson et al. [18] showed that, for reads of 750 bp (e.g. Sanger sequencing), an assembly of Neisseria meningitidis resulted in 59 contigs, 48 of which were >1 kbp, whereas at 70 bp, the assembly consisted of >1800 contigs, of which only a sixth were >1 kbp. Even for relatively long reads (200 bp), the resulting assembly was substantially fragmented (296 contigs). Similar results were obtained by Whiteford et al. [16], who observed a rapid decrease in contig sizes for reads shorter than ~50 bp.
To overcome some of the challenges posed by repeats, Sundquist et al. [19] proposed a hierarchical sequencing strategy called SHRAP (SHort Read Assembly Protocol) wherein a genome is first sheared into a collection of large fragments [e.g. bacterial artificial chromosome (BAC) clones], each of which is sequenced by SRS. The reads are used to infer a tiling of the BAC clones along the genome, and an assembly is constructed by pooling together reads originating from localized regions within the tiling. The individual assemblies are combined based on the BAC tiling. Tests using simulated data show the SHRAP strategy to be effective in assembling large genomes (several human chromosomes and Drosophila melanogaster); however, read lengths of 200 bp or longer are necessary for good quality assemblies.
In general, assembly tools originally developed for Sanger sequencing data cannot be directly applied to SRS technologies, partly because of specific algorithmic choices that rely on long read lengths and partly because of the specific error characteristics of SRS data (e.g. pyrosequencing technologies are characterized by high error rates in homopolymer regions). Many of these tools would also encounter performance limitations because of the vastly larger number of reads generated by SRS projects; for example, 8 times coverage of a mammalian genome (3 Gbp in length) requires 30 million Sanger reads but 750 million Illumina reads.
Several assembly programs have been developed for de novo assembly of SRS data. Newbler (roche-applied-science.com) is distributed with 454 Life Sciences instruments and has been successfully used in the assembly of bacteria [20]. With sufficiently deep coverage, typically 25–30 times, the resulting assemblies are comparable to those obtained through Sanger sequencing [21]. Note, however, that these results do not account for the additional information provided by mate-pairs – information commonly available in Sanger data but only recently introduced to the 454 technology.
Three recently developed assembly tools tackle the de novo assembly using very short sequences (30–40 bp). SSAKE [22], VCAKE [23] and SHARCGS [24] all use a similar ‘greedy’ approach to genome assembly. Specifically, reads are chosen to form ‘seeds’ for contig formation. Each seed is extended by identifying reads that overlap it sufficiently (more than a specific length and quality cut-off) in either the 5′ or 3′ direction. The extension process iteratively grows the contig as long as the extensions are unambiguous (i.e. there are no sequence differences between the multiple reads that overlap the end of the growing contig). This procedure avoids misassemblies caused by repeats but produces very small contigs. The assembly of bacterial genomes using Illumina data created contigs that are only a few kilobases in length [23,24], in contrast to hundreds of kilobases commonly achieved in Sanger-based assemblies. This fragmentation is caused in part by inherent difficulties in assembling short read data, although future improvements in assembly algorithms should overcome some limitations through more sophisticated algorithms (as was the case when Sanger sequencing was first introduced). These programs have relatively long running times, on the order of 6–10 h for bacterial assemblies [23,24]—at least partly because of the large number of reads generated in an SRS project. By contrast, assemblers for Sanger data can assemble bacterial genomes in just a few minutes.
Another strategy for de novo genome sequencing uses a hybrid of SRS and Sanger sequencing to reduce costs and fill in coverage gaps caused by to cloning biases. Such an approach was followed by Goldberg et al. [25], who used Newbler for an initial assembly of data obtained from a 454 sequencer. They broke the Newbler contigs into overlapping Sanger-sized fragments and used Celera Assembler [26] to combine these fragments with sequence reads obtained from Sanger sequencers. This strategy proved successful in the assembly of several marine bacteria. The addition of 454 data produced better assemblies than those obtained with Sanger data alone, and for two of the genomes, the hybrid assembly enabled the reconstruction of an entire chromosome without gaps.
The assemblers named above follow the standard over-lap-layout-consensus approach to genome assembly, a paradigm that treats each read as a discrete unit during the reconstruction of a genome. An alternative recently proposed by Chaisson and Pevzner [27] uses a deBruijn graph approach, an extension of the authors’ prior work on assembly of Sanger data. Briefly, a deBruijn graph assembler starts by decomposing the set of reads into a set of shorter DNA segments. A graph is constructed that contains the segments as nodes and in which two segments are connected if they are adjacent in one of the original reads. A correct reconstruction of the genome is represented as a path through this graph that traverses all the edges (an Eulerian path). By fragmenting the original reads into smaller segments, this paradigm is less affected by the short read lengths generated by SRS technologies, and it also provides a simple mechanism for combining reads of varied lengths. Chaisson and Pevzner [27] showed their assembler (Euler-SR) is able to generate high-quality assemblies of bacterial genomes from 454 reads and of BAC clones from Solexa reads. They also explored the use of a hybrid assembly approach (Sanger + 454) and interestingly showed that only a small percentage of the longer reads provided information not already represented in the short reads, thus suggesting the need for a careful evaluation of the benefits of hybrid sequencing approaches.
Annotation of genomes sequenced with SRS technology
The highly fragmented assemblies resulting from SRS projects present several problems for genome annotation. The use of SRS technology is so new that few methods have been published describing how current annotation methods can be adapted to account for the various types of sequencing errors that might be present in a genome sequenced with the newer technology.
We can expect that the annotation of genomes sequenced by the new technologies will be reasonably accurate for genes that are found in other species, because the primary annotation methods—sequence alignment programs—are robust in the presence of errors (Box 2). Note that sequencing errors will make some of these genes appear to have in-frame stop codons, such that it might be difficult to distinguish them from pseudogenes. Nonetheless, at least the genes will be found, even if they are fragmented (Figure 3). By contrast, genes that are unique to an organism will be difficult to find with current annotation methods, and many of these might be missed entirely. This problem will be exacerbated by the expected small size of most contigs in assemblies of short-read sequencing projects.
Box 2. Genome annotation methods
The information used to annotate genomes comes from three types of analysis: (i) ab initio gene finding programs, which are run on the DNA sequence to predict protein-coding genes; (ii) alignments of cDNAs and expressed sequence tags (ESTs), if available, from the same or related species; and (iii) translated alignments of the DNA sequence to known proteins. These types of evidence are abundant in various amounts depending on the organism; for less well-studied species, cDNA and EST evidence is often lacking, and annotators depend much more heavily on ab initio prediction programs.
Fortunately, the main bioinformatics programs for aligning cDNAs and protein sequences to genomic DNA are robust in the presence of sequencing errors. Programs for cDNA alignment include GMAP [46], sim4 [47], Spidey [48] and Blat [5]; programs for spliced alignment of proteins to (translated) genomic DNA sequence include DPS [49] and GeneWise [50]. All of these programs must account for the fact that the target genome might not be the same strain or species as the reference cDNA or protein, so they already allow for mismatches. These sequence alignment programs should therefore work well at identifying genes even in the highly fragmented assemblies produced from short reads.
Ab initio gene finders, of which there are many, are not nearly so robust in the presence of errors. Even with near-perfect data, the best pure ab initio methods for human gene finding (those not relying on alignment to other species) only identify 50–60% of exons and 20–25% of genes correctly [51]. Gene finding in smaller eukaryotes tends to be more accurate because of their smaller introns and greater gene density, and gene finders for bacteria, archaea and viruses are very accurate, predicting >99% of protein-coding genes correctly for most genomes [52]. All of these methods assume that the DNA sequence is (mostly) correct, and certain types of errors will lead to erroneous gene predictions. In particular, any sequencing error that introduces an in-frame stop codon is likely to result in a mistaken gene prediction, because ab initio methods organize their searches around open reading frames.
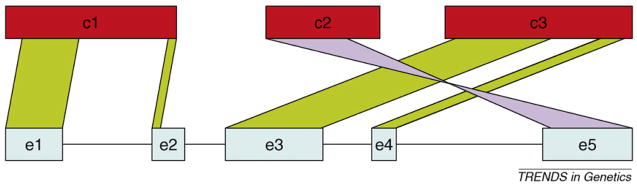
Figure 3.
Fragmentation of a gene caused by fragmented genome assembly. This example shows how a five-exon gene (blue), known from cDNA sequencing or a closely related species, maps to three different short contigs (red). The assembly fails to capture all of exons e2 and e5, which run off the ends of contigs. Exon e5 maps in reverse orientation to contig c2. The cDNA would allow us to order the contigs as c1-c3-c2r (where ‘r’ means reversed), but without a full-length cDNA, we would have no indication of how these contigs were related to one another.
Sequencing of transcripts and regulatory elements
The sequencing of transcribed gene products [expressed sequence tags (ESTs)] has long been a vital tool for the characterization of genes in the human genome and other species. EST sequencing also has an important role in the characterization of splice variants and the identification of regulatory signals in a genome—tasks that are not effectively performed through computational means alone. Transcriptome and regulome sequencing projects have been, perhaps, the most successful application of SRS technologies. 454 technology has been applied successfully to the sequencing of ESTs in Medicago trunculata [28] using a data analysis pipeline initially developed for Sanger data. Specifically, the ESTs were assembled with the TGICL clustering tool [29], and the software PASA [30] was used to characterize splice variants through alignments to the Medicago draft assembly. The 454 technology was also applied, using a customized mate-pair protocol, to dramatically increase the throughput of the paired-end tag (PET) variant of the SAGE technique, wherein tags are sequenced in pairs from both ends of a transcript [31]. A specialized software package, PET-Tool [8], was developed for analysis of these data.
Illumina technology has been used recently for the discovery of protein binding sites [32,33] and histone methylation patterns [34] through chromatin immunopre-cipitation using a protocol named Chip-Seq. Specifically, a protein of interest is cross-linked to the chromatin, which is physically sheared. The bound chromatin segments are precipitated using an antibody, and the DNA is sequenced using SRS. The resulting sequences can either be mapped to a reference genome or used to create short de novo assemblies of the genomic region flanking the binding site. The latter technique is particularly useful in genomes for which high-quality assemblies are not available.
The techniques described above can also be performed using microarray technologies. SRS data, however, provide several advantages. First, sequencing can be applied to species whose genomes have not yet been sequenced or used to discover regulatory patterns in regions not yet present in reference genome assembly. Furthermore, sufficiently deep sequencing can provide quantitative information regarding the interactions being analyzed, as described, for example, in a study of chromatin–chromatin interactions using the chromosome conformation capture technology [35].
Annotation of metagenomics projects
One of the most promising applications of SRS technologies is sequencing of environmental samples, also known as metagenomics. In these projects, DNA is purified from an environment such as soil, water or part of the human body, and the mixture of species is sequenced using a random shotgun technique. The resulting reads might originate from hundreds or even thousands of different species, presenting a much greater assembly challenge than a single genome sequencing project.
Currently, metagenomics projects are focusing on bacterial species, which simplifies the annotation problem somewhat. Because bacterial genomes are gene-rich, a large majority of sequence reads should contain fragments of protein-coding genes. However, the usual annotation approach to bacterial genomes, which relies on (highly accurate) bacterial gene finders, does not work for environmental mixtures. Annotators have thus far relied on a simple but effective BLAST-based strategy: for each read, they use tblastn [36] to translate the sequence in all six frames and search a protein database for matches. For example, this annotation strategy was used for the Sargasso Sea project [37], which sampled the bacterial population of a region of the Atlantic ocean. A BLAST-based strategy can also work for short reads (e.g. deep mine microbiome [38]), although accuracy declines as reads and contigs get shorter.
Huson et al. [39] have developed a method – MEGAN –to enhance this translated BLAST strategy, making it more robust with short-read sequencing data. Rather than providing a detailed annotation of genes, MEGAN attempts to characterize the phylogenetic makeup of a sample, which often is the primary goal of a metagenomics sequencing project. In other words, the goal is to identify the species present rather than the precise identities and locations of all the genes in a mixed sample. Huson et al. applied their system onto a woolly mammoth sample [40], which was sequenced by an early version of the 454 technology, which produced 95-bp average read lengths with a relatively high error rate. Because more than one half of the sample consisted of bacterial contaminants, they treated it as a metagenomics sample and applied the MEGAN system to identify species. They were able to assign ~16% (50 093 of 302 692 total) of the short reads to taxa using their algorithm. Of these, 16 972 were assigned to a variety of bacterial species.
In another recent development, Krause et al. [21] enhanced a translated BLAST approach in an effort to make it more robust to the sequencing errors common in SRS projects. Their CARMA systems combines translated BLAST searches with a postprocessing step that merges protein fragments across frameshifts. They tested their system on a synthetic metagenomic dataset sequenced with 454 technology and were able to identify many of the frameshifts and in-frame stop codons caused by sequencing errors. However, accuracy was substantially lower than a standard bacterial gene finder would obtain on a genome assembled from Sanger sequencing data.
The MetaGene system of Noguchi et al. [41] is designed specifically for metagenomic data from short reads. It uses two dicodon frequency tables, one for bacteria and another for archaea, and applies them based on the GC-content of the sequence fragment. It could reproduce >90% of the gene annotation from the Sargasso Sea project using the contigs generated from that project (which were ~1 kbp in length on average). Tests on simulated short reads from a mixture of 12 bacteria showed that sensitivity remained high for read lengths (or contigs) as short as 200 bp, although it declined rapidly on shorter reads.
Simulated data were also used by Mavromatis et al. [42] to evaluate the performance of annotation pipelines commonly used for single organisms when applied to metagenomic data. They compared fgenesb (www.softberry.com) with a pipeline combining CRITICA [43] and Glimmer [44], using three datasets of varied complexity constructed by randomly selecting shotgun reads from already sequenced isolate genomes. Note that these were Sanger reads and not SRS reads. This study showed a rapid decrease in the accuracy of the annotation produced by all the methods as the assemblies became more fragmented. In a high-complexity environment (containing many mixed species), ~70% of the genes predicted within single reads were correct compared with >90% in a low-complexity environment.
Concluding remarks
Fifteen years of research have shown that, for DNA sequencing technology, longer is better, especially where genome assembly is involved. Someday, perhaps, we will be able to isolate a single chromosome and read it end to end, eliminating the assembly step entirely. At present, however, new short read sequencing (SRS) technologies can sequence so rapidly and so cheaply, that it is clear that SRS is here to stay. Despite their limitations, these still-evolving technologies can replace Sanger sequencing in studies aimed at characterizing the regulatory interactions within genomes. Their wider application in de novo sequencing will require improvements in the length of the reads produced and robust mate-pair protocols. The potential impact of these technologies is underscored by the numerous studies that have successfully applied SRS in biological research, only a few years after their initial availability. Despite these successes, few software tools are available today for the analysis of these data. Continued advances in the application of SRS technologies in biological research will require the development of new algorithms and programs able to handle the specific characteristics of these technologies. In particular, new tools are needed to manage the large amounts of data generated by the SRS technologies and to efficiently perform standard bioinformatics operations (such as alignment) using large numbers of short reads. As SRS technologies start replacing Sanger sequencing, and as they are applied to new analysis tasks, it will be important to begin a critical evaluation of the quality of the data generated through these technologies, both as a means to evaluate SRS experiments and to prioritize future improvements in these technologies. Finally, an open model for the release of both software and data is critical to the success of bioinformatics efforts aimed at the analysis of SRS data. An open-source/open-access model will accelerate progress by allowing the scientific community to join forces in addressing the challenges and promises of the new sequencing technologies.
Footnotes
Publisher's Disclaimer: This article was published in an Elsevier journal. The attached copy is furnished to the author for non-commercial research and education use, including for instruction at the author’s institution, sharing with colleagues and providing to institution administration.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- 1.Check E. Celebrity genomes alarm researchers. Nature. 2007;447:358–359. doi: 10.1038/447358a. [DOI] [PubMed] [Google Scholar]
- 2.Mardis ER. The impact of next generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Miller RD, et al. Efficient high-throughput resequencing of genomic DNA. Genome Res. 2003;13:717–720. doi: 10.1101/gr.886203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl F, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci U SA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber AP, et al. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu KP, et al. PET-Tool: a software suite for comprehensive processing and managing of Paired-End diTag (PET) sequence data. BMC Bioinform. 2006;7:390. doi: 10.1186/1471-2105-7-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huse SM, et al. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore MJ, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickerson DA, et al. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, et al. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koenig R. Tuberculosis. Few mutations divide some drug-resistant TB strains. Science. 2007;318:901–902. doi: 10.1126/science.318.5852.901a. [DOI] [PubMed] [Google Scholar]
- 16.Whiteford N, et al. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 2005;33:e171. doi: 10.1093/nar/gni170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pop M, et al. Comparative genome assembly. Brief Bioinform. 2004;5:237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- 18.Chaisson M, et al. Fragment assembly with short reads. Bioinformatics. 2004;20:2067–2074. doi: 10.1093/bioinformatics/bth205. [DOI] [PubMed] [Google Scholar]
- 19.Sundquist A, et al. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS ONE. 2007;2:e484. doi: 10.1371/journal.pone.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poly F, et al. Genome sequence of a clinical isolate of Campylobacter jejuni from Thailand. Infect Immun. 2007;75:3425–3433. doi: 10.1128/IAI.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause L, et al. Finding novel genes in bacterial communities isolated from the environment. Bioinformatics. 2006;22:e281–e289. doi: 10.1093/bioinformatics/btl247. [DOI] [PubMed] [Google Scholar]
- 22.Warren RL, et al. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeck WR, et al. Extending assembly of short DNA sequences to handle error. Bioinformatics. 2007;23:2942–2944. doi: 10.1093/bioinformatics/btm451. [DOI] [PubMed] [Google Scholar]
- 24.Dohm JC, et al. SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 2007;17:1697–1706. doi: 10.1101/gr.6435207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg SM, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci U S A. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 27.Chaisson M, Pevzner P. Short read fragment assembly of bacterial genomes. Genome Res. 2007 doi: 10.1101/gr.7088808. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung F, et al. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea G, et al. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 30.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng P, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 33.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 34.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 38.Edwards RA, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poinar HN, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi H, et al. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavromatis K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 43.Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 44.Delcher AL, et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medvedev P, et al. Computability and equivalence of models for sequence assembly. Lecture Notes Comput Sci. 2007;4645:289–301. [Google Scholar]
- 46.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 47.Florea L, et al. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheelan SJ, et al. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X, et al. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- 50.Birney E, et al. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guigo R, et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl 1):S21–S31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer DD, et al. Minimus: a fast, lightweight genome assembler. BMC Bioinform. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]