Abstract
The inhibition by the regulatory domain and the interaction with calmodulin (CaM) vary among plasma membrane calcium pump (PMCA) isoforms. To explore these differences, the kinetics of CaM effects on PMCA4a were investigated and compared with those of PMCA4b. The maximal apparent rate constant for CaM activation of PMCA4a was almost twice that for PMCA4b, while the rates of activation for both isoforms showed similar dependence on Ca2+. The inactivation of PMCA4a by CaM removal was also faster than for PMCA4b, and Ca2+ showed a much smaller effect (2- vs. 30-fold modification). The rate constants of the individual steps that determine the overall rates were obtained from stopped-flow experiments in which binding of TA-CaM was observed by changes in its fluorescence. TA-CaM binds to two conformations of PMCA4a, an “open” conformation with high activity, and a “closed” one with lower activity. Compared with PMCA4b (Penheiter et al. (2003) Biochemistry 41:12115–12124), the model for PMCA4a predicts less inhibition in the closed form and a much faster equilibrium between the open and closed forms. Based on the available kinetic parameters, we determined the constants to fit the shape of a Ca2+ signal in PMCA4b-overexpressing CHO cells. Using the constants for PMCA4a, and allowing small variations in parameters of other systems contributing to a Ca2+ signal, we then simulated the effect of PMCA4a on the shape of a Ca2+ signal in CHO cells. The results reproduce the published data (Brini et al. (2003) J. Biol. Chem. 278:24500–24508), and thereby demonstrate the importance of altered regulatory kinetics for the different functional properties of PMCA isoforms.
All eukaryotic cells have in their plasma membrane a Ca2+ pump (PMCA)1, which couples the hydrolysis of ATP to Ca2+ transport from the cytosol to the extracellular space. In humans and rodents, four genes code for the major isoforms of PMCA (named PMCA 1 to 4). The variability of PMCA isoforms is greatly enhanced by alternative splicing affecting the size of the first intracellular loop (splice site A) and the length of the COOH-terminal tail (splice site C)(1). At least in PMCA2, splicing at site A has recently been shown to control the differential targeting of the pump to the basolateral versus apical membrane in polarized cells (2,3). By contrast, splicing at site C affects the COOH-terminal tail (C-tail) in a complex, but still incompletely understood manner (1,4)
The C-tail is unique for PMCAs among the P-type ATPases. It contains many sites important for regulation of the pump, including sites for phosphorylation by protein kinases A and C, proteolysis, and perhaps most importantly, the calmodulin (CaM) binding region (for reviews, see (4–7). CaM binding to the C-tail regulates the activity of the pump (4,8). Because CaM activation is the most-studied mode of PMCA regulation, further studies on CaM binding and activation may also shed light on the mechanism of regulation by other factors. In the absence of CaM, the C-tail acts as an autoinhibitory domain of the pump (9–11). When CaM binds to its binding site, the C-tail dissociates from the catalytic core of the pump and the inhibition is overcome. The C-tail sequence varies between isoforms and alternative splice variants, leading to different kinetics of CaM and Ca2+ activation of these pumps. At the physiological level, this difference in activation kinetics may well form the basis for an adaptive response to different Ca2+ signaling needs in different cells (12).
In a previous paper we studied PMCA4b, and proposed a kinetic model for the activation of this isoform by CaM (13). PMCA4b is the most abundant isoform in red blood cells. Since early studies on PMCA properties and regulation were mostly done in red blood cells, PMCA4b is the best-studied PMCA isoform. By contrast, much less is known about the C-terminal alternative splice variant PMCA4a, which is most abundant in brain, heart and smooth muscle. In this paper, we provide a detailed analysis of the kinetic characteristics of CaM binding and activation of PMCA4a and compare the results with corresponding data on PMCA4b. Data fitting allows us to propose a two-step model for the binding mechanism of CaM to PMCA4a. We also analyze, by means of a simulation based on the kinetic parameters determined here and taken from others, the possible effects of PMCA4a expression on Ca2+ signaling. The results agree with earlier experimental findings and provide a rationale for the apparent greater efficiency of PMCA4a in handling transient Ca2+ spikes compared to PMCA4b.
MATERIALS AND METHODS
Construction of Plasmids
The construction of baculovirus transfer vector pVL1393-PMCA4b has been described (13,14). pVL1393-hPMCA4a was similarly made by subcloning the full-length coding region of hPMCA4a into the pVL1393 vector.
Preparation and Amplification of Recombinant Baculovirus
Recombinant baculovirus was prepared using the Pharmingen BaculoGold Transfection Kit. Briefly, Spodoptera frugiperda (Sf9) cells, in Grace culture medium without fetal bovine serum, were co-transfected with 2 µg transfer plasmid DNA and 5.0 µl of BaculoGold DNA following the manufacturer’s protocol. Plaques expressing the recombinant protein were isolated by plaque assay and screened for PMCA by Western blot. The selected early viral stock was amplified using a multiplicity of infection of 0.1–0.2 following standard procedures and the titer of the amplified stock determined (15). The viral stock was kept at 4°C in the dark.
Expression of PMCA in Sf9 Cells
Sf9 cells suspended in complete Grace medium were infected with the recombinant virus at a multiplicity of infection of 1. After 48 hours of incubation at 27°C, the cells were harvested. A 500-mL culture yielded ~8 × 108 cells. The cells were washed with PBS containing 1 mM EDTA and protease inhibitors, quick frozen, and kept at −80°C until microsome processing.
Microsomal Preparation
Crude microsomal membranes from Sf9 cells, untransfected or transfected with PMCA4a or PMCA4b, were prepared essentially as described for COS cells with some minor modifications (16).
Quantification of PMCA4a in Microsomal Membranes
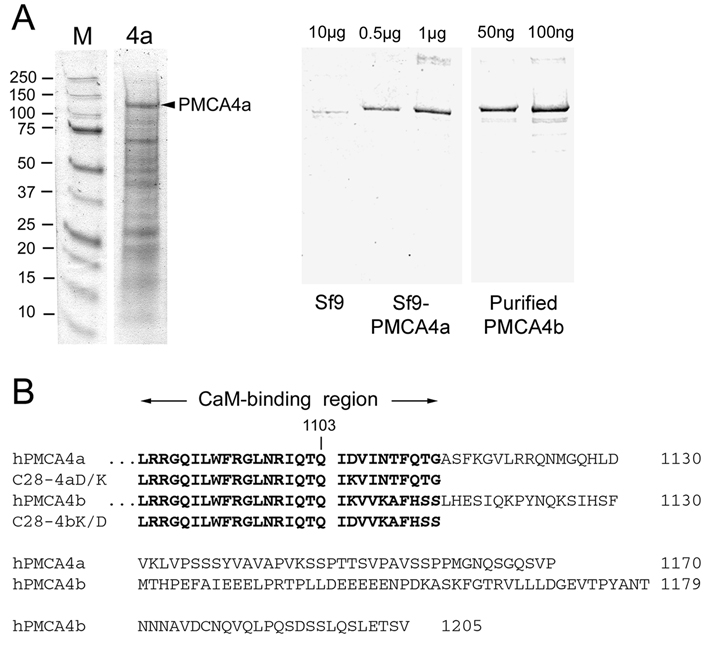
Quantification of the amount of PMCA in microsomal membranes was done by electrophoresis in 7.5% PAGE and subsequent staining with Coomassie Brilliant Blue and densitometry as described (13). A representative gel is shown in Figure 1A (left panel). Alternatively, the amount of expressed PMCA was estimated from Western blots (17) of known amounts of Sf9 cell membranes and standards of purified PMCA4b, using the PMCA-specific antibody 5F10 for detection (Figure 1A, right panel). By this method, PMCA4a was determined to be ~5% of the total microsomal protein from Sf9 cells, which is well within the range of 2.5–10% determined previously for PMCA4b expressed in the Sf9/baculovirus system (13).
FIGURE 1. Quantification of PMCA4a in microsomes from baculovirus-infected insect cells and amino acid sequence comparison of the COOH-terminal tails of human PMCA4a and 4b.
A. Left panel: Coomassie Blue stained gel of microsomal membrane proteins (5 µg) from Sf9 cells expressing PMCA4a. Molecular size standards are shown on the left with their masses given in kDa. PMCA4a (arrowhead) migrates at ~130 kDa. Right panel: Quantification of PMCA4a expression in Sf9 cell membranes by Western blotting. 10 µg of uninfected Sf9 cell membranes were electrophoresed next to 0.5 µg and 1 µg of PMCA4a-expressing Sf9 membranes and 50 ng and 100 ng of purified PMCA4b as standards. Western blotting for PMCA was performed with antibody 5F10 as described (17). PMCA4a amounts to ~5% of the Sf9 microsomal protein as determined by Western blotting and densitometry (13). B. Sequence comparison of human PMCA4a and PMCA4b starting with the CaM binding region. The last common residue (1103) preceding the alternative splice site is indicated. The sequences of the CaM-binding peptides C28-4a and C28-4b, and of the single-residue mutant peptides C28-4aD/K and C28-4bK/D are indicated in bold print.
Peptide Synthesis
Peptides C28-4b, C28-4b(K/D), C28-4a and C28-4a(D/K) were synthesized in the Mayo Protein Core Facility, and purified to homogeneity by HPLC. The sequences of the peptides are given in Figure 1B.
Origin of TA-CaM
2-Chloro-(amino-Lys75)-[6-[4-(N,N-diethylamino)phenyl]-1,3,5-triazin-4-yl] calmodulin (TA-CaM) was prepared, purified and characterized as previously described (13) and was a kind gift from Dr. Katalin Török and Richard Thorogate (St. George Hospital Medical School, London, UK).
Measurements of ATPase Activity
The ATPase activity was measured by continuously monitoring the absorbance at 360 nm, following the procedure outlined before (18). The media contained 120 mM KCl, 30 mM TES-triethanolamine (pH 7.2), 5 mM MgCl2, 200 µM EGTA, 2.5 mM ATP, 0.2 mM 2-amino-6-mercapto-7-methyl purine, 1 U/ml purine nucleoside phosphorylase, 5 mM NaN3, 1 mM dithiothreitol, 0.5 mM ouabain, 4 µg/ml oligomycin, 200 nM thapsigargin, 2 µg/ml aprotinin, 0.5 µg/ml leupeptin, 200 nM CaM, and enough CaCl2 to obtain the concentration of free Ca2+ indicated in the figures. Steady–state measurements of the ATPase activity of PMCA4a expressed in insect cells by using the baculovirus system are in agreement with our previous results in COS-cells (19). This assures the validity of the results obtained with the insect cell system.
Binding of TA-CaM to PMCA
We previously showed that there is no significant amount of CaM-binding proteins in Sf9 cell microsomes other than the expressed PMCA (13). Equilibrium fluorescence measurements were performed in a Varian Cary Eclipse fluorometer. The media contained 30 mM Tes-TEA (pH 7.2), 120 mM KCl, 5 mM MgCl2, 0.2 mM EGTA, and enough CaCl2 to obtain 100 µM free Ca2+. Stopped-flow measurements of changes in fluorescence of TA-CaM were performed in an Applied Photophysics SX.18MV reaction analyzer. The excitation wavelength was 365 nm, and the emitted light was detected using a 390 nm cut-off filter. The syringes contained the concentrations of TA-CaM and PMCA or C28 peptide indicated in the legend of the figures in media containing 30 mM Tes-TEA (pH 7.2), 120 mM KCl, 5 mM MgCl2, 0.2 mM EGTA, and enough CaCl2 to obtain 10 µM free Ca2+. Measurements were performed at 37°C so they were directly comparable to the ATPase activity results.
Data Analysis
Individual curves were fitted by non-linear regression using the GraphPad Prism software. For global analysis different models were fitted by using the Dynafit software (Biokin, Inc.; software free for academic users at www.biokin.com) (20) with local offset option to compensate for possible instrument voltage drift, and corroborated by analysis using the M-LAB platform (21). The script files and data used to generate Figure 8 are included as Supplementary Material.
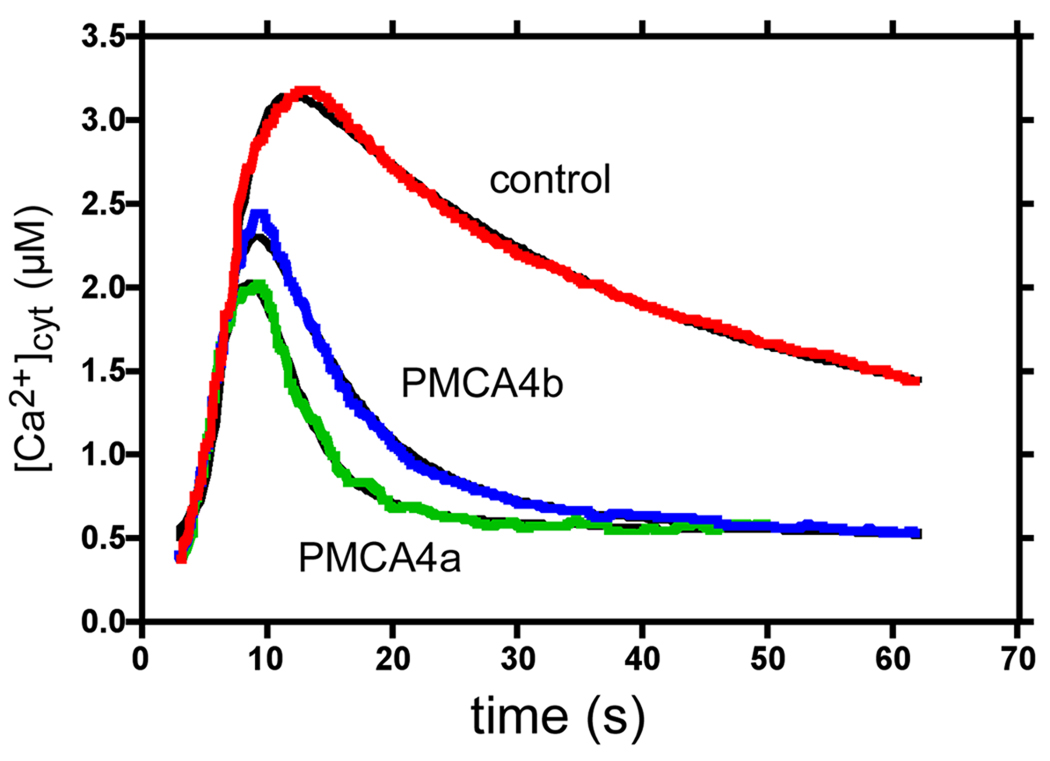
FIGURE 8. Fit of the kinetics of PMCA4a and PMCA4b to the data determined by Brini et al. (22).
Red, blue, and green lines correspond to the change in cytosolic [Ca2+] following the addition of 100 µM ATP to control CHO cells and to CHO cells overexpressing PMCA4b and 4a, respectively (22). The black lines represent the fit calculated by the simulations. The model used for the simulation is explained in the text.
Digitizing
This was done with FindGraph 1.724 (www.uniphiz.com) using Panel C from Figure 8 of Brini et al. (22).
RESULTS
ATPase Measurements
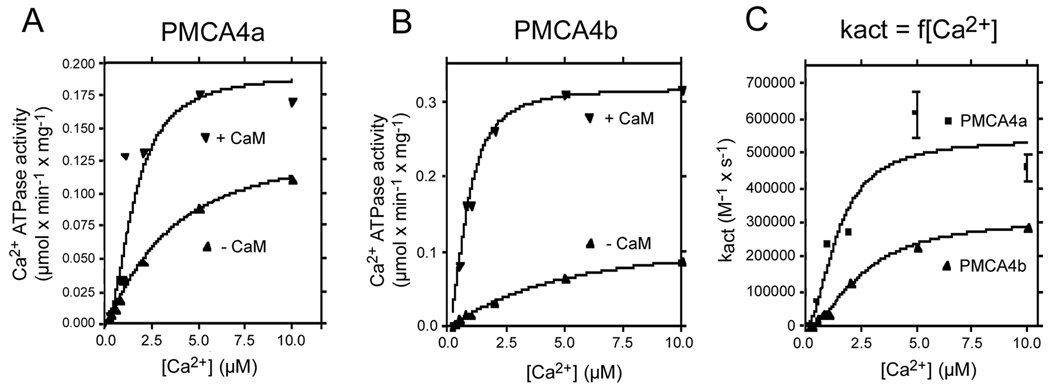
PMCA4a and 4b differ in their C-terminal sequence. Following splice site C (at amino acid position 1103), the sequence of PMCA4a is remarkably different from PMCA4b (see Fig. 1B). We have previously shown that this region in PMCA4b is important in stabilizing its binding to CaM (23). Here we studied the interaction of the corresponding region in PMCA4a with CaM. Figure 2 shows the effect of CaM on the Ca2+ dependence of PMCA4a (panel A) and PMCA4b (panel B). In both cases CaM increases the Vmax and the apparent affinity for Ca2+. However, the effect on Vmax is less pronounced in PMCA4a (i.e., in the absence of CaM, PMCA4a has a higher basal activity). Continuous monitoring of ATPase activity allowed us to measure the rate at which the activation by CaM occurs. In panel C this rate is plotted as a function of the Ca2+ concentration. At all Ca2+ concentrations, the rate of activation by CaM of PMCA4a was faster than that of PMCA4b. Ca2+ increased the apparent rate constant for CaM activation (kact) along a curve with a KCa of about 2 µM in both cases. The maximal kact was 5.4 ± 0.98 × 105 s−1M−1 for PMCA4a and 3.1 ± 0.14 × 105 s−1M−1 for PMCA4b. This value for 4b is close to the value determined previously (2.2 × 105 s−1M−1 (24)). The Ca2+ dependence of the rate of activation by CaM suggests that formation of the Ca2+-CaM complex is required for the activation of both isoforms of PMCA. The results also confirm our previous observation that PMCA4a is activated faster by CaM than PMCA4b (18), and extend this observation to all Ca2+ concentrations tested.
FIGURE 2. Ca2+ dependence of the ATPase activity of PMCA4a (A), PMCA4b (B) and of the rate constant for activation by CaM (kact) (C) for PMCA4a and PMCA4b.
The ATPase activity was measured as described in Materials and Methods.
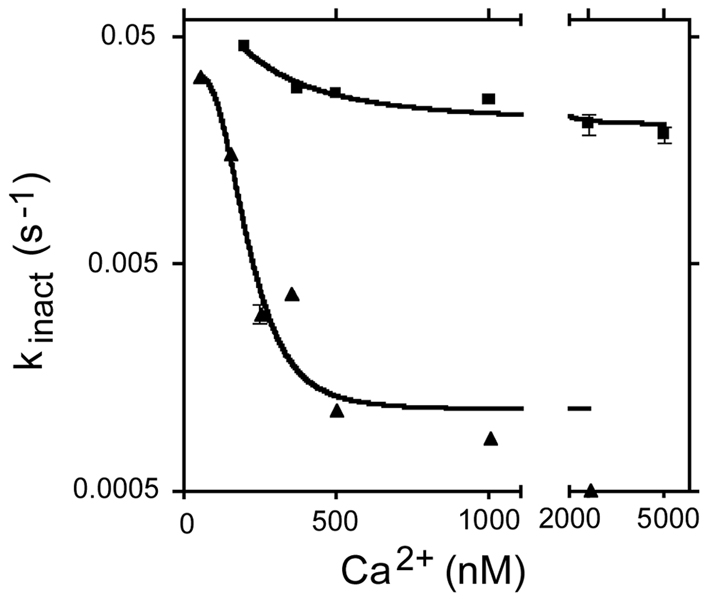
Figure 3 shows the effect of Ca2+ on the rate of inactivation of PMCA4a and PMCA4b by CaM removal. Ca2+ has a profound effect on the inactivation rate of PMCA4b, decreasing the apparent rate constant by a factor of about 100. By contrast, inactivation of PMCA4a by CaM dissociation was faster than for PMCA4b at all Ca2+ concentrations tested. Surprisingly, the effect of Ca2+ on the rate of inactivation of PMCA4a was much less evident than for PMCA4b. Increasing [Ca2+] from 0.2 µM to 2 µM decreased kinact from 0.043 s−1 to 0.02 s−1 (2.1 fold change). The same increase in Ca2+ concentration for PMCA4b decreased kinact from 0.015 s−1 to 0.0005 s−1 (~30 fold change). This result suggests that binding of Ca2+ to its four sites in CaM stabilizes the complex between CaM and PMCA4b in a much tighter manner than between CaM and PMCA4a.
FIGURE 3. Rate of inactivation of PMCA4a (filled squares) and PMCA4b (filled triangles) by CaM removal as a function of the Ca2+ concentration.
CaM removal was carried out as described (18,24) by the addition of 10 µM of a CaM-binding peptide derived from the sequence of myosin light chain kinase. Note that the y-axis is in log scale to show all the experimental points in the same figure.
Fluorescence Measurements
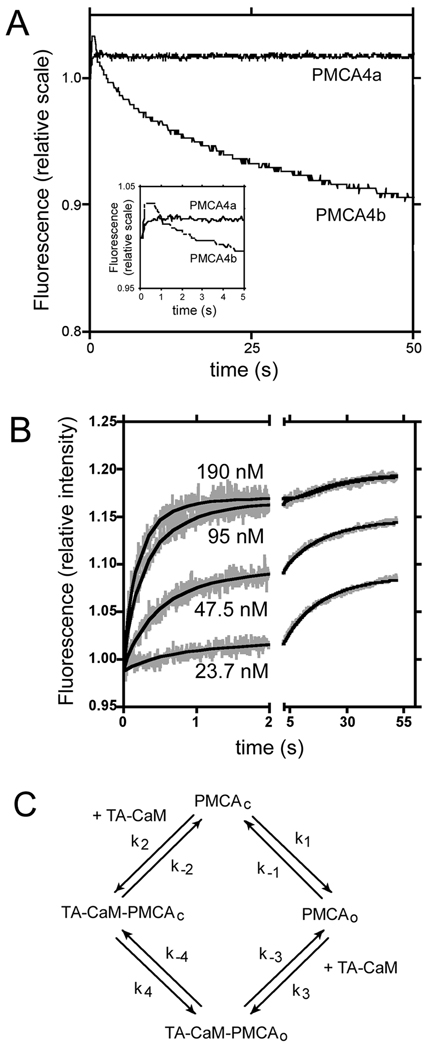
To investigate the regulation of PMCA4a at a more detailed level, we studied the binding of a fluorescently labeled CaM, TA-CaM. We measured the time course of the change in fluorescence of 17 nM TA-CaM upon mixing with Sf9 cell microsomes expressing PMCA4a or PMCA4b. The microsome concentration was adjusted so the medium contained 50 nM of PMCA in each case. Results are shown in Figure 4, panel A. As demonstrated previously, upon mixing with PMCA4b, TA-CaM shows a rapid increase in fluorescence, followed by a slow decrease. In contrast, when mixed with microsomes expressing PMCA4a, TA-CaM shows the initial increase, but not the slow decrease in fluorescence. Panel B of Fig. 4 shows an experiment in which TA-CaM was mixed with different concentrations of microsomes expressing PMCA4a. The curves can be described by a relatively rapid initial increase, and a small and slow but significant further increase in fluorescence. We attempted to fit the data with several models by using global fitting. The best fit was obtained with the kinetic model shown in Figure 4C. In this model PMCA4a can exist in two conformations, open and closed. TA-CaM can bind to both conformations, but prefers the open conformation. The model is similar to the one used to fit the binding of TA-CaM to PMCA4b (13), except that there is no stabilization step. The rate constants for the fitting shown in Fig. 4B are shown in Table I.
FIGURE 4. Binding of PMCA4a to TA-CaM.
A: Changes of fluorescence in TA-CaM upon mixing with microsomes containing 50 nM of either PMCA4a or PMCA4b. Inset: Traces of the first 5 s of the reaction. B: Changes of fluorescence in TA-CaM upon mixing with microsomes containing the concentration of PMCA4a indicated in the figure. The experimental data are shown in grey. The fit to the model depicted in C is shown in continuous black lines. TA-CaM concentration was 17 nM in all cases. C: Kinetic model that represents the binding of CaM to PMCA4a.
Table I. Parameters that fit the data in Figure 4.
The model is described in Figure 4C. When the standard error of a parameter was higher than 100% of the parameter, the upper limit for such parameter is listed.
| Parameter | Fit | Error |
|---|---|---|
| k1 (s−1) | 0.07631 | 0.03443 |
| k−1 (s−1) | 0.8952 | 0.3861 |
| k2 (s−1 nM−1) | <0.0003 | |
| k−2 (s−1) | <0.09 | |
| k3 (s−1 nM−1) | 0.06945 | 0.0303 |
| k−3 (s−1) | 0.09559 | 0.02423 |
| k4 (s−1) | 0.01949 | 0.1111 |
| k−4 (s−1) | 0.01829 | 0.02461 |
| rTA-CaM-PMCAc | 0.073145 | 0.005103 |
| rTA-CaM | 0.0581061 | 3.776e-005 |
| rTA-CaM-PMCAo | 0.0692477 | 0.0001402 |
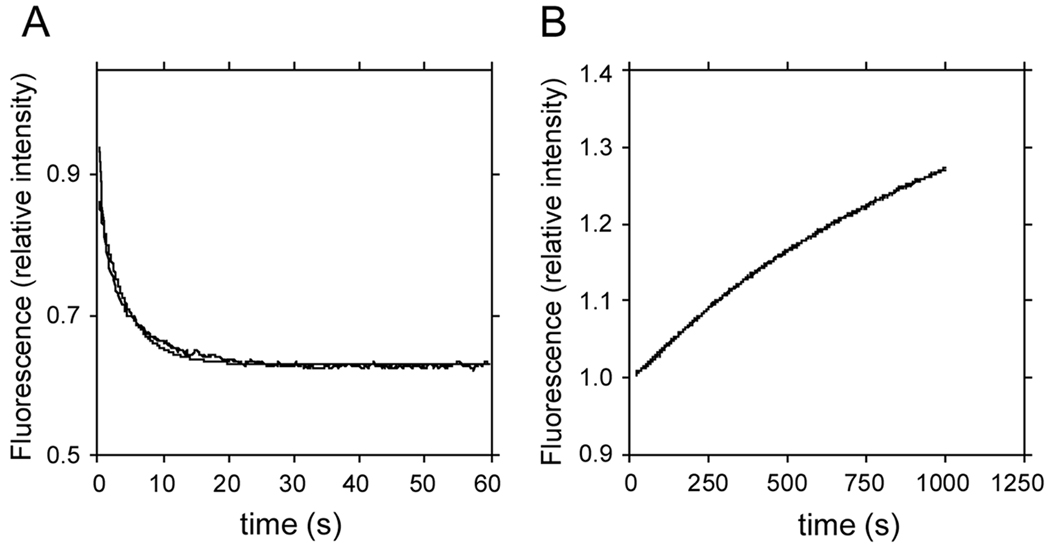
In the experiment in Figure 5, we measured the dissociation of TA-CaM from PMCA4a (panel A) and PMCA4b (panel B). As expected, since association with PMCA4a produced an increase in TA-CaM fluorescence, dissociation produced a decrease in TA-CaM fluorescence, and the opposite was true for PMCA4b. A comparison of panels A and B shows that dissociation of TA-CaM from PMCA4a is very much faster than from PMCA4b. The rate constants for this process were 0.23 ± 0.02 s−1 for PMCA4a and 0.00095 ± 0.00002 s−1 for PMCA4b. These values are consistent with the kinetic models we have proposed for PMCA4a and PMCA4b.
FIGURE 5. Dissociation of TA-CaM from PMCA4a (A) and PMCA4b (B).
Dissociation was started by adding 1 µM CaM to 17 nM TA-CaM saturated with 200 nM of either PMCA4a or 4b. All concentrations represent final values.
Binding of TA-CaM to Peptides
To investigate whether the kinetic properties of CaM binding to PMCA4a are solely due to the interaction of CaM with its CaM-binding region, or are the result of more complex interactions with the whole PMCA molecule, we tested the binding of TA-CaM with C28-4a, a 28-residue synthetic peptide that comprises the sequence of the putative CaM-binding region of PMCA4a (see Fig. 1B). This peptide is analogous to the peptide that we previously called C28, but with the 4a sequence instead of the 4b sequence (note that the 19 N-terminal residues of these two peptides are identical). To avoid confusion we will call C28 C28-4b in this paper. Figure 6A shows the titration of TA-CaM with C28-4a. Fluorescence decreases at low concentrations of C28-4a, while at concentrations higher than 200 nM, the fluorescence starts to increase. The resulting curve is described by the existence of two binding sites, a high affinity site with a Kd of 75 nM and a low affinity site with a Kd of 1000 nM. For comparison, in the same panel we show the titration of TA-CaM with C28-4b, which shows a single site with a Kd of 18 nM. Figure 6B shows the results of an experiment in which TA-CaM was mixed with 25, 50, 100, 200 and 400 nM C28-4a, and the changes in TA-CaM fluorescence monitored over time. In all cases, the curves show a very small, barely detectable increase, followed by a decrease in fluorescence. Figure 6C shows the results of an experiment in which 17 nM TACaM were mixed with 2 µM of either C28-4a or C28-4b and the time course of the fluorescence change was monitored. After an initial decrease in fluorescence, there was a slow increase in the case of C28-4a, but not in the case of C28-4b. Using this information, we fitted the model described in Fig. 6D to the data in Fig. 6B. Several models, including the one used previously to describe the binding of C28-4b to TA-CaM (13) were tried, but they did not give as good a fit as the one shown in Fig. 6D. The parameters of the fit of this model to the experimental data are shown in Table II. The model is basically a two-step binding reaction (intermediate product TA-CaM-C, and final product TA-CaM-C*) with the possibility to bind with low affinity a second molecule of peptide to TA-CaM, presumably to the N-terminal lobe (denoted as TA-CaM-C# in Fig. 6D). The species with two peptides bound is called TA-CaM-C2 in the model. Binding of a second peptide with low affinity is supported by the low affinity component detected in the titration curve, and by the slow rise in fluorescence observed during time-course experiments when using peptide concentrations of 1 µM or above (see Fig. 6C). Given its low affinity, the physiological significance of binding of a second peptide is dubious, but it is interesting from the point of view of the binding mechanism.
FIGURE 6. Kinetics of peptide binding to CaM.
A: Titration of TA-CaM with peptides C28-4a and C28-4b. TA-CaM concentration was 34 nM, other conditions were as described in Methods. B: Time course of fluorescence changes upon mixing 17 nM TA-CaM with different concentrations of C28-4a, from 25 nM up to 400 nM. The data were fitted with the model shown in panel D. C: Time course of fluorescence changes upon mixing 17 nM TA-CaM with 2 µM of either C28-4a or C28-4b. D: Kinetic model fitted to the data in panel B. The addition of C28-4a peptide is denoted by (+ C). TA-CaM-C# and TA-CaM-C* represent different conformational species of the CaM-peptide complex. The model is also consistent with the results shown in panels A and C.
Table II. Parameters of the fit of the model in Figure 6D to the data in Figure 6B.
| Parameter | Fit | Error |
|---|---|---|
| k1 (s−1nM−1) | 0.1292 | 0.03075 |
| k−1 (s−1) | 32.211 | 2.334 |
| k2 (s−1) | 2.611 | 1.473 |
| k−2 (s−1) | 3.0977 | 0.1648 |
| k3 (s−1nM−1) | 0.1743 | 0.03602 |
| k−3 (s−1) | 2.513 | 0.5582 |
| k4 (s−1nM−1) | 0.02999 | 0.005997 |
| k−4 (s−1) | 6.705 | 1.148 |
| k5 (s−1nM−1) | 2.436 | 0.3555 |
| k−5 (s−1) | 4.548 | 1.493 |
| rTA-CaM | 0.129711 | 0.0001535 |
| rTA-CaM-C | 0.16029 | 0.01239 |
| rTA-CaM-C* | <0.0001 | |
| rTA-CaM-C# | <0.0001 | |
| rtA-CaM-C2 | 0.096469 | 0.001041 |
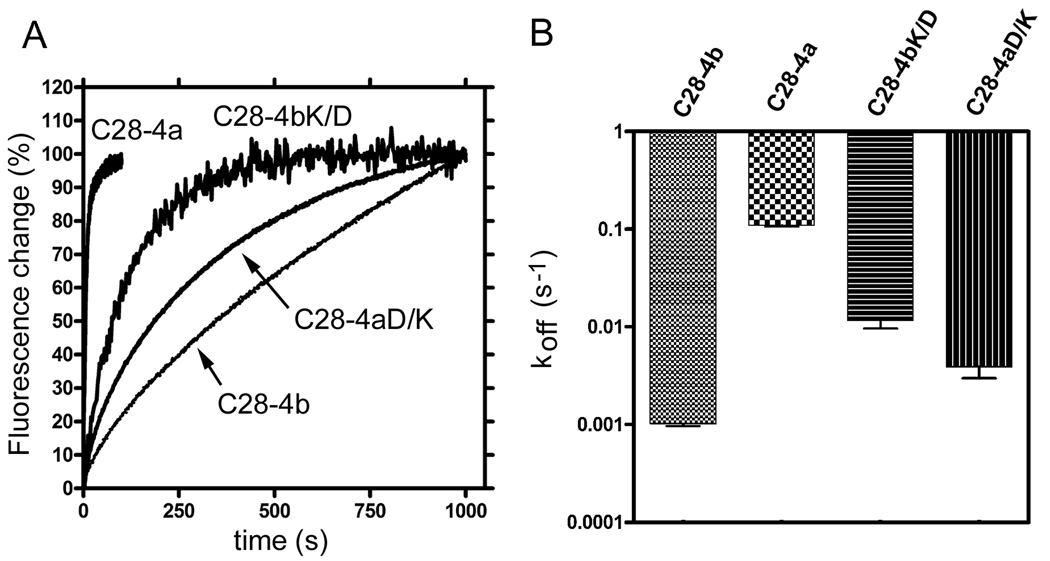
One of the most prominent differences between the sequences of the CaM-binding region of PMCA4a and PMCA4b is the charge reversal at position 1105 (D in PMCA4a and K in PMCA4b; see Fig. 1B). Since the most striking difference between the kinetic characteristics of C28-4a and C28-4b was in the rate of CaM dissociation, we tested the effect of mutating K1105 to D in C28-4b, and the effect of mutating D1105 to K in C28-4a on the rates of TA-CaM dissociation. Figure 7A shows the time course of fluorescence changes when TA-CaM was dissociated from each of the peptides analyzed. The rate of dissociation of C28-4b from TA-CaM was 0.0010 ± 0.00005 s−1, and for C28-4a was 0.110 ± 0.004 s−1 (Fig. 7B). These values were very close to the rate constants for dissociation of TA-CaM from PMCA4b and PMCA4a, respectively, indicating that the dissociation of TA-CaM from its complexes with the peptides is likely to mimic the dissociation of TA-CaM from the intact proteins. Changing K1105 to D in C28-4b (peptide C28-4bK/D) increased the rate constant for TA-CaM dissociation to 0.012 ± 0.002 s−1. Conversely, changing D1105 to K in C28-4a (peptide C28-4aD/K) reduced the dissociation rate constant to 0.0039 ± 0.0009 s−1 (Fig. 7B). The results thus clearly indicate that the charge at position 1105 is very important in regulating the rate of dissociation of CaM.
FIGURE 7. Dissociation of peptides C28-4a, C28-4b, C28-4aD/K and C28-4bK/D from TA-CaM.
A: TA-CaM was pre-mixed with the peptide indicated and dissociation was then initiated by mixing with unlabeled CaM. Final concentrations: TA-CaM, 17 nM; peptide, 200 nM; CaM, 1 µM. All other conditions were as described in Materials and Methods. B: Comparison of the dissociation rates (koff) of the peptides from TA-CaM as determined from the data in panel A. Note that most of the difference in dissociation rates between C28-4a and C28-4b appears to be due to the difference in a single charged residue (D in 4a and K in 4b) as illustrated by the dissociation rates of the reciprocal mutant peptides C28-4aD/K and C28-4bK/D.
Modeling of a Cellular Ca2+ Signal
We were interested to see how the kinetic properties determined for PMCA4a could affect Ca2+ signaling in comparison to PMCA4b. We designed a kinetic model taking into account the main systems contributing to a Ca2+ signal in an hypothetical cell, and analyzed how changing the kinetic parameters from PMCA4b to PMCA4a would affect the shape of a Ca2+ peak.
The model assumes release of Ca2+ from the endoplasmic reticulum (ER) upon binding of an agonist (A) to a receptor (B) in the plasma membrane, followed by Ca2+ re-uptake into the ER via the SERCA pump as well as expulsion through the plasma membrane by the PMCA. The aim of this model is not to comprehensively explain all aspects of the control of Ca2+ signaling, but rather to determine the place of PMCA in controlling the Ca2+ signal. For this purpose, it is possible to simplify the description of the contribution of some of the other controllers of the Ca2+ signal. The sequence of reactions involved is as follows:
| K1, klr |
| K2 |
These steps comprise agonist binding to extracellular receptor, leading to Ca2+ release from the ER (originally from (25), but allowed to fit).
| k3 |
This is Ca2+ entry through plasma membrane channels (C), e.g., triggered by store Ca2+ release (store-operated Ca2+ entry); allowed to fit freely.
| k4, k4r |
| k5 |
These steps comprise CaM-independent Ca2+ expulsion by the PMCA (from (19,26)).
| k6, k6r |
| k7, k7r |
These two steps represent the binding of Ca2+ to the two pairs of Ca2+-binding sites in CaM. CaiCaMh is CaM half-loaded with 2 Ca2+, and (CaiCaM) is CaM fully-loaded with 4 Ca2+. These two steps were allowed to fit.
| k8, k8r |
| k9, k9r |
| k10 |
| k11, k11r |
| k12 |
These steps comprise CaM activation of PMCA and CaM-activated PMCA activity. The rate constants for these five reactions were taken from (13,19,24) and the results in this paper.
| k13, k13r |
| k14, k14r |
These reactions correspond to activation of SERCA and SERCA-mediated Ca2+ uptake into the ER (rate constants from (27); k14r was allowed to fit).
In this scheme, A denotes the agonist, B stands for the receptor whose activation leads to Ca2+ mobilization, C is a Ca2+ channel in the plasma membrane, CaER, Cai, and Caex represent Ca2+ in the ER, cytosol and extracellular space, respectively, P represents the PMCA, CaM represents calmodulin, and S represents SERCA2b, which is the isoform present in most non-muscle cells. Constants k1 and k1r, etc. are the forward and reverse rate constants for the reactions shown. The fit of this model to the experimental results in Figure 8 of the paper by Brini and co-workers (22) is shown in Figure 8. The values for the constants used are listed in Table III. To get this fit, the best value for the rate constants for the steps regarding Ca2+ release from the ER and Ca2+ entry through the plasma membrane were obtained from the best fit for the Ca2+ curves in control and PMCA4b-overexpressing cells (the red and blue curves, respectively, in Fig. 8). Control cells were assumed to express a small amount of PMCA4b. Then, with the rest of the parameters fixed, the amount of PMCA4a was freed and the curve for PMCA4a (the green curve in Fig. 8) was fitted to the data. Thus, the only difference between the PMCA4b and PMCA4a curves was the amount of pump and the kinetic parameters for the respective PMCAs (which were taken from the indicated references and this work, without modification). The results (Fig. 8) clearly show that despite its lower affinity for CaM, PMCA4a is more effective at removing Ca2+ under the conditions assumed in the simulation, mainly because of its faster reaction with CaM, and because of its higher basal activity in the absence of CaM. From the fitting we calculated that PMCA4b was expressed about 19 times over the control, whereas PMCA4a was expressed about 15 times over the PMCA levels of control cells.
Table III. Parameters for the fit of Ca2+ changes shown in Figure 8 for PMCA4b- and PMCA4a-overexpressing cells.
For the control cells all constants were the same as for PMCA4b-overexpressing cells, except [P] = 0.01944 µM.
| Parameter | Fit PMCA4b | Fit PMCA4a |
|---|---|---|
| k1 (s−1µM−1) | 9.296 | |
| k1r (s−1) | 16.93 | |
| k2 (s−1µM−1) | 0.004712 | |
| k3 (s−1µM−2) | 0.6615 | |
| k4 (s−1µM−1) | 10 | |
| k4r (s−1) | 50 | |
| k5 (s−1) | 5.5 | 12 |
| k6 (s−1µM−2) | 2.669 | |
| k6r (s−1) | 2.682 | |
| k7 (s−1µM−2) | 170.4 | |
| k7r (s−1) | 1.551 | |
| k8 (s−1µM−1) | 0.2 | 0.8 |
| k8r (s−1) | 0.0008 | 0.02 |
| k9 (s−1µM−1) | 50 | |
| k9r (s−1) | 10 | |
| k10 (s−1) | 30 | |
| k11 (s−1) | 10 | 6.2 |
| k11r (s−1µM−4) | 0.0007332 | 0.0005141 |
| k12 (s−1) | 0.033 | |
| k13 (s−1µM−1) | 140 | |
| k13r (s−1) | 20 | |
| k14 (s−1) | 70 | |
| k14r (s−1µM−1) | 0.2661 | 0.168 |
| [A] (µM) | 10 | |
| [B] (µM) | 0.1904 | 0.2303 |
| [CaER] (µM) | 2149 | 1687 |
| [Caex] (µM) | 696.5 | |
| [C] (µM) | 1.241 | 0.294 |
| [CaM] (µM) | 1.942 | 1.47 |
| [S] (µM) | 0.002802 | 0.0119 |
| [P] (µM) | 0.3675 | 0.30 |
DISCUSSION
We have previously interpreted the role of the alternatively spliced C-tail of PMCA4 as being of regulatory importance (18,19). Since the catalytic cores of PMCA4a and PMCA4b are identical, one would expect Vmax to be the same for both isoforms. The results of Figure 2 are consistent with this idea; they show that the Vmax of PMCA4b and PMCA4a are comparable within experimental error. Hence, differences in the degree of inhibition and in the rates of activation and inactivation must account for the differences in the properties of the two pumps.
We found that at all Ca2+ concentrations PMCA4a was activated at a faster rate than 4b. The apparent rate constant for this process, kact, increased with Ca2+ along a sigmoidal curve with a KCa of about 2 µM for both PMCA4a and 4b. This suggests that this effect is due to titration of CaM with Ca2+. Interesting differences were also seen in the rates of PMCA inactivation by CaM removal. This rate is faster for 4a, and depends only slightly on the Ca2+ concentration. In stark contrast, Ca2+ decreases the rate of inactivation dramatically in PMCA4b (by about 30-fold from 0.2 to 2 µM Ca2+).
The results of stopped-flow experiments with TA-CaM indicate that binding of CaM to PMCA4a is not only faster than CaM binding to PMCA4b but that there are also mechanistic differences. In both cases, the mechanism can be described as PMCA being available in two conformations, “open” and “closed”. The open conformation has higher Ca2+-ATPase activity than the closed one, and CaM binds with higher affinity to the open conformation. The major difference is that in the case of PMCA4b it was necessary to include a stabilization step (13) that appears to be absent in the case of PMCA4a. Interestingly, although the individual rate constants are faster, the ratio k1/k−1 accounting for the equilibrium between closed and open forms, is similar in PMCA4a and 4b (compare data in Table I with the results for PMCA4b (13)). Therefore, we can deduce that the higher basal activity of PMCA4a compared to PMCA4b in the absence of CaM is due to a higher activity of the PMCA4a closed form, rather than to a difference in the equilibrium between closed and open forms. Stated in other words, the C-terminal tail of PMCA4a is less inhibitory than that of PMCA4b.
The best fit for the binding of TA-CaM to peptide C28-4a was obtained with a model in which the binding proceeds in two steps (unlike C28-4b, whose binding to TA-CaM is best described by a 3-step model (13)). In addition, the model requires the weak binding of a second C28-4a peptide molecule to CaM. This behavior can be rationalized in view of the published data on CaM-peptide complexes. The only known structure of a CaM-peptide complex for PMCA is the structure of CaM-C20 (28). C20 is a shorter version of C28-4b that lacks the residues VVKAF1110HSS after the alternative splicing point (see Fig. 1B). The structure of CaM-C20 shows that W1093 of the peptide acts as an anchor that interacts with the C-terminal lobe of CaM; in this structure there is no interaction between C20 and the N-terminal lobe of CaM. Instead, CaM is in an extended form with its N-terminus distant from C20 (28). On the other hand, x-ray scattering experiments show that CaM collapses into a compact structure when it interacts with a longer peptide resembling C28-4b (29). The CaM-C28-4b complex has a very slow rate of dissociation, whereas the rate of dissociation of the CaM-C20 complex is two orders of magnitude faster (23). In this paper, we show that the rate of dissociation of both the CaM-C28-4a complex and the CaM-PMCA4a complex is fast, suggesting that CaM interacts with this PMCA splice form through only one of its lobes, with a possible weak interaction with the other lobe. Furthermore, the charge of the residue at position 1105 (D in PMCA4a but K in PMCA4b) seems to make a crucial contribution to the interaction with the second lobe of CaM, as determined by the dissociation rates (see Figure 7).
The rate of pump inactivation by CaM removal and the rate of CaM dissociation are similar for PMCA4b (13,18,24). By contrast, the rate of dissociation of TA-CaM from PMCA4a was approximately 10 times faster than the rate of inactivation by CaM removal. The most plausible interpretation is that there is a slow step in PMCA4a (perhaps involving refolding of the C-tail after CaM dissociation) that is required for full auto-inhibition. This refolding time is not apparent in PMCA4b probably because CaM dissociation in this pump isoform is the rate-limiting step. We previously reported that a C-terminally truncated mutant of PMCA4a ending at G1113 (PMCA4a-ct56) showed slightly lower CaM affinity and substantially less autoinhibition than a truncated mutant ending at V1134 (PMCA4a-ct35), which was identical to the wild type. Based on this result, we concluded that the CaM-binding region of PMCA4a extended beyond the 28 residues reported for PMCA4b (30). However, we show here that peptide C28-4a is able to bind CaM with high affinity. A comparison of the results in the previous paper and the results here suggests that the extra residues (from G1113 to V1134) are important for the correct folding of the auto-inhibitory region rather than for binding of CaM itself.
The most relevant information obtained from our studies relates to how the unique kinetic parameters of the different PMCA isoforms affect Ca2+ signaling in a cell. In this paper we present a simplified model of a Ca2+ spike, and show that our constants produce an excellent fit to the agonist-evoked Ca2+ changes measured by Brini et al. (22) in CHO cells overexpressing PMCA4a or 4b. The parameters used for the fits may be divided into several classes: Class 1 includes those that had the same value in all the fits. These are indicated in Table III by the absence of an entry in the “PMCA4a” column. Most of these parameters were taken directly from the literature or from the results of this paper and were treated as constants in all of the fits to cellular Ca2+ signals. Some (k1, k1r, k2, k3, k6, k6r, k7 and k7r) were allowed to vary to determine the fit to the cells expressing PMCA4b, but were then held to the same values in the fit to the cells expressing PMCA4a. Class 2 comprises parameters that were allowed to vary in the fit to the cells expressing PMCA4a, but did not change very much. These are k14r (the reverse rate of SERCA transport), [B] (the concentration of receptors for Ca2+ mobilization, [CaM] (the concentration of calmodulin), [CaER] (the concentration of Ca2+ in the ER lumen) and [P] (the concentration of expressed PMCA). Considering that the CHO cells used by Brini et al. (22) were grown in a defined way and only differed in the expression of PMCA4a vs. PMCA4b, it is reassuring that these free parameters remained in a narrow range. The third class of parameters includes those that were expected to change when the expressed isoform of PMCA4 was changed. These are k5 (the rate of CaM-independent Ca2+ transport of PMCA), k8 and k8r (the rates for binding and dissociation of fully loaded CaM to PMCA), and k11 and k11r (the rates for dissociation/binding of all 4 Ca2+ from PMCA-CaM). Interestingly, [S] (the concentration of SERCA) changed in the fit for the cells expressing PMCA4a. However, this may be readily explained as other studies have shown that overexpression of one component of the Ca2+ signaling toolkit can lead to a change in the expression levels of other components (31,32). One limitation of the software we used for modeling is that it does not take into account variations in compartment size. Nevertheless, the shape of the Ca2+ signal in the PMCA4a-vs. 4b-expressing cells was still reflected accurately by these simulations. In addition, the fact that almost all of the variables in the fitting of the Ca2+ signals remained constant or changed in a rational way greatly increases our confidence in the validity of this approach. The parameters determined here will provide a basis for future studies of Ca2+ signaling in living cells.
In interpreting the differential effect of PMCA4a and 4b on shaping the cellular Ca2+ signal, it is important to note that these two PMCAs have the same turnover number under Vmax conditions, hence all the differences are a consequence of the different response of the isoforms to CaM and the different basal activities. Several interesting features emerge from our simulation of the handling of a Ca2+ signal by these pumps: a) at equal pump levels, PMCA4a is more effective than PMCA4b in lowering cytosolic Ca2+ after a spike; b) 10 times less PMCA4a is required to roughly reproduce the effect of PMCA4b on restoring cytosolic Ca2+. We emphasize that this occurs in spite of PMCA4a having a lower apparent affinity for CaM than PMCA4b. The explanation is that PMCA4a has a faster rate of binding of the Ca2+-CaM complex and thus reacts faster to the increase in cytosolic Ca2+. The data reported by Brini et al. (22) and our model, both show that PMCA4a decreases cytosolic Ca2+ faster than PMCA4b. On the basis of their results, Brini and co-workers concluded that “the availability of calmodulin may not be critical for the modulation of PMCA pumps in vivo” (22). Here we show that a more rigorous analysis of the kinetic properties of CaM activation provides a different explanation for their results. Moreover, our findings offer a possible explanation for the seeming redundancy of expressing different PMCA isoforms in a single cell.
Supplementary Material
a. Script file for fitting Ca2+ peak data for control and PMCA4b-expressing CHO cells.
b. Script file for fitting Ca2+ peak data for PMCA4a-expressing CHO cells.
c. Data file for Ca2+ peak in control cells.
d. Data file for Ca2+ peak in PMCA4b-expressing cells.
e. Data file for Ca2+ peak in PMCA4a-expressing cells.
Footnotes
We thank Katalin Török and Richard Thorogate (Department of Pharmacology and Clinical Pharmacology, St. George Hospital Medical School, London, UK) for the gift of TA-CaM. This work was supported by grants GM28835 and NS51769 from the NIH.
The abbreviations used are: CaM, calmodulin; ER, endoplasmic reticulum; PMCA, plasma membrane Ca2+ pump; SERCA, sarco(endo)plasmic reticulum Ca2+ pump.
REFERENCES
- 1.Strehler EE, Zacharias DA. Physiol. Rev. 2001;81:21–50. doi: 10.1152/physrev.2001.81.1.21. [DOI] [PubMed] [Google Scholar]
- 2.Chicka MC, Strehler EE. J. Biol. Chem. 2003;278:18464–18470. doi: 10.1074/jbc.M301482200. [DOI] [PubMed] [Google Scholar]
- 3.Hill JK, Williams DE, LeMasurier DE, Dumont RA, Strehler EE, Gillespie PG. J. Neurosci. 2006;26:6172–6180. doi: 10.1523/JNEUROSCI.0447-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penniston JT, Enyedi A. J. Membr. Biol. 1998;165:101–109. doi: 10.1007/s002329900424. [DOI] [PubMed] [Google Scholar]
- 5.Wang KKW, Villalobo A, Roufogalis BD. Trends Cell Biol. 1992;2:46–52. doi: 10.1016/0962-8924(92)90162-g. [DOI] [PubMed] [Google Scholar]
- 6.Carafoli E. FASEB J. 1994;8:993–1002. [PubMed] [Google Scholar]
- 7.Monteith GR, Roufogalis BD. Cell Calcium. 1995;18:459–470. doi: 10.1016/0143-4160(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 8.Carafoli E. Physiol. Rev. 1991;71:129–153. doi: 10.1152/physrev.1991.71.1.129. [DOI] [PubMed] [Google Scholar]
- 9.Enyedi A, Vorherr T, James P, McCormick DJ, Filoteo AG, Carafoli E, Penniston JT. J. Biol. Chem. 1989;264:12313–12321. [PubMed] [Google Scholar]
- 10.Falchetto R, Vorherr T, Brunner J, Carafoli E. J. Biol. Chem. 1991;266:2930–2936. [PubMed] [Google Scholar]
- 11.Falchetto R, Vorherr T, Carafoli E. Protein Sci. 1992;1:1612–1621. doi: 10.1002/pro.5560011209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berridge MJ, Bootman MD, Roderick HL. Nature Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 13.Penheiter AR, Bajzer Z, Filoteo AG, Thorogate R, Török K, Caride AJ. Biochemistry. 2003;42:12115–12124. doi: 10.1021/bi027098+. [DOI] [PubMed] [Google Scholar]
- 14.Heim R, Iwata T, Zvaritch E, Adamo HP, Rutishauser B, Strehler EE, Guerini D, Carafoli E. J. Biol. Chem. 1992;267:24476–24484. [PubMed] [Google Scholar]
- 15.O'Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vector. New York: Oxford University Press; 1994. [Google Scholar]
- 16.Enyedi A, Verma AK, Filoteo AG, Penniston JT. J. Biol. Chem. 1993;268:10621–10626. [PubMed] [Google Scholar]
- 17.Filoteo AG, Enyedi Á, Verma AK, Elwess NL, Penniston JT. J. Biol. Chem. 2000;275:4323–4328. doi: 10.1074/jbc.275.6.4323. [DOI] [PubMed] [Google Scholar]
- 18.Caride AJ, Elwess NL, Verma AK, Filoteo AG, Enyedi Á, Bajzer Z, Penniston JT. J. Biol. Chem. 1999;274:35227–35232. doi: 10.1074/jbc.274.49.35227. [DOI] [PubMed] [Google Scholar]
- 19.Enyedi A, Verma AK, Heim R, Adamo HP, Filoteo AG, Strehler EE, Penniston JT. J. Biol. Chem. 1994;269:41–43. [PubMed] [Google Scholar]
- 20.Kuzmic P. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 21.Knott G. A Mathematical Modeling Laboratory: MLAB Applications Manual. Bethesda, MD: Civilized Software, Inc; 1996. [Google Scholar]
- 22.Brini M, Coletto L, Pierobon N, Kraev N, Guerini D, Carafoli E. J. Biol. Chem. 2003;278:24500–24508. doi: 10.1074/jbc.M300784200. [DOI] [PubMed] [Google Scholar]
- 23.Penheiter AR, Filoteo AG, Penniston JT, Caride AJ. Biochemistry. 2005;44:2009–2020. doi: 10.1021/bi0488552. [DOI] [PubMed] [Google Scholar]
- 24.Caride AJ, Penheiter AR, Filoteo AG, Bajzer Z, Enyedi Á, Penniston JT. J. Biol. Chem. 2001;276:39797–39804. doi: 10.1074/jbc.M104380200. [DOI] [PubMed] [Google Scholar]
- 25.Mak D-OD, McBride S, Foskett JK. J. Gen. Physiol. 2001;117:299–314. doi: 10.1085/jgp.117.4.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamo HP, Rega AF, Garrahan PJ. J. Biol. Chem. 1988;263:17548–17554. [PubMed] [Google Scholar]
- 27.Dode L, Andersen JP, Leslie N, Dhitavat J, Vilsen B, Hovnanian A. J. Biol. Chem. 2003;278:47877–47889. doi: 10.1074/jbc.M306784200. [DOI] [PubMed] [Google Scholar]
- 28.Elshorst B, Hennig M, Försterling H, Diener A, Maurer M, Schulte P, Schwalbe H, Griesinger C, Krebs J, Schmid H, Vorherr T, Carafoli E. Biochemistry. 1999;38:12320–12332. doi: 10.1021/bi9908235. [DOI] [PubMed] [Google Scholar]
- 29.Kataoka M, Head JF, Vorherr T, Krebs J, Carafoli E. Biochemistry. 1991;30:6247–6251. doi: 10.1021/bi00239a024. [DOI] [PubMed] [Google Scholar]
- 30.Verma AK, Enyedi A, Filoteo AG, Strehler EE, Penniston JT. J. Biol. Chem. 1996;271:3714–3718. doi: 10.1074/jbc.271.7.3714. [DOI] [PubMed] [Google Scholar]
- 31.Guerini D, Schröder S, Foletti D, Carafoli E. J. Biol. Chem. 1995;270:14643–14650. doi: 10.1074/jbc.270.24.14643. [DOI] [PubMed] [Google Scholar]
- 32.Liu B-F, Xu X, Fridman R, Muallem S, Kuo TH. J. Biol. Chem. 1996;271:5536–5544. doi: 10.1074/jbc.271.10.5536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. Script file for fitting Ca2+ peak data for control and PMCA4b-expressing CHO cells.
b. Script file for fitting Ca2+ peak data for PMCA4a-expressing CHO cells.
c. Data file for Ca2+ peak in control cells.
d. Data file for Ca2+ peak in PMCA4b-expressing cells.
e. Data file for Ca2+ peak in PMCA4a-expressing cells.