Abstract
The C-terminal repeat domain (CTD) of the largest subunit of RNA polymerase II is composed of tandem heptad repeats with consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. In yeast, this heptad sequence is repeated about 26 times, and it becomes hyperphosphorylated during transcription predominantly at serines 2 and 5. A network of kinases and phosphatases combine to determine the CTD phosphorylation pattern. We sought to determine the positional specificity of phosphorylation by yeast CTD kinase-I (CTDK-I), an enzyme implicated in various nuclear processes including elongation and pre-mRNA 3′-end formation. Toward this end, we characterized monoclonal antibodies commonly employed to study CTD phosphorylation patterns and found that the H5 monoclonal antibody reacts with CTD species phosphorylated at Ser2 and/or Ser5. We therefore used antibody-independent methods to study CTDK-I, and we found that CTDK-I phosphorylates Ser5 of the CTD if the CTD substrate is either unphosphorylated or prephosphorylated at Ser2. When Ser5 is already phosphorylated, CTDK-I phosphorylates Ser2 of the CTD. We also observed that CTDK-I efficiently generates doubly phosphorylated CTD repeats; CTD substrates that already contain Ser2-PO4 or Ser5-PO4 are more readily phosphorylated by CTDK-I than unphosphorylated CTD substrates.
The C-terminal domain (CTD)1 of the largest subunit (Rpb1p) of budding yeast RNA polymerase II (RNAPII) is composed of about 26 tandem repeats of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (YSPTSPS). Considering that five of seven of these consensus amino acids are potential phosphoacceptors, it is not surprising that this domain is a substrate for phosphorylation. The extent of CTD phosphorylation correlates with the activity of the polymerase: initiating polymerases have unphosphorylated CTDs, whereas CTD hyperphosphorylation is associated with elongation (1–6). Phosphorylation also affects the protein-protein interactions between the CTD and binding partners such as mRNA processing factors (reviewed in Ref. 7).
In budding yeast, phosphorylation occurs predominantly at serines 2 and 5 of the CTD, and Ser2-PO4 and Ser5-PO4 are thought to have separate and essential roles. Substitution of either Ser2 or Ser5 with alanine or glutamate in each repeat is lethal in yeast (8), and suppressors of Ser2 mutations do not suppress the lethal phenotype of Ser5 mutation (9). Phosphorylation of Ser5 of the CTD occurs when polymerase is at promoters, whereas Ser2 phosphorylation is observed in coding regions (10). CTD-Ser5 phosphorylation is also present in coding regions of yeast genes, although it is detected at lower levels in coding regions than at promoters (11). In metazoa, as in yeast, the CTD becomes increasingly phosphorylated on Ser2 as RNAPII moves along the transcription unit, and Ser5 phosphorylation has been detected both at promoters and in coding regions (12, 13).
Differential phosphorylation of serine residues of the CTD is thought to identify the stage of transcription and help recruit the appropriate factors for that stage (10). For example, phosphorylation of Ser5 near promoters is important for capping enzyme recruitment (14–17) and activity (18), whereas the 3′-end formation factor, Pcf11p, requires Ser2 phosphorylation for binding (19). CTD kinases and phosphatases combine to define the CTD phosphorylation pattern and determine the protein-protein interactions involving the transcribing polymerase. Four transcriptionally relevant Saccharomyces cerevisiae CTD kinases have been identified. The Bur1p-Bur2p kinase is essential for viability and promotes transcription elongation (20–22). This kinase-cyclin pair co-precipitates with and phosphorylates Rpb1p (20). The other essential yeast CTD kinase is Kin28p, whose cyclin is Ccl1p. This kinase associates with the preinitiation complex (PIC) as a part of the TFIIH general transcription factor and phosphorylates the CTD after PIC formation (23) but before productive elongation. The Srb10p-Srb11p kinase-cyclin pair (24) is thought to be a negative regulator of transcription. Mutation of a catalytically important residue or deletion of srb10 restores viability of CTD truncation mutants, and in vitro assays indicate that Srb10p may inhibit transcription by phosphorylating the CTD prior to initiation (23).
CTDK-I, composed of Ctk1p (catalytic), Ctk2p (cyclin-like), and Ctk3p (unknown function), was the first CTD kinase to be characterized (25–27). This kinase cross-links to genes at all regions of a transcription unit (promoters, 5′-end, coding regions, 3′-end) (28). The CTDK-I catalytic subunit shares a high degree of similarity with CDK9, the catalytic subunit of metazoan positive transcription elongation factor (P-TEFb), and CTDK-I and P-TEFb share several functional similarities including stimulation of transcription elongation (29, 30) and involvement in 3′-end formation (11, 31, 32). In addition, CTDK-I has been implicated in various other nuclear processes including splicing (33), chromatin modification (34), and DNA repair (35).
Determining the positional specificity of phosphorylation by a CTD kinase is one approach to revealing the timing and functional consequences of the activity of a kinase. Ser5 kinases, for example, might be expected to act near promoters and be involved in capping or promoter clearance, whereas Ser2 kinases might be involved in elongation or 3′-end formation. Most of the information about which residues of the CTD are phosphorylated by a CTD kinase has been determined by two methods: 1) comparing the extent of in vitro phosphorylation of wild type CTD substrates with phosphorylation of substrates that have mutations in potential phosphoaccepting residues and 2) comparing reactivity of the CTD with phosphorylation-specific antibodies in wild type strains versus strains with a deleted or inactivated CTD kinase. The results obtained with these techniques are consistent with the idea that the Kin28p, Srb10p, and Bur1p kinases phosphorylate Ser5 of the CTD (10, 20, 23).
Although CTDK-I is required for normal CTD phosphorylation in vivo (26, 28, 36), the exact phosphorylation pattern created by CTDK-I has not been completely determined. CTDK-I has been characterized by genetic interactions between its catalytic subunit gene and rpb1 constructs with point mutations in the CTD (37). Implicating CTDK-I in Ser5 phosphorylation is the synthetic lethal interaction between ctk1Δ and a mutant rpb1 construct that has a mix of wild type repeats and repeats with alanine substituted for serine at position 5. On the other hand, ctk1Δ is also synthetic lethal with rpb1 constructs that have mutations at position 2. CTDK-I has also been implicated in Ser2 phosphorylation because ctk1Δ strains have reduced H5 mAb epitopes in coding regions of certain genes (28).
Here we more completely characterized the phosphorylation of the CTD by CTDK-I. Using BIACORE technology, we first tested the specificity of monoclonal antibodies commonly used to determine CTD phosphorylation sites and found that mAb H5 binds to CTD peptides with Ser5 and/or Ser2 phosphorylated. In light of this lack of specificity, we sought to determine the site of CTDK-I phosphorylation by direct chemical analysis. We found that CTDK-I phosphorylates Ser5 if the CTD substrate is not phosphorylated at Ser5, but CTDK-I will phosphorylate Ser2 of a CTD substrate if Ser5 is already phosphorylated. In addition, CTDK-I is more reactive toward substrates that are prephosphorylated at serine 2 or 5 compared with an unphosphorylated CTD substrate; therefore, it efficiently creates doubly phosphorylated CTD repeats.
EXPERIMENTAL PROCEDURES
Materials
Materials and sources were as follows: H5, H14, and 8WG16 antibodies (Covance), DEAE-Sepharose Fast Flow (Amersham Biosciences), MacroPrep CM Support (Bio-Rad), ImmunoPure Immobilized streptavidin (Pierce), protease inhibitor mixture (catalog no. P-2714; Sigma), 20 × 20-cm cellulose TLC plates (Fisher), EZ-Link sulfo-N-hydroxysuccinimide-LC-Biotin (Pierce), SOURCE 15RPC ST 4.6/100 column (Amersham Biosciences), [γ-32P]ATP >4000 Ci/mmol (ICN), BIACore 3000 (BIACore, Inc.), BIACore 3000 (BIACore), streptavidin sensor chip (BIACore), BIAEvaluation software (BIACore), and RP C18 column (PerkinElmer Life Sciences).
Purification of CTDK-I
CTDK-I was purified based on modification of the CTDK-I purification described in Refs. 25 and 27. The enzyme was tracked through purification by activity assays and Western blotting against Ctk1 and Ctk3 protein subunits. 700 g of Fleischmann yeast were broken open in a 1-gallon stainless steel Waring blender with liquid nitrogen by blending six times for 30 s. The resulting yeast powder was resuspended in 800 ml of buffer B1 (25 mm Tris, pH 7.4, 200 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, phenylmethylsulfonyl fluoride (1:1000 dilution of a saturated solution in isopropyl alcohol), 0.5 mm leupeptin), and debris was pelleted by centrifugation at 15,000 × g. Ammonium sulfate was added to the supernatant to 50% saturation, and the precipitate was pelleted by centrifugation at 15,000 × g. The precipitate was resuspended in buffer BH (25 mm HEPES, pH 7.6, 8% glycerol, 0.1 mm EDTA, protease inhibitor mixture, 1 mm dithiothreitol, phenylmethylsulfonyl fluoride) to give a conductivity equal to 0.2 m (NH4)2SO4 and loaded onto 175 ml of DEAE resin. DEAE flow-through was collected, diluted to a conductivity equal to 150 mm KCl, loaded onto a 45-ml CM resin, and eluted with a gradient of KCl in buffer BH. The 300–450 mm KCl fractions contained a peak of kinase activity and CTDK-I proteins. These fractions were further purified on an immuno-affinity resin made with a rabbit antibody raised against and affinity-purified against a peptide (“S2 peptide”; LSRYNDTSFQTSSRYQGSRY) unique to the N terminus of the Ctk1 protein (see Ref. 27). The concentration of CTDK-I in S2 elution fractions was determined based on Coomassie Blue staining with bovine serum albumin standards and quantitative Western blotting with an antibody against Ctk1p.
CTD Substrates
GST-yCTD was purified as previously described (38). Mutant GST-CTD substrate constructs were kindly provided by Jeff Corden (The Johns Hopkins University) and were purified according to Ref. 36. Purified RNAPII (39) was a gift from Aled Edwards (University of Toronto). The nonphospho peptide, 2-phospho peptide, and 5-phospho peptide were kind gifts from Steve Hanes (Wadsworth Center, New York State Department of Health). The biotinylated 2-phospho peptide, biotinylated 5-phospho peptide, and biotinylated 2 + 5-phospho peptide were synthesized and purified by Anaspec (San Jose, CA). The nonphospho peptide was biotinylated according to Pierce instructions and purified on a SOURCE 15RPC ST 4.6/100 FPLC column. Substrates are described in Table I.
TABLE I. Description of CTD substrates.
| Substrate | Sequence | Description |
|---|---|---|
| RNAPII | Ref. 39 | |
| GST-yCTD | GST fusion of yeast CTD (38) | |
| WT16 | (YSPTSPS)16 | GST fusion of 16 consensus repeats |
| A2 14 | (YaPTSPS)14 | GST fusion of 14 S2A repeats |
| A5 15 | (YSPTaPS)15 | GST fusion of 15 S5A repeats |
| E2 15 | (YePTSPS)15 | GST fusion of 15 S2E repeats |
| E5 18 | (YSPTePS)18 | GST fusion of 18 SE5 repeats |
| Nonphosphoa | (YpSPTSPS)3 | Unphosphorylated peptide |
| 2-Phosphoa | (YpSPTSPS)3 | Each repeat has Ser2 phosphorylated |
| 5-Phosphoa | (YSPTpSPS)3 | Each repeat has Ser5 phosphorylated |
| 2+5-Phosphoa | (YpSPTpSPS)3 | Each repeat has Ser2 + Ser5 phosphorylated |
Synthetic peptides with three CTD repeats. pS, phosphoserine.
In Vitro Kinase Activity Assays
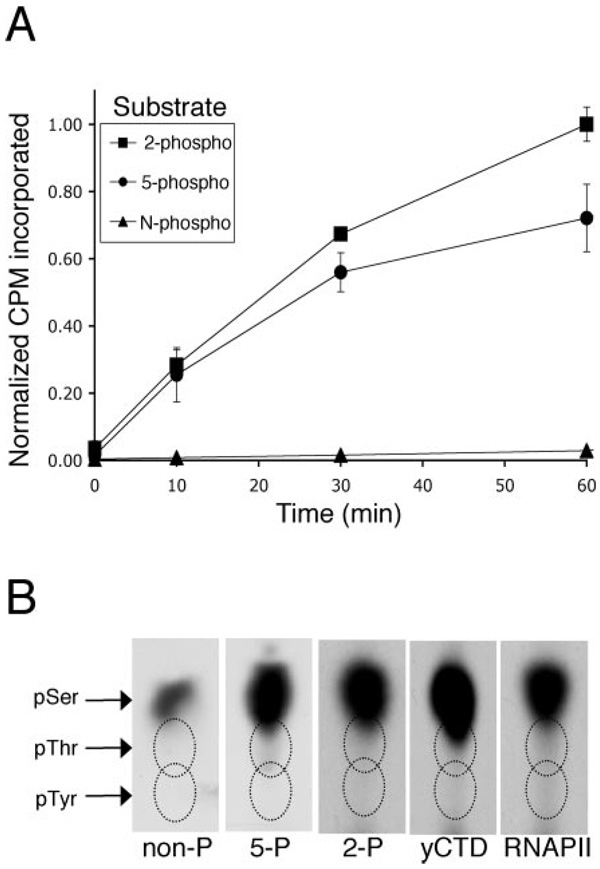
Typically, kinase reactions included 270 µm ATP (10 times the apparent enzyme Km for ATP (25)), 5 µCi of [γ-32P]ATP, 10 mm MgCl2, 25 mm HEPES, 10–50 ng of CTDK-I, and 1.0 µg of GST-CTD substrate or 5.0 µg of synthetic CTD peptide substrate in a 25-µl reaction volume. Reactions were 1 h at 30 °C (unless otherwise specified). For phosphorylation site determination, higher specific activity of radioactive ATP (100 µm ATP and 10 µCi of [γ-32P]ATP) was used to increase radioactive phosphate incorporation. For the time course with biotinylated peptides, reactions were stopped by quenching with 25 mm cold ATP and 100 mm EDTA followed by refrigeration. Biotinylated peptides were precipitated with 25 µl of streptavidin beads and washed in 1 ml of phosphate-buffered saline until radioactivity was undetectable in supernatants (about 10×). Pellets were Cerenkov-counted, and radioactivity incorporation (arbitrarily normalized to a value of 1.0 for the 60-min 2-phospho peptide) was plotted against time. Streptavidin bead precipitation was selected over SDS-PAGE gels, because some phosphorylated peptide species co-migrate with free ATP (data not shown; also see Ref. 40 and Fig. 3B), whereas streptavidin pellets should bind all peptide species independent of their phosphorylation status.
FIG. 3. Phosphorylation of CTD peptides by CTDK-I in a time course.
A, 5.0 µg of biotinylated peptides described in Table I were phosphorylated in vitro with CTDK-I and [γ-32P]ATP. Peptides were pelleted with streptavidin beads and washed extensively in phosphate-buffered saline, and radioactivity incorporation was measured by Cerenkov counting. Radioactivity incorporation (counts/min in streptavidin bead pellet) is plotted against time with S.E. bars for n = 2 time courses. The 2-phospho and 5-phospho CTD peptides were the most heavily phosphorylated by CTDK-I. The nonphospho CTD peptide was also phosphorylated but to a lesser extent compared with the two prephosphorylated peptide substrates. The 2 + 5-phospho CTD peptide was a very poor CTDK-I substrate (data not shown). B, the nonphospho, 5-phospho, and 2 + 5-phospho CTD peptide substrates were phosphorylated by CTDK-I in vitro for 60 min, and HPLC-purified products were analyzed for the presence of phosphoserine, phosphotyrosine, and phosphothreonine after acid hydrolysis. GST-yCTD and purified RNA-PII were also analyzed for phosphoamino acid content. Only phosphoserine was detected in all products analyzed
Purification of Phosphorylated CTD Peptide Products
Kinase reactions (25 µl) were diluted to 100 µl in 0.1% trifluoroacetic acid and loaded onto a C18 HPLC column. After 60 min of washing with 0.1% trifluoroacetic acid at 1 ml/min, products were eluted with an acetonitrile gradient (0–100% over 30 min at 0.2 ml/min). Peaks of phosphorylated products were identified by Cerenkov counting.
Phosphoamino Acid Analysis
CTD peptide substrates were phosphorylated in an in vitro kinase assay (as described above). Phosphorylated products were purified by HPLC on a C18 column and incubated in 6 m HCl for 1 h at 110 °C. Hydrolyzed products, along with phosphoamino acid standards (Ser(P), Tyr(P), and Thr(P)), were separated by thin layer chromatography in one dimension on cellulose plates (41). Phosphoamino acids were visualized by autoradiography on Kodak Biomax MR film.
Phosphorylation Site Determination
CTD peptides were phosphorylated in vitro (as described above), and phosphorylated products were separated by HPLC on a C18 column. Peptide immobilization and Edman sequencing were done as described in Ref. 42. Radioactivity released in each cycle was counted by Cerenkov counting.
Determination of Antibody-CTD Affinities Using Surface Plasmon Resonance Measurements (BIACORE)
For immobilization of CTD peptides to the streptavidin sensor chip surface, biotinylated CTD peptides (Table I; nonphospho, 2-phospho, 5-phospho, and 2 + 5-phospho peptide) were diluted to ~5 ng/ml in phosphate-buffered saline with 0.05% Tween 20. For low density immobilization, about 150 response units (RU) of each CTD peptide were immobilized to the sensor chip by injecting peptides over the surface at 5 µl/min, whereas about 600 RU of each peptide were immobilized to prepare the high density surface. Raw ascites fluid containing monoclonal antibody H5, H14, or 8WG16 was diluted in phosphate-buffered saline with 0.05% Tween 20 to the concentrations indicated in the figure legends. Antibodies were interacted with CTD peptides (on the low and high density surfaces) for 300 s at 50 µl/min. Dissociation time was 200 s, followed by surface regeneration with 2 m MgCl2 (100 µl at 50 µl/min).
For monocl onal antibodies H5 and H14, apparent equilibrium and kinetic association and dissociation constants were determined by fitting curves to the binding sensorgrams using the Langmuir binding model (A + B = AB) and using the BIAEvaluation software. Using BIAEvaluation software, the goodness of fit was assessed by examining residual plots and χ2 values. In cases where a simple Langmuir model gave a poor fit, additional parameters such as base-line drift or mass transfer were added (as indicated in the figure legends). An acceptable fit could not be obtained for the 8WG16 mAb, but equilibrium constants were determined using Scatchard analysis. Concentrations of monoclonal antibodies in ascites fluid were estimated based on manufacturer literature and stained SDS-polyacrylamide gels. The stock raw ascites fluids were determined to contain about 3 µm H14, 3 µm H5, and 33 µm 8WG16.
RESULTS
Specificity of Monoclonal Antibodies 8WG16, H5, and H14
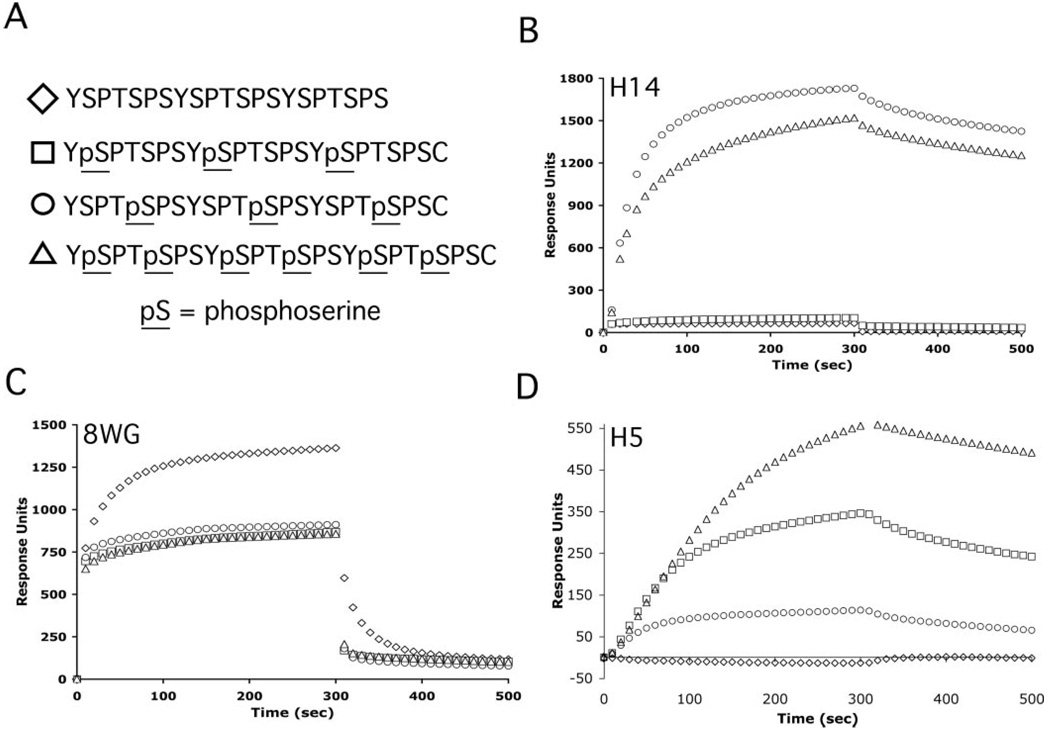
Commercially available monoclonal antibodies that are regarded as phosphorylation specific have commonly been employed to determine in vivo CTD phosphorylation patterns and the positional specificity of phosphorylation by CTD kinases. The specificity of these antibodies has previously been characterized based on binding to wild type and mutant CTD substrates in Western blots, pull-downs, and enzyme-linked immunosorbent assay (19, 28, 36). Because we hoped to use the available mAbs to analyze CTD phosphorylation by CTDK-I, we further characterized their binding properties. We were particularly interested in following up on previous indications that mAb H5 is not specific for serine 2 phosphorylation (19, 28, 36). Toward this end, surface plasmon resonance measurements (BIACORE) were used to investigate the binding of these antibodies to a set of CTD peptides (Fig. 1A). Because it is currently not possible to chemically synthesize or enzymatically generate long CTD substrates with rigorously known phosphorylation patterns, we used three-repeat synthetic peptides with Ser-PO4 in exactly known positions for the binding analyses. In these experiments, diluted ascites fluid containing either mAb H5, H14, or 8WG16 was interacted with CTD peptides that were immobilized on the surface of a streptavidin sensor chip. In the BIACORE sensorgrams, binding is represented by response units (on the y axis) as a function of time (on the x axis).
FIG. 1. Specificity of binding of H5, H14, and 8WG16 antibodies to CTD peptides using BIACORE (surface plasmon resonance).
Biotinylated CTD peptides were coupled to the surface of streptavidin sensor chips, and the specificity of binding of monoclonal antibodies to these peptides was determined using BIACORE technology. A, sequences of biotinylated peptides that were immobilized on streptavidin sensor chips with symbols used in graphs in B–D. B, sensorgrams for the binding of mAb H14 (39 nm) to CTD peptides show that it binds to the 5-phospho and 2 + 5-phospho peptides but not the nonphospho or 2-phospho peptides. C, sensorgrams for the binding of mAb 8WG16 (4 mm) to CTD peptides show that it binds to the nonphospho CTD peptide with the highest steady state RU level. D, sensorgrams for the binding of mAb H5 (1.2 nm) to CTD peptides show that it binds to the 2-phospho, 5-phospho, and 2 + 5-phospho CTD peptides but not to the nonphospho peptide. Sensorgrams shown here are from the lower peptide density chip but are also representative of the specificities observed from the higher peptide density chip.
Monoclonal antibody H14 bound to the 5-phospho and 2 + 5-phospho CTD peptides (Fig. 1B) but not the nonphospho or 2-phospho peptides, consistent with specificity for serine 5 phosphorylation. Also in agreement with previous characterization, mAb 8WG16 bound to the nonphospho CTD peptide with the highest steady state RU level. Binding to the 2-phospho, 5-phospho, and 2 + 5-phospho CTD peptide displayed very rapid dissociation kinetics, and much of the signal response was probably due to nonspecific binding (Fig. 1C). The H5 antibody did not display the specificity that it has previously been thought to possess (Fig. 1D). This antibody bound to CTD peptides that are phosphorylated at serine 5 and/or serine 2 (2-phospho, 5-phospho, and 2 + 5-phospho peptides).
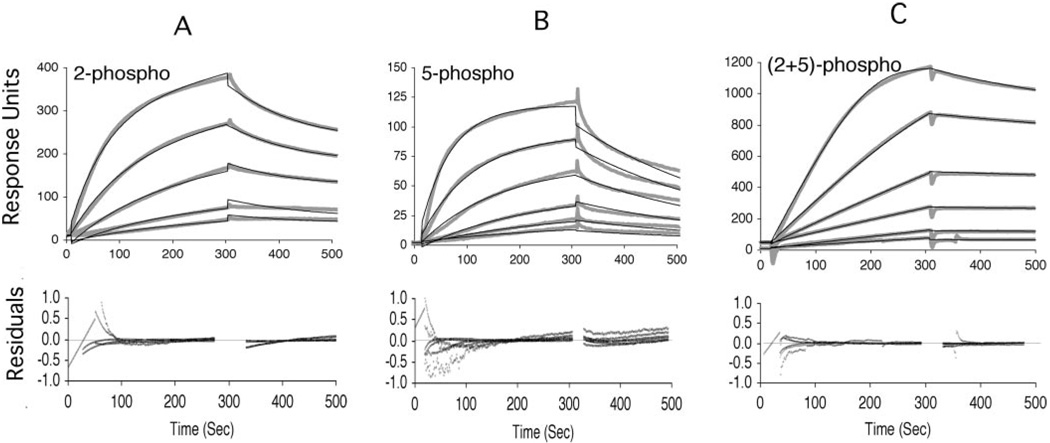
To determine apparent association constants for the interaction between each antibody and each CTD peptide, serial 2-fold dilutions of ascites fluid containing each antibody were titrated over the CTD peptides. Sensorgrams for the H5 mAb-peptide interactions are shown in Fig. 2. Antibodies H14 and 8WG16 were also titrated (data not shown). Although the H5 antibody has been considered specific for Ser2 phosphorylation, these sensorgrams show that the H5 mAb bound to 2-phospho (Fig. 2A), 5-phospho (Fig. 2B), and 2 + 5-phospho (Fig. 2C) peptides at antibody dilutions as high as 1:44,800, and binding response increased with increasing concentrations of antibody.
FIG. 2. Titration of mAb H5 over CTD peptides.
Serial dilutions of mAb H5 were titrated over the four CTD peptides immobilized on the BIACORE surface. Sensorgrams for binding of mAb H5 to the 2-phospho (A), 5-phospho (B), and 2 + 5-phospho CTD peptides (C) indicate that binding to each of these CTD peptides is proportional to the concentration of antibody. Thick light curves are actual binding sensorgrams, whereas thin dark curves were fit to the binding sensorgrams using BIAEvaluation software. For the 2-phospho and 5-phospho peptides, a simple Langmuir two-component model (A + B = C) was used, whereas the 2 + 5-phospho required the addition of a mass transfer parameter. Inclusion of a mass transfer parameter did not improve the fit for Fig. 2, A or B. Curve fits were used to determine apparent kinetic and equilibrium association and dissociation constants (Table II). Residual plots describe how well the fitted curves match the actual binding sensorgrams (see methods). The top sensorgrams in 2-phospho and 5-phospho plots are a 1:2800 (1.2 nm) dilution of mAb H5 with serial 2-fold dilutions of mAb H5 from top to bottom. Top sensorgram for 2 + 5-phospho plots is a 1:1700 (1.9 nm) dilution of mAb H5 raw ascites fluid with serial 2-fold dilutions of mAb H5 from top bottom. For each peptide, background binding to the nonphospho CTD peptide was subtracted as background. Sensorgrams shown here are generated from titrations on either the low or high peptide density chip but are also representative of the sensorgrams generated on the other chip.
The apparent affinities of each antibody for CTD peptides were calculated by curve fitting to data from the mAb titrations as described under “Experimental Procedures.” Measured affinities of mAb H5 for the phosphorylated CTD peptides were relatively high, with dissociation constants in the low nanomolar range. The H5 antibody bound to the 5-phospho and 2-phospho peptides with similar high affinities, but this antibody bound to the 2 + 5-phospho peptide with the highest affinity (Fig. 1D and Table II). Curve-fitting analyses also indicated that the H14 antibody bound to the 2 + 5-phospho and the 5-phospho peptides with very similar high affinities (see legend to Table II).
TABLE II. Apparent kinetic and affinity constants for antibody H5 binding to CTD peptides.
Apparent antibody binding constants were determined with BIACORE by titrating antibodies over two separate surfaces with a high or low density of immobilized CTD peptides. Apparent kinetic (kon and koff) and equilibrium (KA and KD) constants were determined by curve fitting based on a two-component binding model (A + B = C). For some interactions, a better fit to the simple model was observed when base-line drift or mass transfer parameters were added. For mAb H5, χ2 values ranged from 3.4 to 34.4 for a global fit to sensorgrams for at least five antibody titrations, with the exception of the H5–2+5P interaction on the low density chip, which we were unable to fit as well (χ2 value of 83.3 for a global fit to three titrations). Average values for each binding constant with S.E. (for n = 2 global fits) are reported. Curve fitting was also attempted for the H14 antibody based on a two-component binding model. Although these fitted curves gave higher χ2 values (range 16.6–116), we estimate affinity to be in the low nanomolar range for the interactions between the H14 antibody and the 5-phospho and 2+5-phospho peptides (data not shown). Curve fitting was unsuccessful for the 8WG16 antibody, but Scatchard analysis indicated a low micromolar KD for the interaction between 8WG16 and the nonphospho peptide (data not shown). Fitted curves and residual plots for the H5 antibody are shown in Fig. 2.
| Interaction | kon | koff | KA | KD |
|---|---|---|---|---|
| µm−1 s−1 | s−1 | nm−1 | nm | |
| H5–2P | 15 (9) | 0.0015 (0.0003) | 10 (4) | 0.13 (0.06) |
| H5–5P | 11 (1) | 0.0030 (0.0003) | 3.7 (0.1) | 0.27 (0.01) |
| H5–2+5P | 23 (2) | 0.0010 (0.0001) | 24 (5) | 0.045 (0.009) |
CTD Substrates That Are Already Phosphorylated Are More Efficient Substrates for CTDK-I
In light of the BIACORE results, which showed that H5 bound to CTD peptides that are prephosphorylated at serine 5 and/or serine 2, we sought to determine the residues phosphorylated by CTDK-I without utilizing this antibody. We also wanted to use CTD substrates with wild type sequence (no amino acid substitutions from consensus sequence). We used biotinylated three-repeat CTD peptides (Table I and Fig. 1A) that are either unphosphorylated or phosphorylated at position 2 and/or 5 to mimic CTD phosphorylation patterns that CTDK-I could encounter in vivo. These CTD peptide substrates were phosphorylated in a 60-min time course by CTDK-I, and the extent of radioactive phosphate incorporation was measured in streptavidin bead precipitates (Fig. 3A). Compared with the nonphospho peptide, the 2-phospho and 5-phospho peptides were far better CTDK-I substrates. After 60 min, the 2-phospho peptide and 5-phospho peptides were 14- and 8-fold (respectively) more phosphorylated than the nonphospho CTD peptide. The 2 + 5-phospho peptide was a very poor CTDK-I substrate (data not shown).
CTDK-I Phosphorylates Serines of the CTD
Each consensus heptapeptide repeat of the CTD contains five potential targets for phosphorylation by a CTD kinase. To determine which amino acid(s) of the CTD are phosphorylated by CTDK-I, CTD substrates were phosphorylated in vitro by purified CTDK-I, and phosphorylated products were analyzed by phosphoamino acid analysis (Fig. 3B). Substrates analyzed include the nonphospho, 2-phospho, and 5-phospho peptides, as well as GST-yCTD fusion protein (38) and RNAPII (39). Although each repeat contains one tyrosine, one threonine, and three serines, all substrates tested were only phosphorylated on serine residues by CTDK-I (Fig. 3B).
CTDK-I Can Phosphorylate Ser2 or Ser5 of the CTD
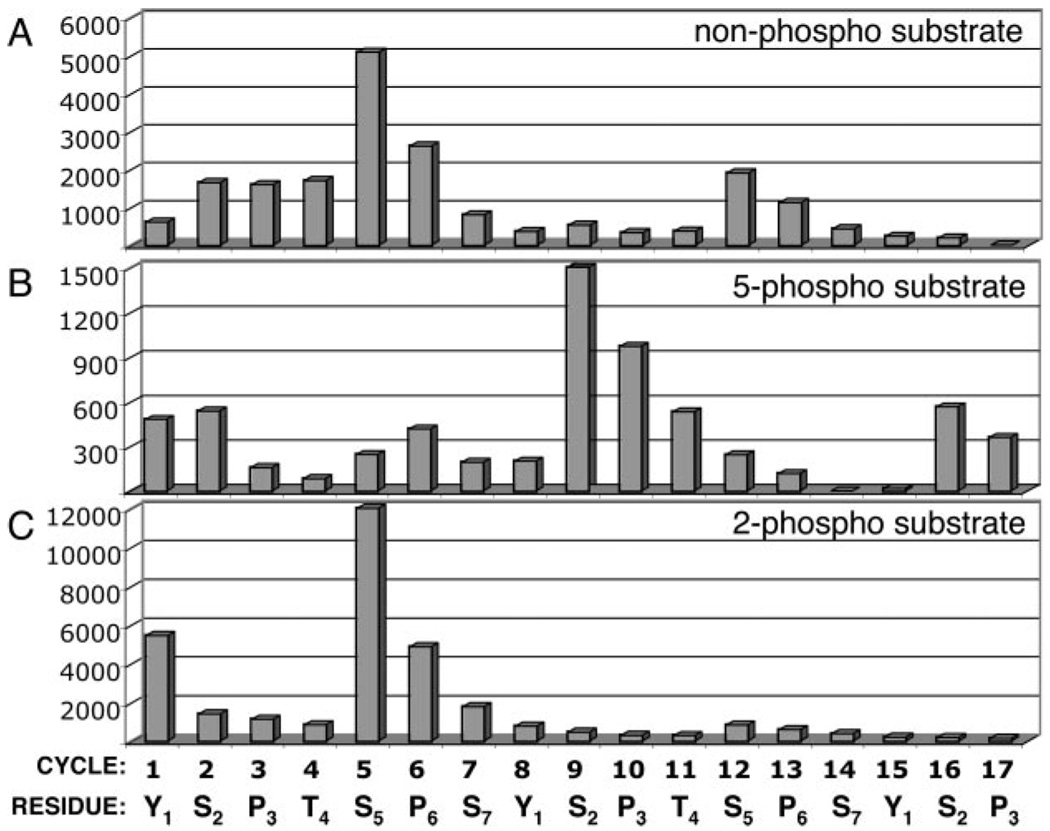
Since each CTD repeat has three serines, we sought to determine whether CTDK-I could phosphorylate serine 2, serine 5, or serine 7. CTD peptides were phosphorylated in vitro with CTDK-I and [γ-32P]ATP. Phosphorylated products were subjected to Edman degradation to sequentially remove individual amino acids from the N terminus, and the amount of radioactivity released in each Edman cycle was measured by Cerenkov counting.
Although Ser5 phosphorylation by CTDK-I has not been reported previously, Edman degradation of the nonphospho peptide product (Fig. 4A) showed a large peak of activity released in the cycle corresponding to Ser5 and a smaller peak in the 12th cycle (Ser5 of the second repeat). The main peaks of radioactivity release corresponded to serines 5 and 12, but radioactivity release continued in subsequent cycles (i.e. proline 6) because of inherent inefficiency in amino acid cleavage at each step.
FIG. 4. Determination of phosphorylation sites by Edman degradation of CTD substrates phosphorylated by CTDK-I.
CTD peptides were phosphorylated by CTDK-I in vitro for 60 min with [γ-32P]ATP, and products were HPLC-purified. Residues were sequentially removed from the N terminus by Edman degradation (for 17 cycles), and radioactivity (32P) release was measured for each cycle. Radioactivity release (Cerenkov-counted counts/min) from the peptide is plotted for each corresponding residue. A, Edman degradation of the nonphospho peptide phosphorylated by CTDK-I shows peaks of radioactivity release corresponding to serines 5 and 12 (equivalent to serine 5 of the first and second heptad repeats). B, Edman degradation of the 5-phospho peptide phosphorylated by CTDK-I shows peaks of radioactivity release corresponding to serines 9 and 16 (equivalent to serine 2 of the second and third heptad repeats). C, Edman degradation of the 2-phospho peptide phosphorylated by CTDK-I shows a peak of radioactivity release corresponding to serine 5 of the first heptad repeat.
Edman analysis was also used to investigate the positional specificity of phosphorylation of prephosphorylated CTD substrates by CTDK-I. For the 5-phospho peptide phosphorylated by CTDK-I (Fig. 4B), a peak of radioactivity was released in the cycle corresponding to Ser2 of each of the consensus repeats, with the largest peak corresponding to Ser2 of the second repeat (serine 9). Like the nonphospho-CTD peptide substrate, the 2-phospho-CTD peptide was phosphorylated at Ser5 by CTDK-I (Fig. 4C). Together these results show that CTDK-I phosphorylates Ser5 of CTD substrates that are not phosphorylated at Ser5, but it phosphorylates Ser2 if Ser5 is already phosphorylated.
CTDK-I Phosphorylates Mutant CTD Substrates to Different Degrees
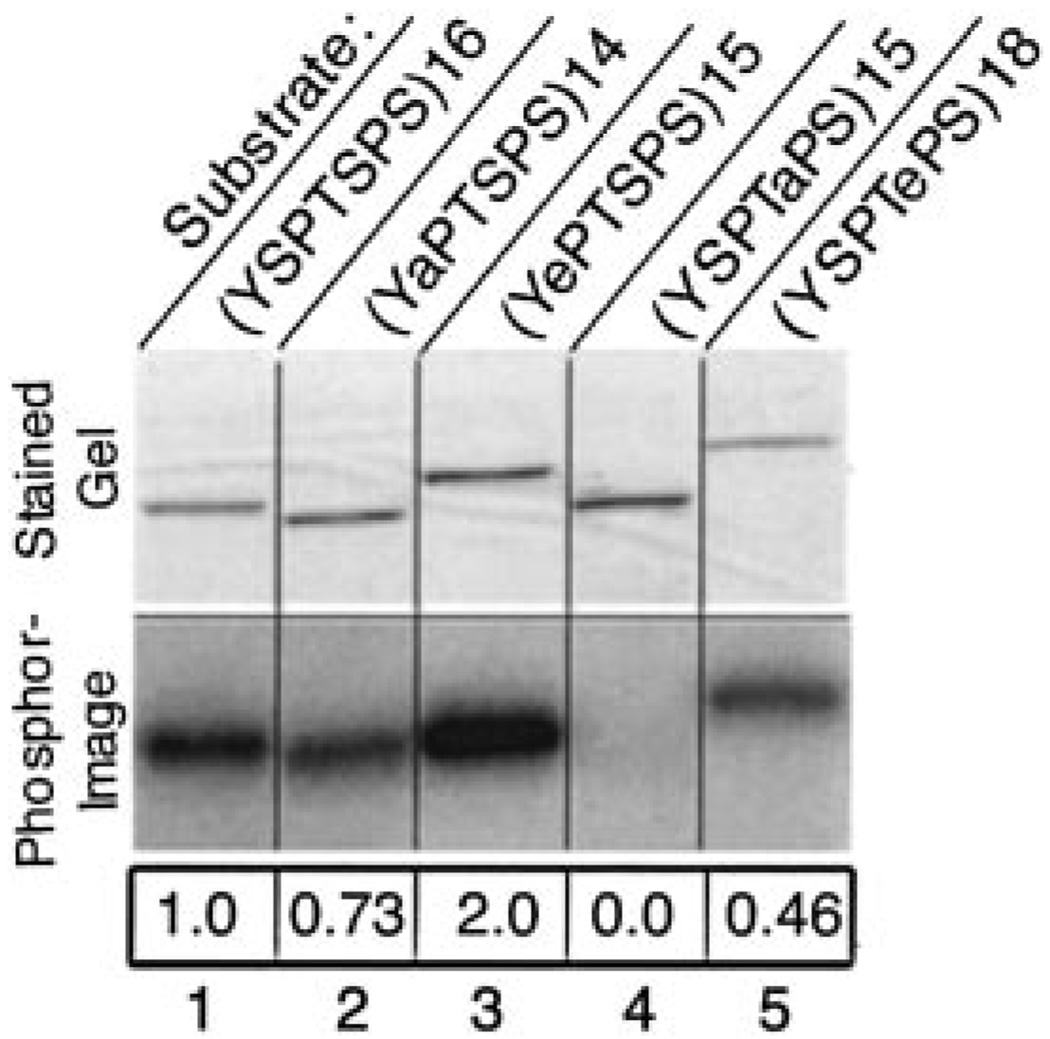
We sought to compare the activity of CTDK-I toward three-repeat CTD peptides with its activity toward longer CTD substrates. Because, as mentioned above, long CTD substrates with rigorously known phosphorylation patterns cannot be generated synthetically or enzymatically, we used bacterially expressed GST-CTD fusion proteins carrying 15 or more wild-type or Ser-substituted repeats as substrates for CTDK-I. These constructs, with alanine (eliminates potential phosphoacceptor) or glutamate (resembles phosphosereine) substitutions at serine 2 or 5 of each repeat, are described in Table I. Purified GST-CTD fusion proteins were reacted in vitro with CTDK-I and [γ-32P]ATP, and phosphorylated products were subjected to SDS-PAGE and visualized using a PhosphorImager (Fig. 5). The results obtained with mutant CTD substrates reconciled well with the Edman sequencing data (Fig. 4). The S5A mutant (Fig. 5, lane 4) was a very poor substrate for CTDK-I. This activity is consistent with the finding that CTDK-I phosphorylated serine 5 of an unphosphorylated CTD (Fig. 4A), but it is also a formal possibility that the S5A mutation interferes with phosphorylation of another residue. Also consistent with serine 5 phosphorylation by CTDK-I, the S2A mutant (Fig. 5, lane 2) was a good CTDK-I substrate (73% of wild type). The reduction in phosphorylation of the S2A mutant compared with the wild type substrate could indicate that serine 2 is important for substrate recognition.
FIG. 5. Phosphorylation of mutant GST-CTD substrates by CTDK-I.
1.0 µg of each purified GST-CTD fusion protein was phosphorylated by CTDK-I in vitro for 60 min. Reaction mixtures were analyzed by SDS-PAGE on a 4–15% Ready Gel and stained with Coomassie Blue. Phosphorylated products were visualized using a PhosphorImager, and the degree of phosphorylation of each substrate was quantified using ImageQuant software. The extent of phosphorylation of each mutant GST-CTD substrate is normalized to a value of 1.0 for the wild type GST-CTD fusion protein (WT16). Substrates are described in Table I. Lane 1, wild type GST-CTD fusion protein (1.0); lane 2, A214 (YSPTSPS)14 (0.73); lane 3, E215 (YePTSPS)15 (2.0); lane 4, A515 (YSPTaPS)15 (0.0); lane 5, E518 (YSPTePS)18 (0.46).
Since glutamate resembles phosphoserine, CTD mutants with glutamate substituted for serine were also tested for phosphorylation by CTDK-I (Fig. 5, lanes 3 and 5). Whereas the S5A CTD mutant (Fig. 5, A515, lane 4) was a very poor CTDK-I substrate, the S5E (Fig. 5, E518, lane 5) was a reasonably good substrate for CTDK-I (46% of wild type). This finding is consistent with the result in Fig. 5B, where Edman degradation shows that CTDK-I is able to phosphorylate serine 2 if serine 5 is already phosphorylated.
The S2E mutant (Fig. 5, E215, lane 3) appeared to be a better substrate for CTDK-I than the wild type substrate (200% of wild type), supporting the idea that phosphorylation of Ser2 may enhance the activity of this kinase toward its CTD substrate. This finding is consistent with the time course in Fig. 3A, which showed that the CTD peptide already phosphorylated at serine 2 is the best CTDK-I substrate. Together these results indicate that CTDK-I can phosphorylate Ser5, but this kinase phosphorylates Ser2 when Ser5 is already phosphorylated.
DISCUSSION
Brief Summary of CTDK-I Phosphorylation Specificity
We have used direct chemical analysis of reaction products to demonstrate that CTDK-I can phosphorylate serine residues at two different positions in the heptameric repeats of the CTD (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7). On unphosphorylated repeats and repeats with Ser2-PO4, CTDK-I adds phosphates preferentially to Ser5. On repeats already containing Ser5-PO4, CTDK-I adds phosphate groups preferentially to Ser2, and it does so with a much higher activity than for nonphosphorylated repeats. Thus, CTDK-I is better at generating doubly phosphorylated repeats than at creating singly phosphorylated repeats, a property we believe is relevant to its roles in vivo.
Specificity of Anti-CTD Monoclonal Antibodies
We were induced to use a chemical method of product analysis because we confirmed that the monoclonal antibody frequently employed to assess Ser2 phosphorylation of the CTD actually reacts with multiple forms of phosphorylated CTD. We performed BIACORE (surface plasmon resonance) analysis on the binding of three commercially available CTD-directed mAbs to synthetic three-repeat peptides with no phosphates, phosphates on Ser2 residues, phosphates on Ser5 residues, or phosphates on both Ser2 and Ser5 residues. BIACORE analysis of the specificity of monoclonal antibodies H14 and 8WG16 was largely consistent with their designated specificities; in contrast, analysis of the H5 mAb highlighted the relevance of the reactivity of this antibody toward the 5-phospho CTD peptide. In BIACORE, at much more dilute concentrations (1:44,800) than those recommended for immunoprecipitations or Western blots (1:100 to 1:500), mAb H5 bound to peptides that were phosphorylated at position 5 and/or 2 (Fig. 2). The effect of Ser5 phosphorylation on H5 reactivity has previously been overlooked, although mAb H5 displayed cross-reactivity toward Ser5 phosphorylation in enzyme-linked immunosorbent assays (28) and pull-downs (19).
The specificity of binding of antibodies H14 and 8WG16 as determined with BIACORE was consistent with expectations. Monoclonal antibody 8WG16 preferentially bound unphosphorylated repeats, and it showed no affinity for repeats containing Ser2-PO4 (Fig. 1C), consistent with the previous conclusion that the unphosphorylated Ser2 side chain is critical for its binding (28, 36). Additionally, BIACORE sensorgrams showed that the 8WG16 antibody has a very fast dissociation rate (compare Fig. 1C, 300–500 s with the same time period in Fig. 1, B and D). This rapid dissociation rate presumably reflects the fact that the 8WG16-RNAPII complex is easily disrupted with low concentrations of salt and polyols (43). Phosphorylated Ser5 was the key feature for H14 reactivity (Fig. 1B), whereas the phosphorylation state of Ser2 was essentially irrelevant. This antibody bound to the 5-phospho peptide with a very similar affinity as it bound to the 2 + 5-phospho peptide.
Despite its lack of specificity, mAb H5 can be useful when used in combination with other antibodies. For example, if mAb H5 epitopes are detected while H14 epitopes are not detected, it is likely that Ser2 is the only phosphorylated residue accessible to the antibodies. This example reflects the antibody pattern seen in coding regions of some yeast genes, although detection of H14 epitopes in coding regions can vary with the batch of H14 used (10, 11). In other cases, the combined use of these mAbs will not be sufficient to determine the phosphorylation pattern generated by a particular CTD kinase. For example, if both H5 and H14 react, whether H5 reacts with Ser2 or Ser5 phosphorylation will be indeterminate. For these reasons, we used a protein sequencing technique to directly determine sites of phosphorylation by CTDK-I.
Positional Specificity of Phosphorylation by CTDK-I
Our results revealed that CTD substrates that are already phosphorylated are much more efficiently phosphorylated by CTDK-I than unphosphorylated CTD substrates. Notably, CTDK-I readily phosphorylates Ser2 of CTD substrates that are already phosphorylated at Ser5. These experiments and in vivo cross-linking experiments (28) together indicate that an important in vivo role of CTDK-I may be to phosphorylate Ser2 of a CTD substrate that is already phosphorylated at position 5 by another kinase (such as Kin28p). Given our in vitro results (Fig. 3A), it is likely that prephosphorylation on Ser5 prepares the CTD for efficient phosphorylation on Ser2 by CTDK-I (see model in Fig. 6). This idea is consistent with the genetic interaction between ctk1Δ and mutant alleles of the serine 5 kinases, bur1 and kin28 (20, 37).
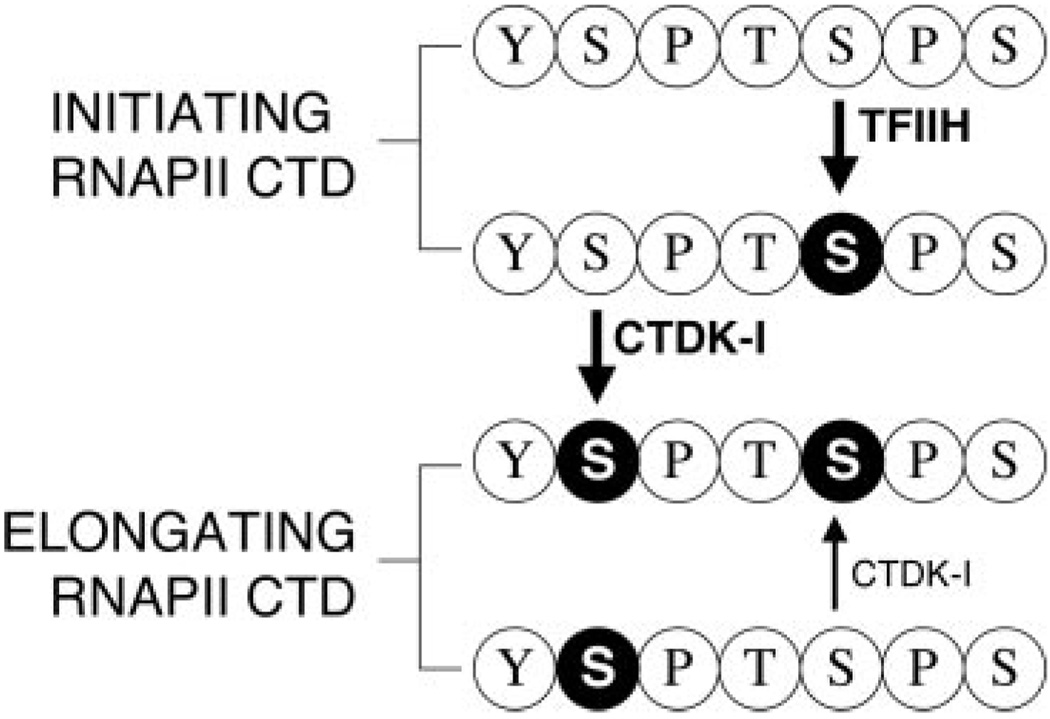
FIG. 6. Model for CTD phosphorylation by CTDK-I during transcription elongation.
After initiation, the TFIIH-associated kinase (Kin28p) phosphorylates the CTD at Ser5. The CTD then becomes a better substrate for phosphorylation by CTDK-I at Ser2. CTDK-I can also efficiently generate doubly phosphorylated CTD from a Ser2-phosphorylated CTD (generated by a CTD phosphatase?) by phosphorylating Ser5. Black circle with white letter, phosphoserine.
We also found that CTDK-I phosphorylates Ser5 of certain CTD substrates (Fig. 4, A and C), an activity previously unreported for CTDK-I. The involvement of CTDK-I in Ser5 phosphorylation is not well characterized, although the genetic interaction between ctk1Δ and an rpb1 construct with alanine substituted for serine at position 5 may imply a physiologically important role for CTDK-I in Ser5 phosphorylation (37). Ctk1p is found associated with genes at promoters where Ser5 phosphorylation is observed, although deletion of CTK1 does not abolish Ser5 phosphorylation at promoters (28). Determining the in vivo relevance of Ser5 phosphorylation by CTDK-I is complicated by the presence of other Ser5 kinases.
Phosphorylation of both serines 2 and 5 by a single CTD kinase is not unprecedented. For example, the Srb8,9,10,11 complex has been shown to phosphorylate CTD substrates at both Ser2 and Ser5 (40). CDK9, a proposed metazoan counterpart to Ctk1p, is the catalytic subunit of P-TEFb that stimulates transcription elongation from the human immunodeficiency virus type 1 promoter by overcoming the effects of negative elongation factors (reviewed in Ref. 30). In the presence of the human immunodeficiency virus Tat protein, P-TEFb can phosphorylate Ser2 and Ser5 of the CTD, but without Tat it only phosphorylates Ser2 (44). The catalytic subunits of CTDK-I and P-TEFb (Ctk1p and CDK9, respectively) share extensive sequence similarity, and both kinases stimulate transcription elongation by acting at a postinitiation step. Like CTDK-I, P-TEFb is also thought to act after CTD phosphorylation by the TFIIH-associated kinase (45). Our new finding that CTDK-I also shares with P-TEFb the ability to phosphorylate CTD repeats on both Ser2 and Ser5 further implicates Ctk1p as the functional yeast homolog of CDK9, although the Bur1p-Bur2p kinase also shares many common characteristics with P-TEFb (20, 21, 46).
Doubly Phosphorylated CTDs in Yeast?
How does the ability of CTDK-I to generate doubly phosphorylated repeats fit with extant information on CTD phosphorylation patterns in vivo? Chromatin-immunoprecipitation (ChIP) experiments in budding yeast (10, 11) showed that mAb H14 epitopes (Ser5- PO4) are prevalent at promoters but decrease as RNAPII moves into coding regions. In contrast, mAb H5 reactivity (epitope was “Ser2-PO4” but see above) increases as the polymerase transcribes into coding regions. These results are frequently interpreted to mean that the CTD repeats of RNAPII at or near the promoter carry Ser5-PO4, whereas most repeats on actively elongating yeast RNAPII carry Ser2-PO4 but not Ser5-PO4. However, there are other findings consistent with the idea that elongating RNAPII can have CTD repeats with both Ser2 and Ser5 phosphorylated. Chromatin immunoprecipitation on the yeast PMA1 gene, for example, detects about 30–40% as much Ser5 phosphorylation in the coding region compared with the promoter (11). It is also likely that detection of Ser5-PO4 by mAb H14 in coding regions is reduced because of epitope masking by phospho-CTD-binding proteins. Additionally, in our laboratory we have identified numerous yeast proteins that bind to the hyperphosphorylated CTD generated by CTDK-I in vitro, and most of these proteins bind preferentially to doubly phosphorylated repeats in BIACORE and in pull-down experiments.2 Also favoring the idea of (Ser2 + Ser5)-phosphorylated CTD repeats are ChIP and polytene chromosome immunofluorescence experiments with the active Drosophila hsp70 gene (12). On this gene (and others), the ratio of CTD-Ser5-PO4 to total RNAPII remains constant as the polymerase traverses the transcription unit, whereas the H5 epitope is present throughout and increases toward the 3′-end of the gene.
Techniques for Studying Phosphorylation Specificity of CTD Kinases
As previously noted, determination of the phosphorylation specificity of CTD kinases has often depended on “phosphorylation-specific” monoclonal antibodies or CTD substrates with altered amino acid sequence. In the case of CTDK-I, the information collected with mutant CTD substrates was complementary to the conclusions from direct sequencing, but it also highlighted some of the shortcomings of relying on mutant substrates to determine a kinase’s phosphorylation site. First, these results draw attention the importance of mutating potential phosphoacceptors to both glutamate and alanine. If we had only used alanine mutants, the usual conclusion would have been that CTDK-I is a Ser5 kinase, because it was able to phosphorylate the S2A mutant but not the S5A mutant. The S5E mutant, however, was a good substrate for CTDK-I, indicating that CTDK-I is not just a Ser5 kinase, and Edman degradation confirmed that CTDK-I phosphorylates Ser2 when Ser5 is already phosphorylated (Fig. 4B). In this case, wild type prephosphorylated substrates were essential to determine that CTDK-I can phosphorylate Ser2 and Ser5, depending on the nature of the CTD substrate. Bienkiewicz et al. (47) have shown another complication with using mutant CTD substrates. Mutation of serine residues to alanine, especially Ser5, has consequences beyond just removal of a phosphoaccepting residue; this mutation causes structural changes in the CTD that could interfere with phosphorylation by a CTD kinase even if its normal phosphorylation site is still present. Overall, we could not have determined the actual specificity properties of CTDK-I by solely relying on mutant substrates and available antibodies for characterizations.
Acknowledgments
We thank Jeff Corden for CTD constructs, Aled Edwards for purified polymerase, Kolla Kristjansdottir for technical advice, and Steve Hanes and the New York State Department of Health peptide synthesis facility for CTD peptides.
Footnotes
This work was supported by National Institutes of Health Grant GM40505 (to A. L. G.).
The abbreviations used are: CTD, C-terminal domain of RNA polymerase II large subunit; RNAPII, RNA polymerase II; GST, glutathione S-transferase; RU, resonance units; mAb, monoclonal antibody; ChIP, chromatin immunoprecipitation; PIC, preinitiation complex; HPLC, high pressure liquid chromatography; P-TEFb, positive transcription elongation factor.
H. Phatnani, J. Jones, and A. L. Greenleaf, manuscript in preparation.
REFERENCES
- 1.Cadena DL, Dahmus ME. J. Biol. Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- 2.Laybourn PJ, Dahmus ME. J. Biol. Chem. 1989;264:6693–6698. [PubMed] [Google Scholar]
- 3.Payne JM, Laybourn PJ, Dahmus ME. J. Biol. Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- 4.Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 5.Kang ME, Dahmus ME. J. Biol. Chem. 1993;268:25033–25040. [PubMed] [Google Scholar]
- 6.O’Brien T, Hardin S, Greenleaf A, Lis JT. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 7.Proudfoot NJ, Furger A, Dye MJ. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 8.West ML, Corden JL. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuryev A, Corden JL. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komarnitsky P, Cho EJ, Buratowski S. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn SH, Kim M, Buratowski S. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 12.Boehm AK, Saunders A, Werner J, Lis JT. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C, Sharp PA. Mol. Cell. Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho EJ, Takagi T, Moore CR, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Mol. Cell. Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder SC, Schwer B, Shuman S, Bentley D. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho CK, Shuman S. Mol. Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 19.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 20.Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Mol. Cell. Biol. 2001;21:4089–4096. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keogh MC, Podolny V, Buratowski S. Mol. Cell. Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irie K, Nomoto S, Miyajima I, Matsumoto K. Cell. 1991;65:785–795. doi: 10.1016/0092-8674(91)90386-d. [DOI] [PubMed] [Google Scholar]
- 23.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Mol. Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 24.Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJ, Young RA. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee JM, Greenleaf AL. Proc. Natl. Acad. Sci. U. S. A. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JM, Greenleaf AL. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 27.Sterner DE, Lee JM, Hardin SE, Greenleaf AL. Mol. Cell. Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JM, Greenleaf AL. J. Biol. Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 30.Price DH. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skaar DA, Greenleaf AL. Mol. Cell. 2002;10:1429–1439. doi: 10.1016/s1097-2765(02)00731-1. [DOI] [PubMed] [Google Scholar]
- 32.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 33.Morris DP, Greenleaf AL. J. Biol. Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 34.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostapenko D, Solomon MJ. Eukaryot. Cell. 2003;2:274–283. doi: 10.1128/EC.2.2.274-283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patturajan M, Conrad NK, Bregman DB, Corden JL. J. Biol. Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- 37.Lindstrom DL, Hartzog GA. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris DP, Lee JM, Sterner DE, Brickey WJ, Greenleaf AL. Methods. 1997;12:264–275. doi: 10.1006/meth.1997.0478. [DOI] [PubMed] [Google Scholar]
- 39.Edwards AM, Darst SA, Feaver WJ, Thompson NE, Burgess RR, Kornberg RD. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2122–2126. doi: 10.1073/pnas.87.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 41.Jelinek T, Weber MJ. BioTechniques. 1993;15:628–630. [PubMed] [Google Scholar]
- 42.MacDonald JA, Mackey AJ, Pearson WR, Haystead TA. Mol. Cell Proteomics. 2002;1:314–322. doi: 10.1074/mcp.m200002-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Thompson NE, Aronson DB, Burgess RR. J. Biol. Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- 44.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Mol. Cell. Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Kipreos ET. Mol. Biol. Evol. 2000;17:1061–1074. doi: 10.1093/oxfordjournals.molbev.a026387. [DOI] [PubMed] [Google Scholar]
- 47.Bienkiewicz EA, Moon Woody A, Woody RW. J. Mol. Biol. 2000;297:119–133. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]