Abstract
The zinc finger protein growth factor independent-1 (Gfi1) is a transcriptional repressor that is critically required for normal granulocytic differentiation. GFI1 loss-of-function mutations are found in some patients with severe congenital neutropenia (SCN). The SCN-associated GFI1-mutant proteins act as dominant negatives to block granulopoiesis through selective deregulation of a subset of GFI1 target genes. Here we show that Gfi1 is a master regulator of microRNAs, and that deregulated expression of these microRNAs recapitulates a Gfi1 loss-of-function block to granulocyte colony-stimulating factor (G-CSF)–stimulated granulopoiesis. Specifically, bone marrow cells from a GFI1-mutant SCN patient and Gfi1−/− mice display deregulated expression of miR-21 and miR-196B expression. Flow cytometric analysis and colony assays reveal that the overexpression or depletion of either miR induces changes in myeloid development. However, coexpression of miR-21 and miR-196b (as seen in Gfi1−/− mice and a GFI1N382S SCN patient) completely blocks G-CSF–induced granulopoiesis. Thus, our results not only identify microRNAs whose regulation is required during myelopoiesis, but also provide an example of synergy in microRNA biologic activity and illustrate potential mechanisms underlying SCN disease pathogenesis.
Introduction
MicroRNAs (miRs) are small, noncoding RNAs (∼ 22 nucleotides in length) that are generated from endogenous hairpin-shaped transcripts encoded in the genomes of humans, animals, plants, and viruses.1 miRs act as negative regulators of gene expression by targeting mRNA through translational repression or mRNA cleavage of more than 30% of protein coding genes.2 Pluripotent hematopoietic cells express a variety of lineage-specific miRs that may enforce lineage commitment by blocking the expression of transcripts from alternative lineages.3 Based on their function, various studies have suggested a role for microRNAs in normal growth, development, and differentiation.4–7 Recent investigations have found that many microRNAs are deregulated in primary human tumors,8–12 they are located at genomic regions linked to various cancers,13–15 and they are also up-regulated in response to cellular stress.16 Overall, these findings strongly implicate the potential link between microRNAs and pathogenesis of human disease.17
Severe congenital neutropenia (SCN) is a rare hematologic disease characterized by maturation arrest of granulopoiesis at the promyelocyte stage resulting in low levels of mature neutrophils (< 0.5 × 109/L).18 Recently, the molecular basis underlying SCN has been linked to genes coding for the ELA2, GFI1, WAS, and HAX1 proteins.19–22 The most common cause of SCN is an autosomal dominant mutation of ELA223; however, autosomal dominant mutations in WAS19 and in GFI122 as well as recessive mutations in HAX121 have also been described.
Gfi1 is a transcriptional repressor protein that is critically required for normal myelopoiesis.24,25 Gfi1 loss-of-function mutations in mice and humans arrest myeloid differentiation as well as block the formation of terminally differentiated neutrophils.22,24–26 Gfi1−/− mice dramatically accumulate granulocyte-monocyte progenitors (GMPs) and abnormal promyelocytes with monocytic characteristics.24,26 Gfi1 expression is normally induced during differentiation from common myeloid progenitors (CMPs) to GMPs.25,26 Subsequently, Gfi1 functions as a rate-limiting granulopoietic molecular switch.26 Mutations in GFI1 found in SCN patients encode proteins that function as dominant negatives to deregulate a subset of GFI1 target genes. For example, Gfi1−/− mice and humans with mutant GFI1 show abnormal levels of the monopoietic cytokine CSF1 and its receptor.26 However, given the severity of the phenotypes engendered by Gfi1 loss of function, it is anticipated that Gfi1 integrates a transcriptional network of target genes.27,28
Here we show that myelopoiesis is controlled by a novel transcriptional program in which the transcriptional repressor Gfi1 regulates the expression of miR-21 and miR-196b. Moreover, whereas forced expression or down-regulation of either miR alone alters myeloid colony formation, simultaneous deregulation of miR-21 and miR-196b synergizes to potently block granulocyte colony-stimulating factor (G-CSF)–stimulated granulopoiesis, similar to the neutropenic phenotypes observed with Gfi1 loss-of-function.
Methods
MicroRNA microarray and quantitative PCR (TaqMan)
Gfi1−/−24 mice were backcrossed onto C57Bl/6 or Balb/c backgrounds for 8 generations. Total RNA was isolated using Trizol (Sigma-Aldrich, St Louis, MO) from 3 C57Bl/6 Gfi1+/+ and 3 C57Bl/6 Gfi1−/− mice as well as 2 Balb/c Gfi1+/+ and 2 Balb/c Gfi1−/− mice. Labeling RNA and hybridization on microRNA microarray chip was done as previously described29 and performed by The Ohio State University Comprehensive Cancer Center (OSUCCC) Microarray Shared Resource, and the data were deposited in the National Center for Biotechnology Information (CBI) Gene Expression Omnibus (accession GSE15077).30 The steady-state level of mature microRNAs was determined using miR-specific TaqMan MicroRNA assay kits (Applied Biosystems, Foster City, CA). Relative expression was calculated using the comparative 2ΔΔCt method. RNU6B was used as an internal control for normalization. The TaqMan results are represented as means plus or minus SD from 5 independent experiments. The Cincinnati Children's Hospital Medical Center Institutional Animal Care and Use Committee reviewed and approved the use and manipulation of mice in these studies.
Plasmids
Genomic fragments containing the miR-21 and miR-196B precursors were amplified from pCMV-miR-21 plasmid and U937 cell RNA, respectively. The XhoI-XhoI and XhoI-EcoRI polymerase chain reaction (PCR) fragment was cloned into MSCV-Puro31 retroviral construct and the miRNA expression was confirmed by TaqMan analysis. Mouse Gfi1 was also cloned into MSCV-Puro vector and the viral particles were generated as described previously.26
Cell culture and bone marrow lineage depletion
HL-60 cells were grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal calf serum (FCS), 50 U/mL penicillin, and 50 μg/mL streptomycin. Cells were maintained at 37°C in a 5% CO2/95% air atmosphere and were used for experiments during the exponential phase of growth. Cells were induced to differentiate along the granulocytic or monocytic pathway by culturing in the presence of 5 μM all-trans retinoic acid (ATRA) for up to 5 days or 10 ng/mL phorbol myristate acetate (PMA), respectively.
Bone marrow cells were isolated from both 6- to 8-week-old C57Bl/6 WT mice and Gfi1 mutant littermates as described previously,26 and lineage-negative cells were isolated using the mouse Lineage Cell Depletion kit (Miltenyi Biotec, Auburn, CA) followed by separation on an AutoMacs magnetic sorter (Miltenyi Biotec). Cells were maintained in serum-free StemSpan medium (StemCell Technologies, Vancouver, BC) supplemented with IL-3 (10 ng/mL), IL-6 (20 ng/mL), SCF (25 ng/mL), and TPO (25 ng/mL; PeproTech, Rocky Hill, NJ). After 48 hours of cytokine expansion, the cells were subjected to retroviral transduction using a spinfection protocol.26 GMP populations from genotype ROSA-CreERT2 Gfi1fex4-5/fex4-5 (Hameyer et al32) were sorted from Lin− cells26 and incubated overnight with or without 1 μM tamoxifen (Sigma-Aldrich).
CD34+ bone marrow cells from a GFI1N382S patient and 3 healthy donors were isolated using CD34 progenitor cell isolation kit (Miltenyi Biotec) followed by separation on an AutoMacs magnetic sorter.
Viral transduction
The Lin− cells were subjected to retroviral transduction after 48 hours of expansion with cytokine as outlined.26 For the Gfi1 knockdown experiments, HL60 cells were transduced with either a nontargeting shRNA that has been bioinformatically qualified by the vendor (Open Biosystems, Huntsville, AL) not to target any open reading frame in the human or mouse genomes, or Gfi1 targeting shRNA vectors (Sigma-Aldrich). The transduced cells were cultured in RPMI media with 10% FBS and puromycin (5 μg/mL).
Immunoblot
Protein extracts were obtained from cell lines or total bone marrow by lysing cells directly in the Complete-M lysis buffer (Roche, Indianapolis, IN) with protease inhibitor. Samples were resolved on 10% SDS–polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to PVDF membranes (Immobilon-P; Millipore, Billerica, MA). Immunoblot analysis was performed using antibodies against GFI1 (2.5D.17) and β-ACTIN (Sigma-Aldrich) with HRP-conjugated goat anti–mouse or anti–mouse secondary antibody (Amersham Biosciences, Piscataway, NJ) with a ECL-PLUS detection kit (Pierce, Rockford, IL).
Chromatin immunoprecipitation
Human HL60 cells (108 cells) were cross-linked with 1% formaldehyde for 10 minutes on ice and terminated with 0.125 M glycine as described previously.26 Soluble chromatin was prepared by resuspending in cell lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA [pH 8.0], 10% glycerol, 1% SDS, 1× Complete protease inhibitor) and sonicated to generate 200- to 800-bp DNA fragments using a Sonicator 3000 cup horn (Misonix, Farmingdale, NY). Chromatin-protein complexes were immunoprecipitated using antibodies against Gfi1 (2.5D1.7) and control normal mouse IgG (NA931V; GE Healthcare, Little Chalfont, United Kingdom). Recovered chromatin was PCR-amplified with the following oligos to the human loci: miR-21 site “A” (5′-GCCTTGCCTAATCCACCTAC-3′ and 5′-AACATTAACACAGATACGACAGAG-3′), site “B” 21 (5′-TGCAAGTGGATGGTTTGGTA-3′ and 5′-ATTCCTCAGCTCTTCGGTGA-3′), as well as miR-196B site “A” (5′-GAATTGCCAATCTTGTTTTAAGC-3′ and 5′-ACGCACAGCAGCAATACAAT-3′), site “B” (5′-ACCAGAACTGGTCGGTGATT-3′ and 5′-GCAGAGGTACCTGGAGACGA-3′), and site “C” (5′-TCTTCCGTCTCTGCCAGATT-3′ and 5′-ACCTCTACTTGAGCCGCAGA-3′). β-ACTIN was used as an internal control for nonspecific enrichment (5′-AGCGCGGCTACAGCTTCA-3′ and 5′-CGTAGCACAGCTTCTCCTTAATGTC-3′). The representative DNA gels are shown from at least 3 independent experiments with similar results.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed by incubating in vitro transcribed and translated Gfi1 protein with radiolabeled oligonucleotides and then subjecting them to electrophoresis as described previously.26 For supershift assay, 0.5 μg polyclonal Gfi1 antibody (neutralizing antibody) or control IgG was added to the reaction before oligonucleotide addition. Oligonucleotides containing the consensus binding site for Gfi1 and mutated Gfi1 binding site used for competition were as follows: miR-21 5′-TTTTCTTGAGCGTTTTGATTTTTTACTTT TA-3′ (site 1), 5′-TTTTATTCTTAGTGTGATTTTTTTCCATT-3′ (site 2), 5′-TTTTCTTGAGCGTTTTTAGTTTTTACTTTTA-3′ (mutant), and miR-196B 5′-GGCTTCTAATCTTAAATCAGAATAAATTAATA-3′ (site 1), 5′-GGCTTCTACTATTAACTGAGAATAAATTAATA-3′ (site 1 mutant), 5′-GGCAGAACTGGTCGGTGATTTAGGTAGTTT-3′ (site 2), 5′-GGCAGAACTGGTCGGTTAGTTAGGTAGTTT-3′ (site 2 mutant). Letters in italics represent the core Gfi1 binding site, or highlight mutations to the core sequence.
AntagomiRs and hematopoietic progenitor assays
The sequence complementary to miR-21, 5′-GUCAACAUCAGUCUGAUAAGCUA-3′, control miR-21 5′-GUCAACUUCAGUCAGAAAAGGUA-3′, miR-196B 5′-CCCAACAACAGGAAACUACCUA-3′, and control miR-196b 5′-CCCAAGAACAGGUA AGUACGUA-3′ was synthesized with 2′-OMe modified bases, phosphothioate on the first 2 and last 4 bases, and a 3′ cholesterol modification through a hydroxyprolinol linkage. AntagomiR oligonucleotides were deprotected, desalted, and purified by high-performance liquid chromatography (Dharmacon, Lafayette, CO).
For methylcellulose assays, 5000 cytokine-expanded and retrovirally transduced cells were plated on MethoCult GF M3534 (StemCell Technologies). For antagomiR experiments, cells were incubated for 30 minutes with 100 nM antagomiR in 100 μL media and then plated in MethoCult GF M3534. Hematopoietic colonies containing more then 50 cells were scored at day 7 or 8 and differentiated based on their morphology. Results are displayed as the mean percentage of colony-forming unit granulocytes (CFU-Gs), CFU-macrophages (CFU-Ms), and CFU–granulocytes-macrophages (CFU-GMs) from 3 independent experiments plus or minus SD with total number of colonies plus or minus SD.
G-CSF liquid culture and flow cytometry
For liquid cultures, wild-type Lin− bone marrow cells were retrovirally transduced and the following day the cells were plated in Iscove DMEM with 10% FBS, 2.5 μg/mL puromycin and stimulated for 4 days with 5 ng/mL murine G-CSF (PeproTech). Then the cells were either cytospun, followed by Giemsa staining, or analyzed by flow cytometry. Cytospins were photomicrographed at 32× magnification using the Axiovert 200M microscope (Carl Zeiss, Rochester, NY) with an Axiovision (version 3.1.1.1) acquisition software. For flow cytometry, cells were stained with antibodies to 7/4 (clone 7/4; Serotec, Raleigh, NC) and F4/80 (clone CI:A3-1; Serotec), then analyzed on an LSRII flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo software (TreeStar, Eugene, OR). Statistical analyses of flow plots from at least 3 independent experiments are depicted with averages plus or minus SD.
Statistics
To determine significance between groups, comparisons were made using the Student t test. For all statistical tests, the .05 level of confidence was accepted for statistical significance.
Results
Gfi1 is a master regulator of microRNA expression in hematopoietic stem and progenitor cells
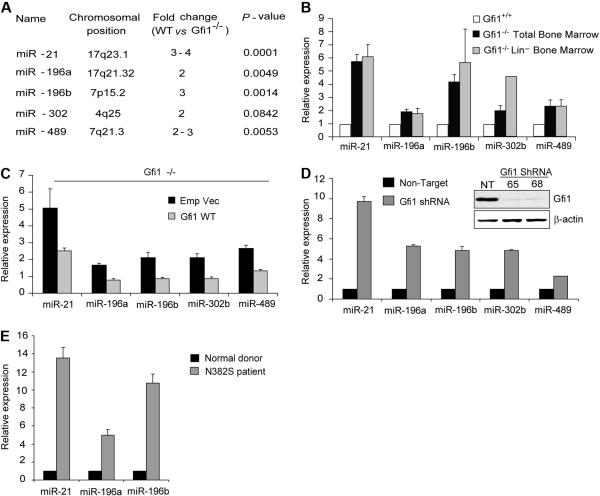
To determine whether Gfi1 regulates microRNA genes, we performed genome-wide oligonucleotide hybridization–based array. First, we extracted RNA from low-density bone marrow cells from Gfi1+/+ and Gfi1−/− littermates.24 To minimize strain-specific variables (independent of Gfi1 deletion), the array was performed on both C57/Bl6 (n = 6) and Balb/c (n = 4) background Gfi1−/− mice and their Gfi1+/+ littermates. Statistical analyses showed a significant deregulation of miR-21 (3- to 4-fold), miR-196a (2-fold), miR-196b (3-fold), and miR-489 (2- to 3-fold) associated with loss of Gfi1 (Figure 1A). The expression of miR-302b also showed a trend toward deregulation (2-fold; Figure 1A) but did not achieve significance. To validate the array data, we monitored the steady-state accumulation of the mature and functional miR through a fluorescent probe–coupled polymerase chain reaction assay (PCR; TaqMan). Similar to the array data, these analyses of unfractionated low-density bone marrow RNA showed a dramatic deregulation of miR-21 (5- to 6-fold), miR-196a (2-fold), miR-196b (4-fold), miR-302b (2-fold), and miR-489 (2-fold; Figure 1B black bars). As a control, we also examined miR-224, which was not significantly deregulated in either hybridization or PCR-based assays (data not shown).
Figure 1.
Gfi1 is a master regulator of microRNA expression. (A) MicroRNA microarray profile of total BM cells from both C57Bl/6 and Balb/c Gfi1+/+ and Gfi1−/− littermates. (B) Quantitative real-time (TaqMan) miR analysis of RNA from panel A and Lin− BM cells. (C) TaqMan analyses of steady-state miR expression in Gfi1−/− Lin− BM cells transduced with MSCV empty vector or MSCV encoding Gfi1. (D) TaqMan analyses of miR expression in HL60 cells transduced with Gfi1-targeting lentiviral shRNA (65 and 68) and nontargeting control (NT). Immunoblot showing Gfi1 knockdown (inset). (E) TaqMan analyses of microRNA in human CD34+ bone marrow cells from healthy donors (n = 3) versus GFI1N382S mutant patient. TaqMan results are represented as means ± SD from at least 3 independent experiments.
Hematopoiesis in Gfi1−/− mice is abnormal and leads to the accumulation of an aberrant promyelocytic cell.24,25 To eliminate the potential contribution of Gfi1−/− promyelocytes to the analyses, we performed an automated depletion of cells expressing lineage markers (Lin−) and repeated the latter analyses on these Lin− bone marrow cells with essentially the same result (Figure 1B gray bars). Interestingly, the expression of miR-302b was significantly more deregulated in Gfi1−/− Lin− cells (Figure 1B). Gfi1−/− deregulation is specific to the miR genes, as the expression of genes nearby to the miR-21 locus (Tmem49) or miR-196b locus (HoxA10) is not significantly different in Gfi1+/+ versus Gfi1−/− Lin− bone marrow cells (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Thus, Gfi1 regulates the expression of multiple microRNAs.
To assess the ability of Gfi1 to control the expression of these miRs, we rescued the expression of Gfi1 in Gfi1−/− Lin− bone marrow cells by transduction of these cells with Gfi1-expressing MSCV-puro retroviral vectors or an empty vector control. Puro selection was performed after 48 hours of retroviral transduction and then miR expression was assayed. Compared with empty vector controls, expression of Gfi1 in Gfi1−/− Lin− bone marrow cells dramatically decreased the expression of miR-21, miR-196a, miR-196b, miR-302b, and miR489 (Figure 1C), but not miR-224 (data not shown). We next used 2 different GFI1-targeting shRNA-expressing lentiviral vectors to knock down expression of human GFI1 in the HL60 cell line (Figure 1D inset). We found that the steady-state levels of the mature miR-21, miR-196a, miR-196b, miR-302b, and miR-489 were significantly higher when GFI1 protein expression was reduced (Figure 1D). Therefore it is possible that miR-21, miR-196a, miR-196b, miR-302b, and miR489 are directly regulated by Gfi1.
Finally, we analyzed miR expression in CD34+ human bone marrow cells from healthy donors compared with CD34+ cells from an SCN patient with the GFI1N382S mutation. Steady-state levels of the mature miR-21, miR-196a, and miR-196b were higher in the patient than in donor with wild-type GFI1 (Figure 1E). Given these data, we focused further analyses upon miR-21 and miR-196B as they may directly correlate to human disease phenotypes.
Gfi1 physically binds to the miR-21 and miR-196B loci
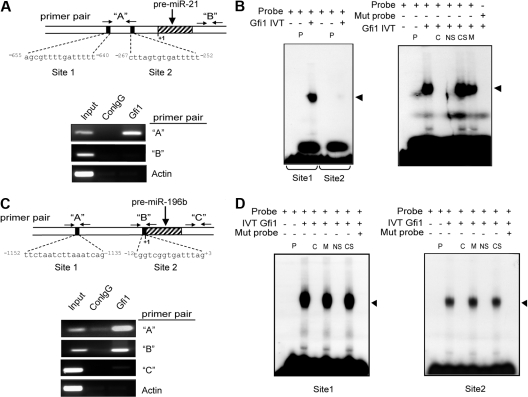
To determine whether GFI1 directly regulates miR-21 and miR-196B, we performed chromatin immunoprecipitation (ChIP) in a human promyelocytic cell line (HL60) using a monoclonal antibody specific for GFI1.33 Bioinformatic profiling was performed using a web-based tool34 to identify potential GFI1 binding sites that are conserved from mouse to human. Immunoprecipitated chromatin fragments were amplified using primers specific to these potential GFI1 binding sites or to negative control sites either within the miR loci that lack GFI1 binding sites or to β-ACTIN gene sequences. The results indicate that GFI1 directly and specifically binds to DNA sequences surrounding miR-21 (Figure 2A) and miR-196B (Figure 2C) in living cells.
Figure 2.
Gfi1 physically binds to miR-21 and miR-196B loci. (A) Diagrammatic representation of putative Gfi1 binding sites in the human miR-21 locus relative to the start of the pre-miR transcript on human chromosome 17, along with primer pairs (“A” and “B”; arrows) used for ChIP analyses (top). ChIP analyses using a Gfi1-specific monoclonal antibody (Gfi1) and isotype control mouse IgG (ConIgG; bottom) in human HL60 cells. Primers amplifying a region of the Beta-actin gene (Actin) and primer pair “B” to the miR-21 locus serve as negative controls. (B) EMSA analyses with in vitro transcribed and translated Gfi1 and oligonucleotides encoding wild-type (Probe) or Gfi1-site mutant (Mut probe) miR-21 locus sequences depicted in panel A. (C) Diagrammatic representation of putative Gfi1 binding sites in the human miR-196B locus relative to the start of the pre-miR transcript on chromosome 6, along with primer pairs (“A,” “B,” and “C”) used for ChIP analyses (top). ChIP analyses using a Gfi1-specific monoclonal antibody (Gfi1), isotype control mouse IgG (ConIgG; bottom). Primers amplifying a region of the Beta-actin gene (Actin) and primer pair “C” to the miR-196b locus serve as negative controls. (D) EMSA analyses with in vitro transcribed and translated Gfi1 and oligonucleotides encoding wild-type (probe) or Gfi1-site mutant (Mut probe) miR-196B locus sequences depicted in panel C. P indicates control IVT protein (luciferase); C, cold competition with wild-type miR-21 or miR-196B oligo; NS, neutralizing Gfi1-specific antibody; CS, nonspecific control antibody; and M, cold competition with Gfi1-site mutant oligo. Results shown are representative of at least 3 independent experiments.
To further dissect the direct Gfi1 binding, we used electrophoretic mobility shift assays (EMSAs) to examine 2 putative Gfi1 binding sites within the amplified ChIP target for miR-21 (Figure 2A) and miR-196B (Figure 2C) loci. First, oligonucleotides encoding either miR-21 site 1 or site 2 (Figure 2A) were labeled with 32P and incubated with in vitro transcribed and translated Gfi1. Although Gfi1 retards the mobility of both oligonucleotides, the affinity of Gfi1 for site 1 is dramatically higher (Figures 2B, S2). Next, we dissected binding specificity by performing competition with a nonradiolabeled oligonucleotide. Binding of Gfi1 to the radiolabeled oligonucleotide is disrupted by an excess of an identical but unlabeled oligonucleotide, but not by an oligonucleotide encoding a mutation in the core Gfi1 binding sequence (Figure 2B right panel, “C” vs “M”). Moreover, the complex was disrupted by preincubation with a Gfi1-specific antibody (neutralizing antibody), but not an isotype-matched antibody (Figure 2B right panel, “NS” vs “CS”). In contrast, similar EMSA analyses on miR-196b site 1 and site 2 indicated that Gfi1 specifically bound to both potential Gfi1 binding sites (Figure 2D). Overall, the results from both ChIP and EMSA experiments indicate that Gfi1 physically binds in vitro and in vivo to the miR-21 and miR-196B loci.
miR-21 and miR-196b are regulated by Gfi1 during myelopoiesis
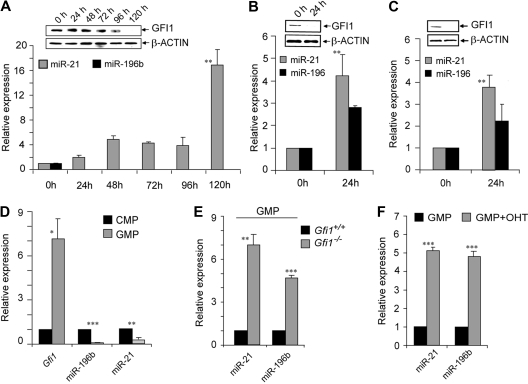
To delineate the regulation of miR-21 and miR-196B during human myeloid differentiation, the expression profile of miR-21, miR-196B, and GFI1 was analyzed during chemically induced differentiation of the HL60 promyelocytic cell line. HL60 cells were treated with 5 μM all-trans retinoic acid (ATRA) to induce granulocytic differentiation. We found that GFI1 protein decreases upon terminal differentiation (Figure 3A immunoblot inset). As GFI1 protein levels diminished, expression of miR-21 increased (Figure 3A). Expression of miR-196B was lost within 24 hours of inducing differentiation (Figure 3A), and so must be dominantly controlled by other mechanisms in this model. To examine monopoiesis, we treated both HL60 and U937 leukemia cell lines with 10 ng/mL phorbol myristate acetate (PMA). Again, GFI1 protein decreased (Figure 3B,C immunoblot insets), whereas miR-21 and miR-196B expression increased in both cell lines (Figure 3B,C). Thus, in models of granulocytic or monocytic differentiation, expression of miR-21 and miR-196B is consistently reciprocal with the expression of GFI1.
Figure 3.
Gfi1 regulates miR-21 and miR-196B during myelopoiesis. TaqMan analysis of miR-21 and miR-196B in the human HL60 cell line treated with (A) ATRA or (B) PMA, or (C) the human U937 cell line treated with PMA. Immunoblot of Gfi1 or Beta-actin (inset). (D) TaqMan analyses of Gfi1, miR-21, and miR-196b expression in sorted wild-type CMPs and GMPs. (E) TaqMan analyses of miR-21 and miR-196b expression in sorted Gfi1+/+ and Gfi1−/− GMPs. (F) TaqMan analysis of miR-21 and miR-196b expression in phenotypically wild-type sorted GMPs, 36 hours after in vitro OHT-induced Cre-ERt2 activation to mediate deletion of floxed Gfi1 alleles. *P < .05; **P < .01; ***P < .001.
Gfi1 expression is induced in the transition between CMPs and GMPs.26,34 To determine a normal physiologic context for regulation of miR-21 and miR-196b by Gfi1, we sorted CMPs and GMPs from wild-type mice and determined the expression of Gfi1, miR-21, and miR-196b. The approximately 7-fold induction of Gfi1 from CMPs to GMPs was accompanied by a 6-fold reduction in miR-21 and 8- to 9-fold reduction in miR-196b steady-state levels (Figure 3D). Thus, it is possible that Gfi1 down-regulates miR-21 and miR-196b expression during the transition from CMPs to GMPs. In agreement with this hypothesis, sorted GMPs from the bone marrow of Gfi1+/+ and Gfi1−/− littermates showed deregulation of miR-21 and miR-196b in Gfi1−/− GMPs (Figure 3E). To delineate a causal relationship between Gfi1 induction and miR-21 and miR-196b suppression during myelopoiesis, we sorted GMPs from a mouse model in which a ubiquitously expressed but tamoxifen (OHT)–induced Cre (ROSA-Cre-ERT2)32 mediates deletion of floxed Gfi1 alleles (Gfi1fex4-5).26 Sorted ROSA-Cre-ERT2 Gfi1fex4-5/fex4-5 GMPs were cultured in vitro with cytokines with or without 1 μM OHT. RNA was extracted after 36 hours and used to analyze miR-21 and miR-196b expression. Conditional deletion of Gfi1 in GMPs resulted in an approximately 5-fold increase in miR-21 and miR-196b (Figure 3F). Notably, treatment of wild-type sorted GMPs with OHT had no effect upon miR-21 or miR-196b levels (Figure S3). Thus, Gfi1 is critically required to control miR-21 and miR-196b expression during normal myeloid differentiation.
miR-21 and miR-196b differentially control myelopoiesis
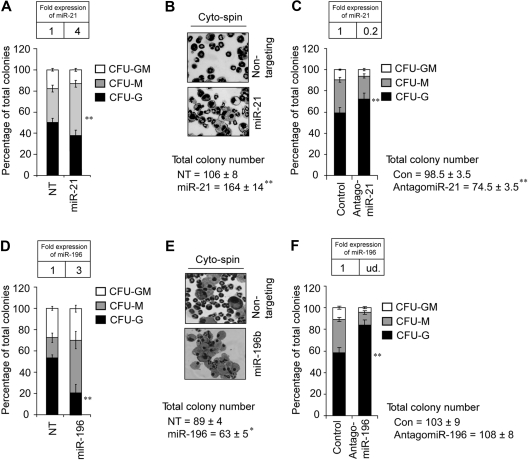
Next, we wanted to determine the downstream sequelae of miR-21 or miR-196b deregulation in wild-type bone marrow progenitors. To dissect the impact of miR-21 and miR-196b on myelopoiesis, we used MSCV vectors that express puromycin resistance and miR-21, miR-196b, or a nontargeting shRNA control. Transduced Lin− bone marrow cells were selected in puromycin overnight, counted, and assayed for myeloid colony-forming units (CFUs) in methylcellulose with SCF, IL3, IL6, and puromycin. Four-fold overexpression of miR-21 (Figure 4A inset) resulted in approximately 50% increased total CFUs (Figure 4B). The additional colonies constituted a significant increase in monocytic colonies (Figure 4A), as confirmed by cytospin (Figure 4B). Next, we used chemically engineered “antagomirs,” which efficiently and specifically silence endogenous miRs.36 Wild-type Lin− bone marrow cells were treated with 100 nmol antagomiR-21 or a control version with 4–base pair substitutions. The treated cells were then assayed for CFUs as described in Figure 4A. AntagomiR-21 reduced miR-21 expression approximately 80% (Figure 4C inset). Addition of the control antagomiR had no effect on CFU numbers or differentiation (data not shown). In contrast to the increase in total CFUs induced by miR-21 overexpression, antagomiR-21 treatment decreased total CFUs by approximately 25% (Figure 4C). Interestingly, reciprocal to miR-21 overexpression, the antagomiR-21–mediated decrease is explained by a significant loss of monocytic colonies. Thus, miR-21 appears to be a promonopoietic effector. Next, 3-fold overexpression of miR-196b (Figure 4D inset) reduced the total number of CFUs, with a significant loss of CFU-Gs (Figure 4D cytospin 4E). Conversely, antagomiR treatment ablated miR-196b (undetected compared with the expression in WT BM cells; Figure 4F inset) and significantly increased CFU-Gs (Figure 4F). Notably, antagomiR-196b exhibited specificity because levels of miR-196a were only mildly affected in treated cells (data not shown). Thus, miR-196b antagonizes granulopoiesis. Collectively, these data suggest that miR-21 and miR-196b control the number of CFU-Ms and CFU-Gs, respectively.
Figure 4.
miR-21 or miR-196b is a partial phenocopy of Gfi1−/−. (A) Methylcellulose colony assay from Lin− bone marrow cells transduced with MSCV encoding miR-21 or a nontargeting shRNA control (NT) reveals significantly increased CFU-Ms. (B) Cytospins from the colony assay in panel A. (C) Colony assay from Lin− bone marrow cells treated with control antagomiR or antagomiR-21 reveals significantly decreased CFU-Ms. (D) Methylcellulose colony assay from Lin− bone marrow cells transduced with MSCV encoding miR-196b or a nontargeting control (NT) reveals significantly decreased CFU-Gs. (E) Cytospins from the colony assay in panel D. (F) Colony assay from Lin− bone marrow cells treated with control antagomiR or antagomiR-196b reveals significantly increased CFU-Gs. Results are displayed as percentage of CFU-Gs, CFU-Ms, and CFU-GMs (n = 3, mean ± SD). TaqMan results are represented as means ± SD from at least 3 independent experiments. *P < .05; **P < .01.
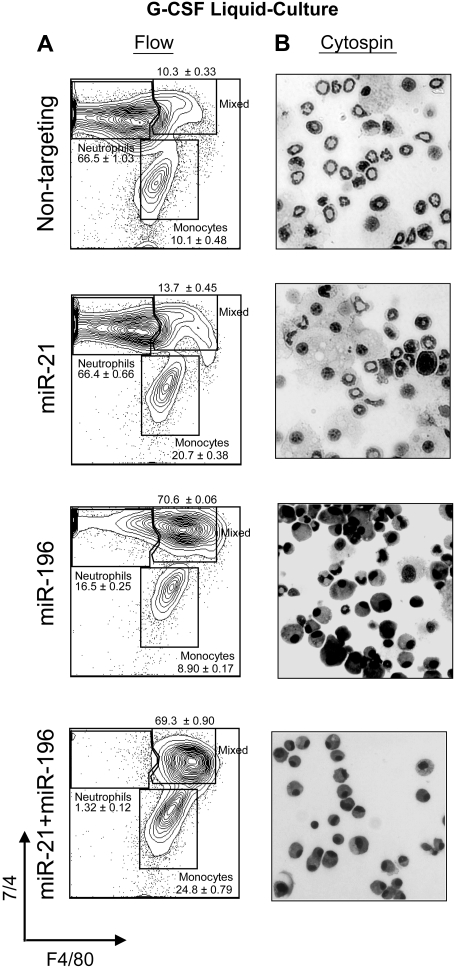
miR-21 and miR-196b block G-CSF–induced granulopoiesis
The latter experiments suggest that miR-21 is promonopoietic and that miR-196b is antigranulopoietic. However, it is possible that IL3, IL6, and SCF are insufficient to facilitate normal myeloid differentiation from these cells. Thus, these cells may simply require instructive cytokine signaling. On the other hand, the forced expression of either miR recapitulated different aspects of Gfi1−/−- and GFI1N382S-blocked myelopoiesis, which cannot be ameliorated by G-CSF signaling.22,26 Therefore, we assembled G-CSF–stimulated liquid cultures and analyzed their morphology and cell surface markers. WT Lin− bone marrow cells were transduced with retrovirus encoding a nontargeting shRNA (control), miR-21, miR-196b, or miR-21 and miR-196b. Liquid cultures were performed with G-CSF and their morphology and cell surface markers were analyzed. Cells transduced with nontargeting vector differentiate mainly into 7/4+ F4/80− granulocytes (Figure 5A), as evidenced by cytospin (Figure 5B). Similarly, cells overexpressing miR-21 responded to G-CSF–instructed granulopoiesis, but also had twice as many 7/4− F4/80+ monocytes as the control (Figure 5A, compare 10.1% with 20.7%). Thus, although miR-21 does not appear to block G-CSF signaling, miR-21 increases monocytic cell numbers even in the context of G-CSF signaling. In contrast, miR-196b overexpression blocked granulopoiesis, even in the context of G-CSF instructive signaling. Notably, there was a corresponding increase in the number of mixed lineage (7/4+ F4/80+) cells (Figure 5A), indicating that G-CSF signaling was not completely blunted (because the cells did not simply become monocytic as in the CFU assays). However, the overexpression of both miR-21 and miR-196b dramatically blunts G-CSF instructive signaling. Cultures expressing both miRs consisted mainly of small cells of mixed lineage. We conclude that deregulated expression of 2 Gfi1 target genes (miR-21 and miR-196b) recapitulates the Gfi1−/− and GFI1N382S-mediated block to myelopoiesis, in that the block is severe and not overcome by G-CSF signaling.
Figure 5.
Overexpression of both miR-21 and miR-196b blocks granulopoiesis in vitro. (A) Flow cytometric analysis with antibodies to 7/4 and F4/80 denoting neutrophil, mixed, and monocytic phenotypes of liquid cultures from Lin− bone marrow cells transduced with MSCV vector encoding miR-21, miR-196b, miR-21, and miR-196b or a nontargeting shRNA control. (B) Cytospin from 4-day G-CSF–stimulated liquid cultures of panel A. Results are displayed as percentage of neutrophils, monocytes, and mixed (n = 3, mean ± SD).
Discussion
Gfi1 is a master regulator of microRNA (miR) expression in hematopoietic cells. Our data reveal Gfi1 transcriptional control of miR-21, miR-196a, miR-196b, miR-302b, and miR-489 in both primary murine cells and a human promyelocytic cell line. MicroRNAs are expressed and play a crucial role in the establishment, maintenance, and function of hematopoietic lineages.3,37 For example, miR-17-5p-20a-106a controls monopoiesis,38 miR-181 directs lymphoid progenitors toward B-lymphoid development, whereas miR-146 and miR-223 appear to favor T lymphopoiesis.3 Recent studies have shown that hematopoietic transcription factors interact with the promoters of miRs and thereby regulate their expression.39–41 For instance, c-MYC activates miR-17-92 clusters,40 CEBPA activates miR-223,39 PU.1 activates miR-424,42 Stat3 activates miR-21,43 and NFI-A represses miR-223 during granulopoiesis.39 We illustrate a novel transcriptional-epigenetic network underlying granulopoiesis. Our work links Gfi1 repression of multiple miRs to the synergistic biologic activity of miR-21 and miR-196b in myelopoiesis downstream of Gfi1.
Gfi1 directly regulates miR-21 and miR196b during myelopoiesis. In this study, we identified the normal down-regulation of miR-21 and miR-196b in the transition between CMPs and GMPs. Gfi1 expression is reciprocally induced in GMPs.35 In Gfi1−/− mice, the expression of miR-21 and miR-196b is elevated in Lin− bone marrow and GMPs. Moreover, conditional deletion of Gfi1 in isolated GMPs increases miR-21 and miR196b expression. miR-21 expression is reciprocal to Gfi1 protein levels during ATRA-stimulated granulocytic differentiation of a human promyelocytic cell line. Both miR-21 and miR-196b expression is reciprocal to GFI1 expression in PMA-induced monocytic differentiation of 2 different human myeloid cell lines. Finally, molecular analyses (ChIP and EMSA) confirm direct interaction between Gfi1 and miR-encoding loci both in vitro and in vivo. Thus, Gfi1 transcriptional programming in myelopoiesis includes control of miR-21 and miR-196b.
Deregulated expression of miR-21 and miR-196b distorts myelopoiesis. Neutrophil development proceeds in a sequential fashion with defined morphology and gene expression to terminal differentiated mature, functional neutrophils.44 Gfi1−/− mice lack terminally differentiated neutrophils and show an accumulation of abnormal myeloid cells with the mixed characteristics of immature granulocyte and macrophage precursors.24,25 Similarly, the combined expression of miR-21 and miR-196b induced the dramatic accumulation of cells with a mixed lineage phenotype. Moreover, similar to Gfi1−/−- and GFI1N382S-blocked granulopoiesis, these changes were not overcome by G-CSF instructive signaling. Interestingly, manipulating levels of either miR induced complimentary aspects of Gfi1 loss of function. Manipulating miR-21 expression specifically and significantly controlled monocytic colony numbers in methylcellulose, and forced miR-21 expression even doubled the small number of monocytic cells in G-CSF–stimulated cultures. Thus, miR-21 acts as a positive regulator of monopoiesis. Manipulating miR-196b expression specifically and significantly controlled granulocytic colony numbers, and significantly (but not completely) blocked G-CSF–stimulated granulopoiesis. Thus, miR-196b acts as a negative regulator of granulocytic differentiation. Because Gfi1 loss of function increases monocytic differentiation and blocks granulopoiesis, the biologic phenotypes engendered by combined deregulated expression of miR-21 and miR-196b (as Gfi1 target genes) represent an important and new epigenetic component to the Gfi1 transcriptional network.
Dysregulation of miRs leads to diseases ranging from cancer to vascular diseases.15,17,45,46 Several lines of evidence have established the importance of miR-2143,47–50 and miR-196b.51 In severe congenital neutropenia patients with one mutant GFI1 allele, the GFI1N382S protein fails to bind DNA and functions as a dominant negative mutant by sequestering GFI1 cofactors thus derepressing a subset of GFI1 target genes.26 Recently, it has been shown that the PRDM5 transcription factor regulates miR-21 and miR-196b (among other miR genes) and physically interacts with Gfi1.52 More work is necessary to understand the interplay between Gfi1 and PRDM5 in miR-21 and miR-196b gene regulation; however, based on our results we can now include miR-21 and miR-196b in this list of Gfi1 targets that must be repressed for proper myelopoiesis. GFI1 function is required for normal expression of miR-21 and miR-196b in healthy individuals; however, miR-21 and miR-196b are deregulated in the bone marrow cells of a GFI1N382S (mutant) SCN patient. As such, the posttranscriptional programs modulated by GFI1-repressed miRs make them an interesting candidate for GFI1-mutant SCN phenotypes. In fact, G-CSF–instructed granulocytic differentiation is disrupted by enforced expression of miR-21 and miR-196b. This suggests that the higher steady-state levels of miR-21 and miR-196b propagate the dominant-negative effect of the GFI1N382S protein. Similar phenotypes engendered by Gfi1 loss of function and miR-21 and miR-196b overexpression provide a compelling argument for a common pathway controlled by these factors. For most SCN patients (most of which have ELA2 mutations) recombinant G-CSF therapy is the current standard; however, not all patients respond to G-CSF, G-CSF does not rescue normal granulopoiesis, and G-CSF therapy might predispose to oncogenic transformation.53,54 Perhaps the potent effect of miR-21 and miR-196b targeting agents (such as antagomiRs) could be harnessed for clinical intervention into hematologic diseases such as SCN, or bone marrow transplantation in which manipulating neutrophil counts is desirable. However, we note that the level of Gfi1 is reduced upon monocytic or granulocytic differentiation of leukemia cell lines, followed by reciprocal induction of miR-21 and miR-196b. Thus, it is possible that these miRs must be repressed to restrict lineage potential, but participate in maturation once lineage commitment is fixed. More research is necessary to reconcile the complex regulatory networks engaged by miR-21, miR-196b, PRDM5, and Gfi1. Taken together our data implicate miR-21 and miR-196b as potential therapeutic targets in neutropenia and other myeloid disorders. However, more work is clearly needed to delineate the molecular control of miRs in normal granulopoiesis and the pathobiology of SCN.
Acknowledgments
The authors thank Michael Jansen for microarray analysis, Terry Camerlengo (OSUCCC Biomedical Informatics Shared Resource), and Jeff Palatini (OSUCCC Microarray Shared Resource) for help with microarray data upload to GEO, A. Berns for Rosa-CreERT2+ mice, B. Cullen for a miR-21 expression plasmid, and T. Bourdeau and M. Meadows for excellent technical assistance as well as A. Jegga, B. Aronow, A. Chaubey, S. Horman, J. Phelan, and M. Rojas for helpful discussions.
This work was partially supported by HL079574, CA112405, and a Scholar award from the Leukemia & Lymphoma Society (White Plains, NY) (H.L.G.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.S.V. designed and performed experiments, analyzed results, interpreted data, generated figures, and wrote the paper; A.M.B. performed the EMSAs; and H.L.G. designed experiments, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: H. Leighton Grimes, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave MLC 7038, Cincinnati, OH 45244; e-mail: lee.grimes@cchmc.org.
References
- 1.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 4.Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 7.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 2007;427:193–213. doi: 10.1016/S0076-6879(07)27011-9. [DOI] [PubMed] [Google Scholar]
- 11.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 16.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 18.Boxer LA. Severe congenital neutropenia: genetics and pathogenesis. Trans Am Clin Climatol Assoc. 2006;117:13–32. [PMC free article] [PubMed] [Google Scholar]
- 19.Ancliff PJ, Blundell MP, Cory GO, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–2189. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- 20.Aprikyan AA, Liles WC, Boxer LA, Dale DC. Mutant elastase in pathogenesis of cyclic and severe congenital neutropenia. J Pediatr Hematol Oncol. 2002;24:784–786. doi: 10.1097/00043426-200212000-00025. [DOI] [PubMed] [Google Scholar]
- 21.Klein C, Grudzien M, Appaswamy G, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease). Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 22.Person RE, Li FQ, Duan Z, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hock H, Hamblen MJ, Rooke HM, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 25.Karsunky H, Zeng H, Schmidt T, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 26.Zarebski A, Velu CS, Baktula AM, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–380. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 28.Duan Z, Horwitz M. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc Natl Acad Sci U S A. 2003;100:5932–5937. doi: 10.1073/pnas.1031694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Center for Biotechnology Information. Gene Expression Omnibus (GEO). [Accessed February 2009]; http://www.ncbi.nlm.nih.gov/geo.
- 31.Hawley RG. High-titer retroviral vectors for efficient transduction of functional genes into murine hematopoietic stem cells. Ann N Y Acad Sci. 1994;716:327–330. doi: 10.1111/j.1749-6632.1994.tb21724.x. [DOI] [PubMed] [Google Scholar]
- 32.Hameyer D, Loonstra A, Eshkind L, et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 33.Kazanjian A, Wallis D, Au N, et al. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 2004;64:6874–6882. doi: 10.1158/0008-5472.CAN-04-0633. [DOI] [PubMed] [Google Scholar]
- 34.Jegga AG, Chen J, Gowrisankar S, et al. GenomeTrafac: a whole genome resource for the detection of transcription factor binding site clusters associated with conventional and microRNA encoding genes conserved between mouse and human gene orthologs. Nucleic Acids Res. 2007;35:D116–D121. doi: 10.1093/nar/gkl1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 37.Georgantas RW, III, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p–20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 39.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 41.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU. 1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 44.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 2002;21:3377–3390. doi: 10.1038/sj.onc.1205324. [DOI] [PubMed] [Google Scholar]
- 45.Nervi C, Fazi F, Rosa A, Fatica A, Bozzoni I. Emerging role for microRNAs in acute promyelocytic leukemia. Curr Top Microbiol Immunol. 2007;313:73–84. doi: 10.1007/978-3-540-34594-7_5. [DOI] [PubMed] [Google Scholar]
- 46.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 47.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 48.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 50.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 51.Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 52.Duan Z, Person RE, Lee HH, et al. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol. 2007;27:6889–6902. doi: 10.1128/MCB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donini M, Fontana S, Savoldi G, et al. G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood. 2007;109:4716–4723. doi: 10.1182/blood-2006-09-045427. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg PS, Alter BP, Link DC, et al. Neutrophil elastase mutations and risk of leukaemia in severe congenital neutropenia. Br J Haematol. 2008;140:210–213. doi: 10.1111/j.1365-2141.2007.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]