Abstract
Mexico is developing the basis for genomic medicine to improve healthcare of its population. The extensive study of genetic diversity and linkage disequilibrium structure of different populations has made it possible to develop tagging and imputation strategies to comprehensively analyze common genetic variation in association studies of complex diseases. We assessed the benefit of a Mexican haplotype map to improve identification of genes related to common diseases in the Mexican population. We evaluated genetic diversity, linkage disequilibrium patterns, and extent of haplotype sharing using genomewide data from Mexican Mestizos from regions with different histories of admixture and particular population dynamics. Ancestry was evaluated by including 1 Mexican Amerindian group and data from the HapMap. Our results provide evidence of genetic differences between Mexican subpopulations that should be considered in the design and analysis of association studies of complex diseases. In addition, these results support the notion that a haplotype map of the Mexican Mestizo population can reduce the number of tag SNPs required to characterize common genetic variation in this population. This is one of the first genomewide genotyping efforts of a recently admixed population in Latin America.
Keywords: admixture, genetic variation, population genetics, SNP tagging
More than 560 million people live in Latin American countries, and according to U.S. Census Bureau estimates the Latino population reached ≈45.5 million in 2007, representing the largest and fastest-growing minority group in the United States. Mexican Mestizos, as other Latino populations, are a recently admixed population composed of Amerindian, European, and, to a lesser extent, African ancestries. Although the diversity of Latino populations poses several challenges for genetic studies (1), it makes them a powerful resource for analyzing the genetic bases of complex diseases (2). In the past 5 years, Mexico has been committed to develop a human and technological infrastructure for genomics with special emphasis on the development of a national platform of genomic medicine to improve healthcare of Mexicans (3–6). This effort, together with a population of ≈105 million inhabitants including 60 Amerindian groups and a complex history of admixture, makes Mexico an ideal country in which to perform genomic analysis of common complex diseases.
Two current approaches to identify genes influencing complex diseases are genomewide association studies (GWAS) and admixture mapping (AM). GWAS depend on efficient SNP tagging (7, 8), and AM on the availability of panels of genomewide markers with frequency differences between parental populations (9, 10). For populations not comprehensively represented in the HapMap (11), such as Latinos, limitations exist for an efficient tagging and imputation, because of the need of a higher number of markers to achieve the same relative power compared to that for Asians and Europeans (12) and the lack of knowledge about population-specific linkage disequilibrium (LD) patterns (13). In addition, false positives because of population structure are minimized in GWAS by excluding individuals with ancestry differences (7). This is not practical in studies including Latinos such as Mexicans, where >80% of the population consists of Mestizos with known differences in ancestral proportions (2). As for AM, there are a few SNP panels developed for Latino populations (14–16); however, detailed genomewide information from Mestizo and Amerindian populations remains limited (17, 18). Recent studies of Latin American populations have shown differential ancestral contribution patterns between and within groups that correlate with pre-Columbian native population density and with patterns of recent demographic growth (2). These differences should be considered to improve AM panels for Latin American populations.
Historically, admixture patterns throughout Mexico have been influenced by differences in parental population densities and demographic growth (19–21). Genetic heterogeneity between and within Mestizos from different regions has been documented (22–29). However, no genomewide comparison of different Mestizo and Amerindian populations in Mexico is currently available in the public domain. To analyze genomic diversity and LD patterns in Mexicans, we developed the Mexican Genome Diversity Project (MGDP). This resource will be useful to develop strategies for the genetic analysis of Mexican and related admixed populations, such as marker selection for optimal coverage of common genetic variation in GWA and targeted association studies, and also for the adequate application of tagging and imputation approaches (30, 31) and for AM (10) in Mexicans and other Latino populations. Our study is one of the first extensive genomewide genotyping efforts performed in Latin America. The MGDP will contribute to the development of genomic medicine in Mexico and the rest of Latin America.
Results
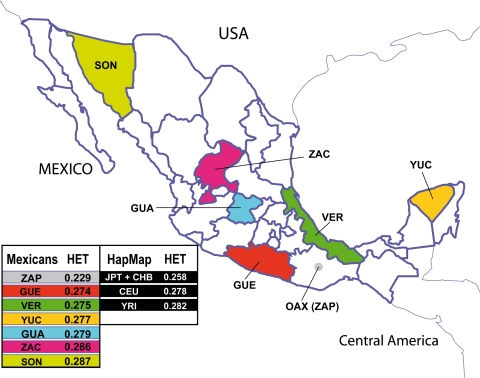
We analyzed data from 300 nonrelated self-identified Mestizo individuals from 6 states located in geographically distant regions in Mexico: Sonora (SON) and Zacatecas (ZAC) in the north, Guanajuato (GUA) in the center, Guerrero (GUE) in the center–Pacific, Veracruz (VER) in the center–Gulf, and Yucatan (YUC) in the southeast. Considering that Zapotecos have been shown as a good ancestral population for predicting Amerindian (AMI) ancestry in Mexican Mestizos (16), we included 30 Zapotecos (ZAP) from the southwestern state of Oaxaca (Fig. 1). For comparative purposes, we included similar data sets from HapMap populations: northern Europeans (CEU), Africans (YRI), and East Asians (EA), including Chinese (CHB) and Japanese (JPT). A HapMap-like database with SNP frequencies in Mexicans and HapMap populations was generated (http://diversity.inmegen.gob.mx).
Fig. 1.
Genetic diversity measured by heterozygosity (HET) in Mexican and HapMap populations. Northern, central, central-Gulf, central-Pacific, and southern regions in Mexico were included. Average HET values are shown for Amerindian Zapotecos (ZAP), 6 Mexican Mestizo subpopulations (GUA, GUE, SON, VER, YUC, and ZAC), and HapMap populations (YRI, CEU, and JPT + CHB).
Analysis of Genetic Diversity in Mexicans.
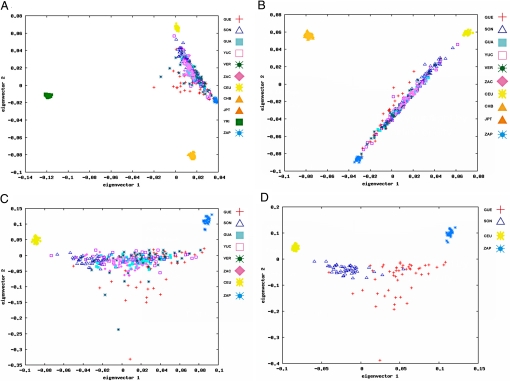
We measured heterozygosity (HET), performed principal components analysis (PCA) (32), and calculated FST statistics using data sets obtained for Mexican and HapMap populations. Mexican Mestizo subpopulations had HET values between 0.274 in GUE and 0.287 in SON. Among HapMap populations, YRI displayed the highest genetic diversity (HET = 0.282) and JPT + CHB the lowest (HET = 0.258), as previously reported (33). Among Mexicans, northern subpopulations (SON and ZAC) had the highest HET values, suggesting more genetic diversity, and the ZAP Amerindian samples had the lowest (HET = 0.229), as expected for an isolated population. For PCA analysis, we used different combinations of data sets and conditions. In all scenarios the 2 most informative eigenvectors for each data set are displayed (Fig. 2 A–D). When included, the HapMap and ZAP populations formed defined clusters, while the Mexican Mestizo subpopulations were widely distributed between the CEU and ZAP samples (Fig. 2 A and B). The ZAP population clustering in the PCA plot suggests the absence of recent admixture in this Amerindian group. As expected, when all groups were analyzed (Fig. 2A), the largest genetic distance exists between the YRI population and the rest of the groups. In the second axis, the ZAP cluster is located between CEU and EA and, in both the first and the second axes, all Mexican Mestizos are spread between CEU and ZAP (Fig. 2 A and B). To better display the distribution of Mexican Mestizos, we generated 2 additional data sets, one leaving out YRI samples (Fig. 2B) and another including only CEU and ZAP. These analyses gave evidence of genetic diversity between and within Mexican Mestizo populations. In addition, a PCA including only CEU, ZAP, and the 2 Mestizo groups with the largest HET difference (SON and GUE) showed that samples from SON were closer to the CEU, and those from GUE were closer to the ZAP (Fig. 2 C and D). In both plots, some individuals were displaced along eigenvector 2, reflecting additional ancestral contributions in Mestizos. To evaluate whether this effect is related to African (AFR) ancestry, we analyzed an additional data set including YRI [supporting information (SI) Fig. S1 A and B]. The distribution of Mestizos in eigenvector 3 (Fig. S1B) indicates that the spread observed in eigenvector 2 (Fig. 2 C and D) reflects AFR ancestral contribution. Interestingly, Mestizos did not organize in a straight line between CEU and ZAP (Fig. 2 C and D). This is most probably because those 2 groups of samples do not fully represent the genetic variability of European and Amerindian ancestral origin present in these Mestizos (2).
Fig. 2.
Principal components analysis. The 2 most informative eigenvectors were plotted in all cases. Four different data sets are presented: (A) all Mexican subpopulations, Mestizo (GUA, GUE, SON, VER, YUC, ZAC) and Amerindian (ZAP) populations, and HapMap populations (YRI, CEU and JPT + CHB); (B) all Mestizos, ZAP, CEU, and JPT + CHB; (C) all Mestizos, ZAP, and CEU; and (D) Mestizo subpopulations showing the largest difference in eigenvector 1 (SON and GUE), ZAP, and CEU.
To measure genetic distances between Mexican subpopulations, and between these populations and those from the HapMap, we performed a pairwise FST statistical analysis (Table 1). Of all Mexican groups, the Amerindian ZAP population showed the highest FST values when compared to all HapMap populations. As expected, the highest value was observed when compared to YRI (23.9), followed by CEU (15.4), JPT (11.9), and CHB (12.0). FST values between ZAP and each Mestizo subpopulation (Table 1) were consistent with their distribution in the PCA plot (Fig. 2C), with GUE and VER closest to the ZAP cluster (FST values 3.2 and 3.8, respectively) and SON at the other end of the distribution (FST of 8.2). Pairwise comparisons between Mexican groups showed that SON when compared to all other Mestizo subpopulations had higher FST values than that observed between CHB and JPT. Moreover, the FST value between SON and ZAP (8.2) was higher than that of any other comparison between any Mestizo subpopulation and non-African HapMap group (Table 1). These results support the presence of considerable genetic heterogeneity between Mexican Mestizo subpopulations and suggest that this diversity is mainly related to a differential distribution of AMI and EUR ancestral components.
Table 1.
FST values between Mexican, Zapoteco Amerindians, and HapMap populations
| GUE | SON | VER | YUC | ZAC | ZAP | CEU | YRI | CHB | JPT | |
|---|---|---|---|---|---|---|---|---|---|---|
| GUA | 0.2 | 1.1 | 0.1 | 0.3 | 0.1 | 4.3 | 5.2 | 15.4 | 6.9 | 6.9 |
| GUE | 1.9 | 0.1 | 0.4 | 0.5 | 3.2 | 6.9 | 15.7 | 7.0 | 7.0 | |
| SON | 1.3 | 1.2 | 0.6 | 8.2 | 2.0 | 13.9 | 7.3 | 7.4 | ||
| VER | 0.2 | 0.2 | 3.8 | 5.8 | 15.7 | 6.9 | 7.0 | |||
| YUC | 0.3 | 4.5 | 5.2 | 15.6 | 7.0 | 7.0 | ||||
| ZAC | 5.3 | 4.0 | 14.5 | 6.8 | 6.9 | |||||
| ZAP | 15.4 | 23.9 | 12.0 | 11.9 | ||||||
| CEU | 15.7 | 11.0 | 11.2 | |||||||
| YRI | 18.4 | 18.5 | ||||||||
| CHB | 0.7 |
Pairwise FST statistics (× 100) between Mexican Mestizos and HapMap populations are shown. Calculations were performed with EIGENSOFT using 99,953 SNPs.
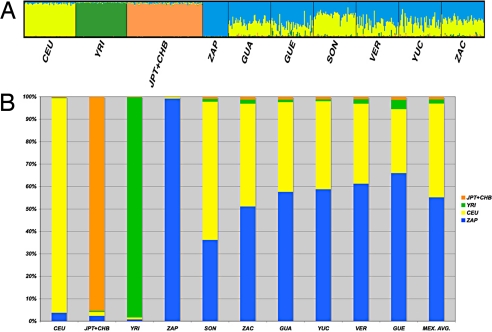
To assess genetic ancestry in Mexicans, we determined individual and population average ancestral proportions using STRUCTURE (34, 35). For this, we used 1,814 ancestry informative markers (AIMs) selected using different criteria to ensure genomewide distribution and minimize LD between SNPs (see Materials and Methods). We used HapMap data and the ZAP population as EUR, AFR, EA, and AMI ancestral sources in the analyses. Our results were most consistent with 4 population groups (K = 4), explaining the major substructure in this set of Mexican Mestizos (Fig. 3 A and B). In this model, their mean ancestries (±SD) were 0.552 ± 0.154 for AMI, 0.418 ± 0.155 for EUR, 0.018 ± 0.035 for AFR, and 0.012 ± 0.018 for EA (Table S1). We observed differences within and between Mestizo subpopulations, mainly in EUR and AMI ancestries (Fig. 3 A and B). The highest and lowest estimates of mean EUR ancestry were 0.616 ± 0.085 for SON and 0.285 ± 0.120 for GUE. Most Mestizo subpopulations displayed statistically significant differences in mean EUR ancestral contribution, and both SON and GUE showed differences when compared to any other Mestizo subpopulation (Table S2). Mestizo groups with similar mean EUR ancestry were those from central and central-coastal regions (VER, YUC, and GUA). In contrast, most Mestizo subpopulations had a similar average AMI ancestral contribution—GUE the highest (0.660 ± 0.138) and SON the lowest (0.362 ± 0.089) (Fig. 3B)—and only subpopulations in the northern states (SON and ZAC) showed statistically significant differences compared with all other Mestizo groups (Table S2). The other 2 ancestries analyzed, AFR and EA, were smaller and almost homogenous among all Mestizo subpopulations. Significant differences in AFR ancestry were observed for SON and ZAC against VER and YUC (Table S2). To evaluate the contribution of ancestry differences to the overall regional genetic diversity between Mestizo subpopulations, we calculated Pearson correlation coefficients between pairwise FST values and differences in AMI, EUR, and AFR ancestral proportions. This analysis revealed a high correlation between overall genetic diversity (FST) and EUR (r = 0.937) and AMI (r = 0.944) ancestry differences. To estimate the size of this effect, we calculated genetic distance between Mexican subpopulations, specifically attributable to differences in the 2 main continental ancestry proportions (Table S3). This analysis revealed that for most pairwise comparisons between Mestizo subpopulations (10 of 15), 50% of the genetic distance between them is attributable to differences in continental ancestry. Interestingly, most comparisons with low contribution of continental ancestry differences to overall genetic distance included the subpopulation of YUC. These samples are the only Mestizos in this study that have a distinctive AMI ancestry (Maya).
Fig. 3.
Population structure analysis using 1814 AIMs. (A) Individual ancestry proportions. (B) Average ancestral contributions in Mexican Mestizos. Significant differences in ancestry proportions were mainly observed for EUR and AMI contributions (Table S2).
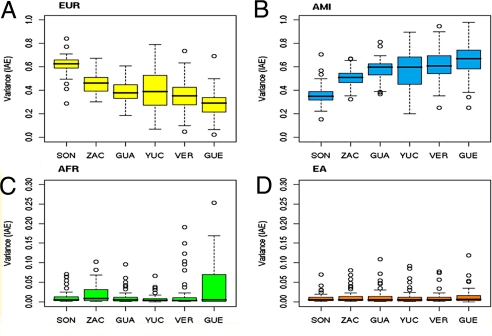
To evaluate intraregional differences in ancestry proportions among Mexican Mestizos, we compared box-plot distributions (Fig. 4) and coefficients of variation (CVs) as normalized measurements of the observed dispersion for each ancestry (Table S4) for each individual ancestral contribution. We observed a wide distribution of CVs, in the range of 0.139–0.421 for EUR, 0.151–0.273 for AMI, 1.236–2.096 for AFR, and 1.264–1.625 for EA. A low-variance distribution was observed for EUR and AMI ancestries in all subpopulations, and the largest CVs for these were observed in GUE (0.421) and YUC (0.273), respectively (Fig. 4 A and B). Outliers with an AFR proportion >15% and intraregional variability in this component were observed in VER and GUE (CVs = 2.096 and 1.501) (Fig. 4C). Although EA contributions were small, a high-variance distribution (CVs = 1.264–1.625) was observed for all subpopulations (Fig. 4D). These results support that population structure in Mexican Mestizos is mainly related to differences in EUR and AMI ancestral contributions, but that other sources of genetic diversity, such as AFR or distinctive AMI, also participate.
Fig. 4.
Boxplot distribution of ancestry estimates. Quantile distributions of ancestry proportions for 6 Mexican Mestizo subpopulations are shown: GUA, GUE, SON, VER, YUC, and ZAC. Panels correspond to parental populations: (A) EUR, (B) AMI, (C) AFR, and (D) EA. The plot represents the minimum and maximum values (whiskers), the first and third quartiles (box), and the median value (midline). Outliers are also displayed. The y-axis represents the variance of the individual ancestral estimate (STRUCTURE).
Private Alleles in Mexican Populations.
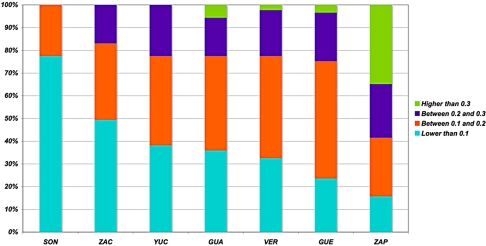
We identified 89 common private alleles that were absent in HapMap populations but present in at least 1 Mexican Mestizo subpopulation and 86 in Mexican Amerindians (ZAP). All alleles private to ZAP were also private to Mestizos, indicating their AMI origin. The number of private alleles was similar in all 6 states, but differences were observed in the proportion of variants with higher frequencies (MAF > 0.20). We did not observe alleles with MAF > 0.20 in SON or with MAF > 0.30 in ZAC or YUC (Fig. 5). These results correlate with our observation that Northern Mexican subpopulations (SON and ZAC) have the highest EUR ancestral contribution and central-coastal region subpopulations (GUE and VER) have the highest AMI ancestries. To analyze this result in the context of continental genetic contributions, we searched for alleles private to each HapMap group compared to the rest and identified 5,660 alleles private to YRI, 1,533 to CEU, and 669 to CHB + JPT. The observation of the highest number of private alleles in AFR and the lowest in AMI is consistent with models of human evolution with an AFR origin reaching the Americas after a series of founder effects (36).
Fig. 5.
Frequency distribution of SNPs private to Mexicans compared to HapMap populations. Private SNPs have a MAF > 0.05 in at least 1 Mexican subpopulation, but are absent in all HapMap populations. Each bar represents the frequency distribution of all private SNPs (n = 89) for each Mexican subpopulation.
To identify genomic regions with intrapopulation differences in Mexico, we first searched for alleles private to a particular Mexican Mestizo subpopulation, but found only 2 SNPs with frequencies >0.05, 1 in SON (rs5973601, MAF = 0.053) and 1 in ZAC (rs3733654, MAF = 0.051). Informativeness for assignment (37) was then used to find a larger subset of SNPs showing geographic variation in allele frequencies between Mexican subpopulations, which resulted in the identification of 14 SNPs with high information content (In > 0.04) (Fig. S2). All were AIMs with δ ≥ 0.27 for at least 1 of the ancestral sources included in previous analyses (Table S5). This result provides additional support to the observed ancestry-related genetic differences among Mestizos and highlights genetic regions with intrapopulation differences in SNP frequencies that could be a source of false positive signals in genetic association studies in Mexicans.
LD Patterns in Mexican Mestizos and HapMap Populations.
Average allele frequency distribution of common SNPs (MAF > 15%) in the Mexican samples was similar to that of HapMap populations (Fig. S3A), indicating no bias in ascertainment. Fewer low-frequency markers (MAF < 0.05) were observed in SON and ZAC than in HapMap populations, indicating less homozygosity in these groups. This result is consistent with SON and ZAC having the highest HET values (Fig. 1). To evaluate the potential size of haplotype blocks in Mexican subpopulations, LD decay plots of highly correlated (r2 > 0.8 and D′ > 0.8) common alleles (MAF > 15%) were compared between Mexicans and HapMap populations. LD decay in Mexicans was similar to that in the non-African HapMap samples (Fig. S4 B and C). To further evaluate genomic structure variability in Mexicans we performed long-range haplotype diversity (LRHD) analysis. When Mexican subpopulations were compared to HapMap populations, most showed decreased diversity, and only SON had a similar LRHD pattern to that of Asians. Of all Mestizo groups, VER and GUE showed the least haplotype diversity (Fig. S4A). On average, 68 haplotypes per megabase accounted for 95% of the chromosomes in Mexicans, while the same coverage required 93, 83, 69, and 70 haplotypes in the YRI, CEU, CHB, and JPT samples, respectively (Fig. S4B). This result indicates reduced haplotype diversity in Mexicans compared to HapMap populations.
Haplotype Sharing (HS) Between Mexican Mestizos and HapMap Populations.
To determine the potential use of HapMap data for targeted and GWA studies in Mexicans, we evaluated the number of common haplotypes (frequency >5%) shared between Mexicans and HapMap populations. This analysis showed that Mexicans share 64% of these haplotypes with YRI, 74% with JPT + CHB, and 81% with CEU and that the proportion of shared haplotypes increased to 96% when the combination of the 4 HapMap populations is used as a reference (Table 2). Although these results show that effective coverage of common genetic variations in Mexicans is feasible using HapMap information, it may be at a high genotyping cost because of the need to include the combined data set for all HapMap populations. To evaluate the potential benefit of using a haplotype map of the Mexican population over that of using only HapMap information, we evaluated HS between Mexican subpopulations. In this analysis either 1 Mexican subpopulation or any possible pair of Mexican subpopulations was used as a reference group. This analysis showed that all Mexican subpopulations share, on average, 86% (84–87%) of the common haplotypes when 1 subpopulation is used as a reference (Table 3) and that the proportion of shared haplotypes increases to an average of 96% (95–97%) when each subpopulation is compared to any pair of subpopulations (Table S6). These results support the idea that a haplotype map of the Mexican Mestizo population may help reduce the number of tag SNPs required to characterize common genetic variation in this population.
Table 2.
Percentages of common haplotypes shared between Mexican and HapMap populations
| Population | CEU | JPT + CHB | YRI | CEU +JPT + CHB | CEU + JPT + CHB + YRI |
|---|---|---|---|---|---|
| GUA | 81 | 75 | 64 | 93 | 96 |
| GUE | 79 | 76 | 65 | 93 | 96 |
| SON | 82 | 71 | 63 | 94 | 97 |
| VER | 80 | 75 | 64 | 93 | 96 |
| YUC | 81 | 74 | 64 | 93 | 96 |
| ZAC | 81 | 73 | 64 | 93 | 97 |
| MEX average | 81 | 74 | 64 | 93 | 96 |
HS was assessed by comparing the frequencies of 5 SNP haplotypes spanning ≈100 kb. The percentages of shared common haplotypes (>5% frequency) between Mexican and HapMap populations are shown.
Table 3.
Percentages of common haplotypes shared between Mexican subpopulations
| Population | GUA | GUE | SON | VER | YUC | ZAC |
|---|---|---|---|---|---|---|
| GUA | 86 | 87 | 87 | 87 | 88 | |
| GUE | 88 | 87 | 88 | 87 | 88 | |
| SON | 83 | 82 | 82 | 83 | 84 | |
| VER | 87 | 86 | 87 | 87 | 88 | |
| YUC | 87 | 85 | 87 | 86 | 87 | |
| ZAC | 85 | 83 | 87 | 85 | 85 | |
| MEX Av | 86 | 84 | 87 | 86 | 86 | 87 |
Haplotype sharing was assessed by comparing the frequencies of 5 consecutive SNP haplotypes spanning ≈100 kb. The percentages of shared common haplotypes (>5% frequency) between Mexican subpopulations are shown.
Discussion
This work is an initial assessment of the potential benefit of generating a haplotype map to optimize the design and analysis of genetic association studies in Mexicans. During the pre-Hispanic period, ethnic groups living in Central and Southern Mexico were more numerous and had stronger political, religious, and social cohesion than ethnic groups from northern regions. African slaves were brought into the region after a notable reduction of the Amerindian population, due to epidemics, between 1545 and 1548 (19). Since then, admixture processes in geographically distant regions have been affected by different demographic and historical conditions, shaping the genomic structure of Mexicans. These factors have generated genetic heterogeneity between and within subpopulations from different regions throughout Mexico (2, 26, 29, 38). Even though participants in our study came from regions corresponding to modern political divisions, they represent different demographic dynamics, human settlement patterns, and Amerindian population densities. Because of known bias of admixture estimates due to socioeconomic stratification in Mexicans (28), Mestizo participants were recruited at state universities, in which most attendees come equally from urban and rural areas and belong to a wide range of socioeconomic strata.
Our results show that genetic differences among Mexican Mestizos from different regions in Mexico are mainly because of differences in AMI and EUR contributions. In most analyses, samples from central regions were closer to ZAP, while samples from northern regions were located closer to CEU, correlating with Amerindian population density in those regions, both in modern days and in the pre-Hispanic period (19). Although our analysis showed that mean AFR ancestry was low (<10%) and mostly homogenous among subpopulations, we observed the presence of individuals with high AFR ancestry in GUE and VER. This is in agreement with historical records indicating these states as the main entry points of Africans during the Colonial period and as residence of African-Mexicans since then (39). Interestingly, samples from the southeastern region (YUC) had the lowest contribution of continental ancestry to genetic distance. Mestizos from Yucatan are the only group in our sample with a distinctive Maya AMI contribution. Mayas are a distinct ethnic group, geographically distant from other AMI groups, with strong cultural, social, and historical differences compared to them (20); thus this result suggests that some of the genetic diversity observed in our Mestizos is related to differential AMI contribution.
Alleles private to Mexican Mestizos have an AMI origin and conservatively represent the genetic variation absent in other continental groups, considering that most SNPs analyzed were identified in populations with genetic backgrounds lacking an AMI contribution (40). Positive detection of SNPs private to AMI is related to the use of a genotyping assay with SNP information from a multiethnic group that included Hispanics/Latinos: http://www.genome.gov/10001552 (40). These SNPs represent variants not covered by the HapMap that may not be captured when tag SNPs are selected using only HapMap information. To better describe SNPs and haplotypes private to Mexicans it is necessary to perform extensive resequencing projects in both Mestizos and Amerindians.
Considering that LD decay patterns of all Mexican groups behaved similarly to those of non-African HapMap populations, average haplotype block size in Mexicans is expected to be similar to that of non-African HapMap populations. The reduced LRHD observed in Mexicans correlates with the AMI contribution, being consistent with the fact that Amerindian populations have significantly reduced haplotype diversity and long-range LD (35) and thus in possible relationship to the progressive decrease in haplotype diversity in human populations migrating out of Africa (41). Shared haplotype analysis was used as an approach to indirectly estimate tag SNP transferability from HapMap to Mexican populations and between Mexican subpopulations. This analysis was performed using different combinations of Mexican and HapMap populations to evaluate the potential benefits of a Mexican haplotype map. Common genetic variation in Mexicans is efficiently covered (96%) only when combined data from all HapMap populations are used, in accordance with previous findings for Latino populations (12), which suggests that selection of tag SNPs exclusively from the HapMap, for studies in Mexicans, will result in a significant increase in costs due to overgenotyping. An indication that a haplotype map for Mexicans could be useful for tag SNP selection is that the use of any combination of two Mexican subpopulations as a reference provided better coverage than using the combination of all HapMap populations. These results support the fact that a haplotype map describing common genetic variability and LD patterns in Mexicans is feasible and useful.
Public availability of data from the MGDP will be important for a more effective design of association studies and resequencing projects in Latino populations. Our study suggests that either genomewide or targeted approaches that use tag SNPs selected with HapMap data may adequately capture 96% of the common genetic diversity in Mexicans. However, it seems possible to generate optimized sets of tag SNPs to improve the efficiency of targeted association studies and help reduce costs without compromising coverage. This is critical for Mexico and other Latin American countries where funding for research is limited. Also, a Mexican haplotype map would help in haplotype tagging and subsequent SNP discovery in Latino populations, improving the search for rare variants associated with common complex diseases.
Imputation is used to improve power and combine data from GWAS that employ different SNP sets (30, 31). However, this approach assumes similar genomewide LD patterns between the analyzed samples and the reference panel (30). Tagging or imputation using HapMap information is not as efficient in Mexicans and other Latinos as it is in other populations because of the presence of a genetic component not captured by HapMap data (13). The MGDP data set will be of great value to test the accuracy of the imputation paradigm in Mexicans and to improve imputation approaches by the inclusion of adequate estimates of individual and local ancestry. The MGDP data will also be useful to optimize existing sets of AIMs (14–16, 29) to perform AM studies in traits and diseases showing ethnicity-based differences in prevalence in Mexicans, such as HDL cholesterol levels (42), gall bladder disease (43), and type 2 diabetes (44).
We are currently increasing the SNP density to ≈1.5 million SNPs per genome using a combination of microarray platforms. Here we present one of the first public genomewide data sets for Mexican Mestizo and Amerindian populations. This effort will contribute to the design of better strategies aimed at characterizing the genetic factors underlying common complex diseases in Mexicans. In addition, this information will increase our knowledge of genomic variability in Latino populations. The scientific and technological infrastructure derived from this project will significantly contribute to the development of genomic medicine in Mexico and Latin America (3, 6).
Materials and Methods
Anonymous blood samples from 300 nonrelated and self-defined Mestizos and 30 Amerindian Zapotecos were collected in 7 states in Mexico: Guanajuato, Guerrero, Sonora, Veracruz, Yucatan, Zacatecas, and Oaxaca (ZAP). The Scientific, Ethics, and Bio-Security Review Boards from the National Institute of Genomic Medicine (INMEGEN) approved this study. An ad hoc process for community consultation and engagement was implemented. Genomic DNA was extracted from blood (QIAGEN). Genotyping was performed according to the Affymetrix 100K SNP array protocol and 99,953 SNPs passed quality control in all populations. Phasing was performed with fastPhase v1.1.4 (45). All genotypes and raw signal intensity files are available (ftp://ftp.inmegen.gob.mx). Average HET was calculated with PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) (46). The PCA was done with EIGENSTRAT (32), and FST with EIGENSOFT (39). Ancestral contributions were assessed with Mann–Whitney U tests, Pearson correlations, box-plot distributions, and their coefficients of variation. For ancestry analysis 1,814 AIMs were used to run STRUCTURE v.2.1 (34, 35). Scripts for informativeness for assignment were kindly provided by N. Rosenberg (37). Alleles private to the Mexican population had a MAF > 0.05 in any of the Mexican subpopulations, but were absent in all HapMap populations. Alleles private to any particular Mexican subpopulation had a MAF > 0.05 in 1 Mexican group and were absent in the other 6. LD calculations, long-range haplotype diversity, and HS analysis were done with Haploview and special-purpose code, as previously described (47, 48). All data analyses were performed at INMEGEN in Mexico City. (see SI Materials and Methods).
Supplementary Material
Acknowledgments.
We thank the Federal Government of Mexico, particularly the Ministry of Health for valuable support throughout the project. Participation of the governments and universities of the states of Guanajuato, Guerrero, Oaxaca, Sonora, Veracruz, Yucatan, and Zacatecas contributed significantly to this work. We thank all volunteers in the study and the National Institute of Genomic Medicine (INMEGEN)'s personnel for important support; Alejandro López, José Bedolla, Alejandro Rodríguez, and Lucía Orozco for their major contributions to the thorough communication strategy; and Blanca Gonzalez-Sobrino for helpful advice on Mexican ethnohistory. This work was supported by funds from the Federal Government of Mexico to the National Institute of Genomic Medicine and by infrastructure donated by the Mexican Health Foundation (FUNSALUD) and the Gonzalo Río Arronte Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903045106/DCSupplemental.
References
- 1.Gonzalez Burchard E, et al. Latino populations: A unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health. 2005;95:2161–2168. doi: 10.2105/AJPH.2005.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez-Sanchez G. Developing a platform for genomic medicine in Mexico. Science. 2003;300:295–296. doi: 10.1126/science.1084059. [DOI] [PubMed] [Google Scholar]
- 4.Hardy BJ, et al. The next steps for genomic medicine: Challenges and opportunities for the developing world. Nat Rev Genet. 2008;9(Suppl 1):S23–S27. doi: 10.1038/nrg2444. [DOI] [PubMed] [Google Scholar]
- 5.Seguin B, Hardy BJ, Singer PA, Daar AS. Genomics, public health and developing countries: The case of the Mexican National Institute of Genomic Medicine (INMEGEN) Nat Rev Genet. 2008;9(Suppl 1):S5–S9. doi: 10.1038/nrg2442. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez-Sanchez G, Silva-Zolezzi I, Hidalgo A, March S. Genomic medicine in Mexico: Initial steps and the road ahead. Genome Res. 2008;18:1191–1198. doi: 10.1101/gr.065359.107. [DOI] [PubMed] [Google Scholar]
- 7.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy MI, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 9.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nat Rev Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 10.Seldin MF. Admixture mapping as a tool in gene discovery. Curr Opin Genet Dev. 2007;17:177–181. doi: 10.1016/j.gde.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bakker PI, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao X, et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian C, et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 18.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 19.Gerhard P. Historical Geography of New Spain, 1519–1821. Mexico City: Universidad Nacional Autonoma de Mexico; 1986. (Spanish) [Google Scholar]
- 20.Gerhard P. La Frontera Sureste de la Nueva España. Mexico City: Universidad Nacional Autónoma de México; 1991. in Spanish. [Google Scholar]
- 21.Gerhard P. La Frontera Norte de la Nueva España. Mexico City: Universidad Nacional Autónoma de México; 1996. in Spanish. [Google Scholar]
- 22.Buentello-Malo L, Penaloza-Espinosa RI, Loeza F, Salamanca-Gomez F, Cerda-Flores RM. Genetic structure of seven Mexican indigenous populations based on five polymarker loci. Am J Hum Biol. 2003;15:23–28. doi: 10.1002/ajhb.10116. [DOI] [PubMed] [Google Scholar]
- 23.Cerda-Flores RM, et al. Gene diversity and estimation of genetic admixture among Mexican-Americans of Starr County, Texas. Ann Hum Biol. 1992;19:347–360. doi: 10.1080/03014469200002222. [DOI] [PubMed] [Google Scholar]
- 24.Cerda-Flores RM, et al. Genetic admixture in three Mexican Mestizo populations based on D1S80 and HLA-DQA1 loci. Am J Hum Biol. 2002;14:257–263. doi: 10.1002/ajhb.10020. [DOI] [PubMed] [Google Scholar]
- 25.De Leo C, et al. HLA class I and class II alleles and haplotypes in Mexican Mestizos established from serological typing of 50 families. Hum Biol. 1997;69:809–818. [PubMed] [Google Scholar]
- 26.Gorodezky C, et al. The genetic structure of Mexican Mestizos of different locations: Tracking back their origins through MHC genes, blood group systems, and microsatellites. Hum Immunol. 2001;62:979–991. doi: 10.1016/s0198-8859(01)00296-8. [DOI] [PubMed] [Google Scholar]
- 27.Lisker R, et al. Gene frequencies and admixture estimates in a Mexico City population. Am J Phys Anthropol. 1986;71:203–207. doi: 10.1002/ajpa.1330710207. [DOI] [PubMed] [Google Scholar]
- 28.Lisker R, Ramirez E, Briceno RP, Granados J, Babinsky V. Gene frequencies and admixture estimates in four Mexican urban centers. Hum Biol. 1990;62:791–801. [PubMed] [Google Scholar]
- 29.Martinez-Marignac VL, et al. Admixture in Mexico City: Implications for admixture mapping of type 2 diabetes genetic risk factors. Hum Genet. 2007;120:807–819. doi: 10.1007/s00439-006-0273-3. [DOI] [PubMed] [Google Scholar]
- 30.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 31.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangel-Villalobos H, et al. Genetic admixture, relatedness, and structure patterns among Mexican populations revealed by the Y-chromosome. Am J Phys Anthropol. 2008;135:448–461. doi: 10.1002/ajpa.20765. [DOI] [PubMed] [Google Scholar]
- 39.Aguirre-Beltran G, editor. La Población Negra de México: Estudio Etnográfico. Mexico City: Fondo de Cultura Economica; 1972. in Spanish. [Google Scholar]
- 40.Matsuzaki H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 41.Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 42.Cossrow N, Falkner B. Race/ethnic issues in obesity and obesity-related comorbidities. J Clin Endocrinol Metab. 2004;89:2590–2594. doi: 10.1210/jc.2004-0339. [DOI] [PubMed] [Google Scholar]
- 43.Everhart JE, et al. Prevalence of gallbladder disease in American Indian populations: Findings from the Strong Heart Study. Hepatology. 2002;35:1507–1512. doi: 10.1053/jhep.2002.33336. [DOI] [PubMed] [Google Scholar]
- 44.Hamman RF, et al. Methods and prevalence of non-insulin-dependent diabetes mellitus in a biethnic Colorado population. The San Luis Valley Diabetes Study. Am J Epidemiol. 1989;129:295–311. doi: 10.1093/oxfordjournals.aje.a115134. [DOI] [PubMed] [Google Scholar]
- 45.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonnen PE, et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38:214–217. doi: 10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- 48.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.