Abstract
Targeted therapy has vastly improved outcomes in certain types of cancer. Extension of this paradigm across a broad spectrum of malignancies will require an efficient method to determine the molecular vulnerabilities of cancerous cells. Improvements in sequencing technology will soon enable high-throughput sequencing of entire genomes of cancer patients; however, determining the relevance of identified sequence variants will require complementary functional analyses. Here, we report an RNAi-assisted protein target identification (RAPID) technology that individually assesses targeting of each member of the tyrosine kinase gene family. We demonstrate that RAPID screening of primary leukemia cells from 30 patients identifies targets that are critical to survival of the malignant cells from 10 of these individuals. We identify known, activating mutations in JAK2 and K-RAS, as well as patient-specific sensitivity to down-regulation of FLT1, CSF1R, PDGFR, ROR1, EPHA4/5, JAK1/3, LMTK3, LYN, FYN, PTK2B, and N-RAS. We also describe a previously undescribed, somatic, activating mutation in the thrombopoietin receptor that is sensitive to down-stream pharmacologic inhibition. Hence, the RAPID technique can quickly identify molecular vulnerabilities in malignant cells. Combination of this technique with whole-genome sequencing will represent an ideal tool for oncogenic target identification such that specific therapies can be matched with individual patients.
Keywords: acute lymphoblastic leukemia, acute myeloid leukemia, chronic myelomonocytic leukemia, molecular diagnosis, personalized medicine
Cancer therapy that is targeted to causative oncogenes has achieved superior outcomes than conventional approaches (1–4). Broad application of this strategy will require a fast diagnosis of the molecular targets involved in cancer pathogenesis in each patient. Tyrosine kinases constitute a gene family that is widely implicated in cancer pathogenesis (5). Hence, a rapid but comprehensive functional screen that identifies specific tyrosine kinases required for survival of malignant cells will be a useful tool for both research and diagnostic purposes.
Aberrant regulation of tyrosine kinases has been found in numerous hematologic malignancies. Chronic myeloid leukemia (CML) is caused by the 9;22 chromosomal translocation, resulting in the BCR-ABL fusion gene in which BCR-ABL deregulates the ABL tyrosine kinase (6). Chronic myelomonocytic leukemia (CMML) has been shown to contain activating mutations in the tyrosine kinases, c-KIT, JAK2, PDGFR, fibroblast growth factor receptor 1 (FGFR1), and colony stimulating factor (CSF)1R in 15–30% of patients, collectively (7–10). One of these mutant alleles (JAK2V617F) also contributes to the pathogenesis of myeloproliferative disorders, including polycythemia vera, primary myelofibrosis, and essential thrombocythemia (11–14). Most acute myeloid leukemia (AML) blast cells exhibit phosphorylation of STAT5, a marker for tyrosine kinase activity (15). Genetic anomalies are known to occur in Fms-related tyrosine kinase (FLT)3, KIT, PDGFR, JAK1, JAK2, and JAK3 in only 35% of AML cases, suggesting that unidentified mechanisms of kinase dysregulation may be operational in the remainder (8, 16–20). Last, several of these same genes, such as BCR-ABL and FLT3, have been implicated in acute lymphoblastic leukemia (ALL). However, the majority of ALL cases remain without explanation of genetic etiology (21, 22). Thus, tyrosine kinases have been shown to have a role in the pathogenesis of numerous hematologic malignancies, and there remain many cases that exhibit abnormal tyrosine kinase activity because of unexplained mechanisms.
In addition to direct mutation of tyrosine kinases, it is also possible to observe mutations in upstream signaling molecules. Recent evidence for this phenomenon has come in the form of mutations in the thrombopoietin receptor, myeloproliferative leukemia virus oncogene (MPL). Initially, a substitution of serine to asparagine at position 505 (MPLS505N) was discovered in patients with familial thrombocytosis (23). Subsequently, a different mutation was found, MPLW515L, that activates wild-type JAK2 in a subset of primary myelofibrosis and essential thrombocythemia cases (24). Also, mutations can occur in phosphatases (e.g., protein tyrosine phosphatase, nonreceptor type 11) that exhibit negative control of tyrosine kinase activation (25). Last, alternative mechanisms such as autocrine signaling loops, activation of downstream signaling cascades, or improper gene splicing can be postulated based on evidence from other malignancies (26–28).
Thus, to better understand the role of aberrant tyrosine kinase signaling in hematologic malignancies, we developed an RNAi-assisted protein target identification (RAPID) technology and applied it to primary patient cells. We previously used this siRNA screen to construct functional profiles of AML cell lines (29). Here, we have expanded this technology to construct RAPID functional profiles of primary samples from patients with hematologic malignancies. After profiling 30 patient samples, we report that ≈1 of 3 exhibits sensitivity to inhibition of 1 or more tyrosine kinases. Using this assay, we have successfully identified known JAK2 (V617F) and K-RAS (G13D) mutations. We have also uncovered disease-specific sensitivity to inhibition of FLT1, CSF1R, PDGFR, receptor tyrosine kinase-like orphan (RO)R1, ephrin type-A receptor (EPHA)4/5, JAK1/3, lemur tyrosine kinase (LMTK)3, LYN, FYN, protein tyrosine kinase (PTK)2B, and N-RAS. Last, we describe the use of RAPID profiling in the discovery of a previously undescribed, activating, insertional mutation in MPL that lies upstream of the tyrosine kinase, JAK2. Broad application of the RAPID functional screen may offer the potential for personalized diagnosis of molecular cancer targets.
Results
RAPID Functional Screening of Patients with Hematologic Malignancies.
To assess the contribution of tyrosine kinases in hematologic malignancies, we performed RAPID screening on 30 patients and 4 normal individuals (Fig. S1). All members of the tyrosine kinase family, as well as N-RAS and K-RAS, were individually silenced with siRNA, and cell viability was evaluated after 4 days in culture. Quality control experiments showed that cells from each type of leukemia tolerated the experimental procedures, and that both individual, as well as pooled siRNAs, exhibited efficient target reduction in primary leukemia cells (Fig. S2). Of the 30 samples, 10 showed sensitivity to individual gene silencing, with 9 showing sensitivity to silencing of 1 or more tyrosine kinases, and 2 showing sensitivity to N-RAS or K-RAS (Table 1). In particular, we detected sensitivity to targeting of JAK2, as well as LYN, EPHA4, and LMTK3, in a patient with JAK2V617F-positive chronic neutrophilic leukemia (CNL). Also, we observed sensitivity to K-RAS inhibition in a patient with ALL. Subsequent sequence analysis revealed the presence of the transforming allele, K-RASG13D. Other targets identified in patients with ALL were ROR1, N-RAS, and CSF1R. Interestingly, overexpression of ROR1 has been observed in several studies of lymphocytic malignancies (30, 31). Also, we identified JAK2 and EPHA5 as targets in patients with CMML, and FLT1, EPHA4, PDGFRα/β, PTK2B, FYN, and JAK1/3 in patients with AML. We reasoned that positive results occurring in subjects without identification of kinase mutations were indicative of oncogenic mutations in upstream signaling proteins. To test this hypothesis, we undertook mechanistic investigation of an individual patient whose malignancy was predicted to have JAK2 dependence by RAPID analysis, but did not have a detectable JAK2 mutation.
Table 1.
| Disease/Patient no. | Targets identified | Sequence abnormalities |
|---|---|---|
| All | ||
| 07-278 | K-RAS, CSF1R | K-RAS G13D |
| 07-091 | N-RAS | |
| 07-112 | ROR1 | |
| CMML | ||
| 07-079 | JAK2 | MPL 1886InsGG |
| 07-172 | EPHA5 | |
| CNL | ||
| 07-214* | JAK2, EPHA4, LYN, LMTK3 | JAK2 V617F |
| AML | ||
| 07-008 | FLT1 | |
| 07-020 | EPHA4 | |
| 07-025 | PDGFRα/β, FYN, PTK2B | PDGFRβ V316M |
| 07-078 | JAK1/3 |
*Not run in triplicate because of limited sample.
Detection of JAK2 Dependence in a Case of ASM with Associated CMML.
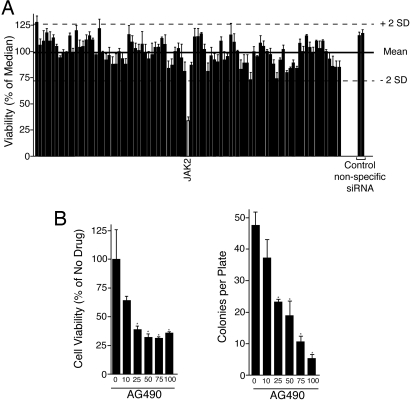
We performed RAPID analysis on peripheral blood cells from a patient with aggressive systemic mastocytosis (ASM) and CMML (patient 07-079). We observed that siRNA targeting JAK2 resulted in significantly reduced viability of the leukemic cells, whereas no other siRNA constructs induced a significant decrease in viability (Fig. 1A; a complete list of viability and statistical values is shown in Table S1). To corroborate this result by an alternative means of target modulation, we assayed whether cells from this patient were sensitive to a selective JAK2 inhibitor, AG490. We performed cell viability assays, as well as colony assays, in the presence of a dose gradient of AG490, and observed significant sensitivity to this JAK2 inhibitor in both settings (Fig. 1B). As noted, sequencing of JAK2 failed to reveal mutations. Recent studies of myeloproliferative disorders have identified an activating mutation in the thrombopoietin receptor, MPLW515L, that signals upstream of JAK2 (24). Therefore, we screened for MPL mutations. Sequence analysis of MPL revealed wild-type sequence at position W515. However, we observed a 2 base pair guanine insertion (hereafter termed MPL1886InsGG) in exon 12 near the end of the ORF (Fig. S3A). This insertional mutation was observed at equal incidence with wild-type sequence in PCR products from genomic DNA and cDNA, indicating heterozygous expression from this locus (Fig. S3A). Also, we sequenced exon 12 of MPL in purified populations of CD3+ or CD33+ cells. Although we observed the insertional mutation in the CD33+ population, we saw no evidence that this mutation occurred in the CD3+ cells, indicating that MPL1886InsGG is a somatic mutation (Fig. S3A). The predicted effect of this 2 base pair insertion is a frameshift that alters the final 6 aa of the coding region and extends the ORF by 46 aa. This insertion produces a higher molecular mass protein with unique C-terminal tail sequence (Fig. S3B). The MPL1886InsGG insertion was not seen in sequence from 75 normal individuals. Of note, genomic DNA from a bone-marrow sample from patient 07-079 was screened for MPL1886InsGG, as well as the mastocytosis-associated allele, KITD816V, by allele-specific quantitative PCR. The MPL1886InsGG mutation was detected, whereas the KITD816V allele was undetectable, suggesting that the MPL1886InsGG allele may have been driving both the ASM and CMML components of the disease. Indeed, thrombopoietin signaling through MPL has been previously shown to support the growth of human mast cells in culture (32).
Fig. 1.
RAPID functional profiling of a patient with ASM with associated CMML. (A) White blood cells were incubated with 1 μM siRNA, individually targeting each member of the tyrosine kinase family. Cells were electroporated and replated into culture media, and cell viability was determined by addition of a tetrazolium salt (MTS assay) at day 4 postelectroporation. Values represent percentage mean (normalized to the median value on the plate) ± SEM (n = 3). (B) Cells from patient 07-079 were treated with a dose curve of AG490. After 3 days, cell viability was measured with an MTS assay (Left). Alternatively, colonies were counted after 10 days (Right). Values represent percentage mean (normalized to untreated controls) ± SEM (n = 3). *, P < 0.05.
Functional Characterization of a Previously Undescribed MPL Mutant Allele.
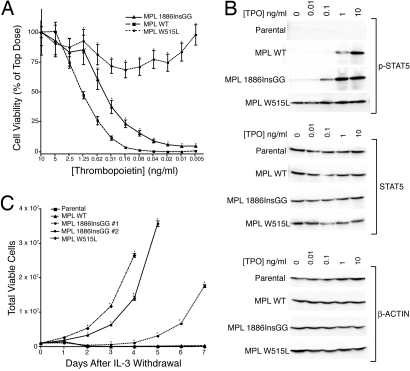
To assess the capacity of MPL1886InsGG for leukemogenesis, we introduced this insertion into the retroviral expression construct, MSCV-IRES-GFP-MPL, and created stable BA/F3 cells expressing both MPLWT and MPLW515L, as well as MPL1886InsGG. To determine the oncogenicity of the 1886InsGG mutation, BA/F3 cells were tested in several ways. First, we conducted proliferation assays in the presence of a dose gradient of thrombopoietin. Results showed increased growth and viability at lower doses of thrombopoietin in MPL1886InsGG compared with MPLWT, indicating hypersensitivity of MPL1886InsGG to ligand (Fig. 2A). Interestingly, MPL1886InsGG conferred an intermediate phenotype compared with the strongly activating MPLW515L allele. We next examined signaling pathways stimulated by MPL in the BA/F3 cells. BA/F3 cells expressing MPLWT exhibited phosphorylation of the downstream signaling molecules JAK2, STAT3, STAT5, ERK, and AKT at high doses of ligand. Consistent with data from proliferation assays, BA/F3 cells expressing MPL1886InsGG exhibited stronger phosphorylation of each of these signaling cascades at ≈10-fold lower doses of thrombopoietin compared with MPLWT (Fig. 2B and Fig. S4). Again, MPL1886InsGG conferred an intermediate phenotype compared with MPLW515L, which showed phosphorylation of signaling pathways even in the absence of ligand. As expected, parental BA/F3 cells that do not express human MPL failed to show any response to thrombopoietin stimulation. Last, deprivation of growth factor showed that 2 independently derived clones of MPL1886InsGG, as well as MPLW515L conferred complete factor-independent growth on BA/F3 cells. MPL1886InsGG exhibited a 1 to 3-day delay in outgrowth compared with MPLW515L, again indicating a more subtle phenotype (Fig. 2C). Immunoblotting of these factor-independent BA/F3 cells, in the absence of ligand stimulation, demonstrated that both MPL1886InsGG and MPLW515L constitutively induce activation of JAK2, STAT5, STAT3, ERK, and AKT. The predicted increase in molecular weight of MPL1886InsGG was confirmed by a shift in electrophoretic mobility of MPL1886InsGG compared with MPLWT or MPLW515L in these BA/F3 cell lysates. Importantly, expression levels of MPLWT and MPL1886InsGG appeared to be equivalent, indicating that this phenomenon is not caused by autoactivation because of elevated expression of MPL in the MPL1886InsGG cell line (Fig. S5). Of note, MPLW515L does appear to have increased expression compared with MPLWT and MPL1886InsGG (Fig. S5B). Also, although both MPL1886InsGG and MPLW515L appear to constitutively activate the same signaling pathways, there are dramatic differences in the relative levels of activation of JAK2, STAT3, and STAT5 by these 2 mutant MPL proteins. There are several possible explanations for this observation, including the divergent expression levels of the 2 transgenes or different mechanistic causes of constitutive activity between MPL1886InsGG and MPLW515L. Last, transformed BA/F3 MPL1886InsGG cells were sensitive to individual and pooled siRNA knockdown of JAK2 in a manner consistent with the observed protein silencing pattern observed in primary CMML cells (Fig. S2B).
Fig. 2.
MPL1886InsGG is hypersensitive to thrombopoietin and transforms BA/F3 cells. (A) BA/F3 cells were plated in a dose gradient of thrombopoietin. After 3 days, cell proliferation was assessed by using an MTS assay. Values represent percentage mean (normalized to 10 ng/mL TPO wells) ± SEM (n = 6). *, P < 0.05. (B) BA/F3 cells were stimulated for 15 min with 0–10 ng/mL thrombopoietin. Whole-cell lysates were subjected to immunoblotting with antibodies specific for total (Middle) or phospho-STAT5 (Top), as well as β-actin (Bottom). (C) BA/F3 cells were plated in medium lacking WEHI-conditioned medium, and total viable cells were counted daily for 1 week. Values represent mean viable cells ± SEM (n = 3). *, P < 0.05.
Confirmation That RAPID Results Predict Targeted Therapy Efficacy.
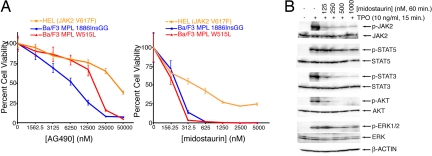
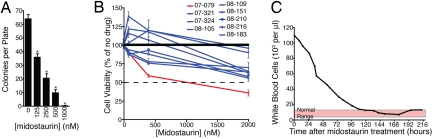
Midostaurin (PKC412) is a derivative of staurosporine, a broad-spectrum tyrosine kinase inhibitor previously shown to bind JAK2 (33). Given the dependence of MPL signaling on JAK2, we determined the sensitivity of factor-independent BA/F3 cells expressing MPL1886InsGG or MPLW515L to AG490 and midostaurin. Both cell lines showed sensitivity to these inhibitors with the IC50 values for midostaurin being ≈200 nM. (Fig. 3A). Also, low doses of midostaurin were sufficient to prevent transformation of BA/F3 MPL1886InsGG cells to factor-independent growth (Fig. S6). Parental BA/F3 cells and those expressing MPLWT or empty vector also depend on JAK2 because of the need for exogenous IL-3 for maintenance of cell viability. Thus, if midostaurin is an effective JAK2 inhibitor, it would also be predicted to block growth of these cells. Consistent with this notion, MPLWT and empty vector BA/F3 cells grown in the presence of IL-3 were also sensitive to midostaurin and AG490 at approximately the same IC50 as the transformed mutant MPL lines (Fig. S7). We also tested the sensitivity of HEL cells, which express the JAK2V617F oncogene, to AG490 and midostaurin (34). Similar to the MPL-dependent BA/F3 cells, HEL cells exhibited sensitivity to both inhibitors with a midostaurin IC50 value of ≈400 nM (Fig. 3A). HEL cells were less sensitive to both drugs than the MPL-expressing BA/F3 cells, likely because of the high dependence BA/F3 cells generally exhibit toward any oncogene conferring factor-independent growth. To further assess the potential of using midostaurin in JAK2-dependent malignancies, we treated BA/F3 MPL1886InsGG cells with midostaurin before stimulation with thrombopoietin. Immunoblot analysis indicated that midostaurin decreased phosphorylation of JAK2 and downstream signaling components in response to thrombopoietin (Fig. 3B). We also tested the in vitro effects of midostaurin on cells from patient 07-079 in a colony assay, and observed a dose-dependent reduction in colony formation (Fig. 4A). To determine whether this effect was a patient-specific drug sensitivity, we compared the effect of midostaurin on cells from patient 07-079, as well as 8 other CMML patients, in a cell viability assay. These additional CMML patients had no known mutations that would induce activation of the JAK2 pathway, and 6 of the 8 were examined by RAPID analysis and found to have no sensitivity to silencing of JAK2. As such, we would predict cells from patient 07-079 to have greater sensitivity to midostaurin as an indication of unique dysregulation of the JAK2 signaling pathway. Indeed, we observed significantly greater sensitivity of cells from patient 07-079 to midostaurin than any other CMML patient. Only cells from patient 07-079 reached an IC50 over the concentration gradient tested (Fig. 4B). Consistent with the in vitro results indicating midostaurin would be effective in JAK2-dependent malignancies such as MPL1886InsGG, investigational treatment of patient 07-079 with midostaurin resulted in normalization of WBC counts over a period of 5 days (Fig. 4C). Thus, the RAPID assay successfully determined an oncogenic lesion and successfully guided targeted therapy for this patient. Because of unrelated medical complications, the durability of this response could not be determined in this patient.
Fig. 3.
Midostaurin is effective against JAK2-dependent cells. (A) HEL cells or BA/F3 cells were treated with a dose gradient of AG490 (Left) or midostaurin (Right). Cell proliferation was determined after 3 days. Values represent percentage mean (normalized to untreated control wells) ± SEM. (n = 3). (B) BA/F3 cells expressing MPL 1886InsGG were incubated for 1 h in 0–1,000 nM midostaurin. Cells were then stimulated for 15 min with thrombopoietin (10 ng/mL), and cell lysates were subjected to immunoblot analysis for phospho or total-JAK2, STAT5, STAT3, AKT, and ERK1/2, as well as β-actin.
Fig. 4.
Midostaurin exhibits patient-specific therapeutic efficacy in patient 07-079. (A) Cells from patient 07-079 were treated with a dose curve of midostaurin. Colonies were counted after 10 days. Values represent mean colonies per plate ± SEM (n = 3). *, P < 0.05. (B) Cells from patient 07-079, as well as 8 other CMML patients, were treated with a dose curve of midostaurin. After 3 days, cell viability was measured with an MTS assay. Values represent percentage mean (normalized to untreated controls) ± SEM (n = 3). (C) Patient 07-079 was treated with midostaurin (50-mg bid), briefly in combination with hydroxyurea, and white blood cell counts were monitored over 9 days. Normal white blood cell counts range from 4 to 12 × 103 per mL.
Discussion
Targeted therapy represents a major step forward in the treatment of cancer. Application of this paradigm to a broad spectrum of malignancies will require fast diagnosis of the molecular “vulnerabilities” of each individual cancer. Here, we demonstrate that RAPID functional screening of primary cells from patients with hematologic malignancies can detect therapeutic targets in 4 days. The screen confirmed a role for tyrosine kinases in oncogenesis of approximately one-third of patients evaluated. Last, as proof-of-principle, we detect JAK2-dependent viability in a case of CMML, elucidate the mechanism underlying this JAK2 dependence (the novel MPL1886InsGG mutation), and demonstrate that a tyrosine kinase inhibitor with activity against JAK2 effectively controlled the disease in this patient. Also, midostaurin is suggested as a potential therapeutic in patients with JAK2V617F and MPLW515L.
MPL1886InsGG was not detected in a screen of DNA from other patients with CMML, as well as AML, indicating it is likely a rare mutation. This point highlights one of the main strengths of the RAPID screen: It detects therapeutic targets regardless of their frequency in patients. The importance of this point is highlighted by several recent large-scale sequencing projects completed without discovery of high-frequency mutations (35–38). One implication of these findings is that many cancer cases may be caused by rare mutations that share common signaling pathways. Indeed, it is possible to postulate that several of the other genes identified in Table 1 may be activated in a similar fashion. For example, an AML patient (07-078) exhibiting sensitivity to JAK1 and JAK3 may have dependence on an upstream cytokine receptor, either because of mutation or autocrine feedback loop. Many of the other targets are, themselves, cell surface receptors. Although most have not exhibited point mutations, there are numerous other mechanistic possibilities for dysregulated activity, including overexpression, mis-splicing, or an autocrine ligand feedback loop. Hence, a functional assay such as the RAPID screen currently represents a faster and more economical approach to identification of molecular vulnerabilities of malignancies compared with a purely genetic, sequencing-based approach.
Although we did uncover the oncogenic mechanism underlying several of the therapeutic targets identified by RAPID screening, other cases remain mechanistically unexplained. This observation represents another benefit of this type of functional screening: Knowledge of the specific genetic abnormality is not a prerequisite for target identification. This point is illustrated by the determination of JAK2-sensitivity in patient 07-079 despite the genetic lesion occurring upstream of JAK2. A detailed understanding of cancer biology will require that the genetic etiology of each case ultimately be unraveled. However, it is likely that activation of tyrosine kinases in cancer may be explained by numerous divergent mechanisms. In some cases, the affected gene will contain a genetic abnormality, whereas other cases will exhibit dependence on tyrosine kinases that are genetically normal. Thus, functional screens may identify target genes beyond those containing genetic abnormalities.
It is likely that the RAPID functional screen does not detect all potential therapeutic targets. This outcome could be because of technical problems such as incomplete target silencing in certain samples. However, these false negatives may also stem from the actual biology of the disease such as a requirement for simultaneous inhibition of multiple targets before impacting cell viability. In this case, it may be necessary to adapt this assay to incorporate simultaneous use of multiple siRNAs or to use the siRNA library in combination with inhibitors against known or common targets. Another potential reason for false-negatives might involve the presence of clonal malignant cells that have undergone differentiation and are no longer responsive to target inhibition. In this case, the specimen will contain a heterogeneous mixture of cells, some of which will depend on a particular oncogene, whereas others may not exhibit that particular oncogene addiction, despite deriving from the same clone. In this case, more advanced purification of cells for specific surface markers before initiation of the RAPID screen may enable detection of targets. Alternatively, flow-cytometry may be incorporated to distinguish effects on different subpopulations of cells within the heterogeneous mixture.
It is also possible that this form of functional screening may detect false-positive results caused by technical aberrations or off-target effects of siRNA. Stringent statistical analyses can help to minimize this limitation. However, it is likely that certain false-positive results will still occur (39). As a result, it will be important to corroborate findings from RAPID functional screening with in vitro small-molecule inhibitor sensitivity studies (Fig. 1B). Also, as a comprehensive database of oncogenic mutations accumulates, DNA sequencing screens for likely genetic abnormalities linked to the identified target may be possible. By combining multiple lines of evidence from these mechanistically divergent means of target identification, it will be possible to minimize false-positive findings.
Last, functional screens such as the RAPID screen will be useful adjuncts to sequencing-based profiling even in the case of known oncogenic mutations identified by sequencing. Numerous examples exist of complex factors mediating sensitivity to target inhibition even with knowledge of an oncogene (40–42). Functional screens, such as RAPID, will predict which of these patients could benefit from targeted therapeutic intervention, and may also lead to new genetic discoveries that will help predict this sensitivity.
The RAPID functional screen represents an advance toward individually tailored cancer therapy. Malignant cells from cancer patients can be economically screened to determine appropriate targeted therapies within a few days. Because the number and availability of selective kinase inhibitors increases, compilation of a large database of oncogenic genetic lesions will be critical in matching the appropriate drugs with the appropriate patients.
Experimental Procedures
All clinical samples were obtained with informed consent with approval by the Institutional Review Boards of Stanford University and Oregon Health and Science University. Patient 07-079 was enrolled on an IRB-approved protocol at Washington University in St. Louis (04-1065; Principal Investigator, Tim Graubert). Blood or bone marrow mononuclear cells were electroporated with 1 μM siRNA (Table S2) at 1110 V (equivalent of 150 V per well), 200 μs, 2 pulses, and 50,000 cells per well were replated into triplicate plates in culture media. Cell viability was determined, and all values were normalized to the median value on the plate. A Student's t test was carried out for each well compared with both single and pooled nonspecific siRNA controls. The mean of the 2-tailed P value was determined for consideration of significance. Data points with value >2 SDs of the mean below the mean value on the plate, and P value <0.05 were considered significant. For additional experimental procedures, see SI Experimental Procedures.
Supplementary Material
Acknowledgments.
This work is supported in part by The Leukemia and Lymphoma Society, the T.J. Martell Foundation, and the Doris Duke Charitable Foundation. J.W.T. is supported by the National Institutes of Health Roadmap for Medical Research, by grants from the National Institutes of Health Cancer Biology Training, and from the William Lawrence and Blanche Hughes Fund, and by the Oregon Clinical and Translational Research Institute Grant UL1 RR024140 from the National Center for Research Resources. M.C.H. is supported in part by a Veterans Affairs Merit Review grant. B.J.D. is an investigator of the Howard Hughes Medical Institute and prinicipal investigator of the Oregon Health & Science University (OHSU) Cancer Center Support Grant 5 p30 CA069533.
Footnotes
Conflict of interest statement: The authors declare a conflict of interest (such as defined by PNAS policy). Oregon Health and Science University (OHSU) and B.J.D. have a financial interest in MolecularMD. Technology used in this research has been licensed to MolecularMD. This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee and the Integrity Program Oversight Council.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903233106/DCSupplemental.
References
- 1.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Smith I, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: A randomised controlled trial. Lancet. 2007;369:29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 5.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 6.Davis RL, Konopka JB, Witte ON. Activation of the c-abl oncogene by viral transduction or chromosomal translocation generates altered c-abl proteins with similar in vitro kinase properties. Mol Cell Biol. 1985;5:204–213. doi: 10.1128/mcb.5.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunby RH, et al. Sensitivity to imatinib but low frequency of the TEL/PDGFRbeta fusion protein in chronic myelomonocytic leukemia. Haematologica. 2003;88:408–415. [PubMed] [Google Scholar]
- 8.Levine RL, et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzo F, et al. Mutational analysis of the KIT gene in myelodysplastic syndrome (MDS) and MDS-derived leukemia. Leuk Res. 2006;30:1235–1239. doi: 10.1016/j.leukres.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Padua RA, et al. RAS, FMS and p53 mutations and poor clinical outcome in myelodysplasias: A 10-year follow-up. Leukemia. 1998;12:887–892. doi: 10.1038/sj.leu.2401044. [DOI] [PubMed] [Google Scholar]
- 11.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 12.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 13.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 14.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Birkenkamp KU, Geugien M, Lemmink HH, Kruijer W, Vellenga E. Regulation of constitutive STAT5 phosphorylation in acute myeloid leukemia blasts. Leukemia. 2001;15:1923–1931. doi: 10.1038/sj.leu.2402317. [DOI] [PubMed] [Google Scholar]
- 16.Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 17.Walters DK, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Xiang Z, et al. Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood. 2008;111:4809–4812. doi: 10.1182/blood-2007-05-090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 20.Yokota S, et al. Internal tandem duplication of the FLT3 gene is preferentially seen in acute myeloid leukemia and myelodysplastic syndrome among various hematological malignancies. A study on a large series of patients and cell lines. Leukemia. 1997;11:1605–1609. doi: 10.1038/sj.leu.2400812. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong SA, et al. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103:3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 22.Clark SS, McLaughlin J, Crist WM, Champlin R, Witte ON. Unique forms of the abl tyrosine kinase distinguish Ph1-positive CML from Ph1-positive ALL. Science. 1987;235:85–88. doi: 10.1126/science.3541203. [DOI] [PubMed] [Google Scholar]
- 23.Ding J, et al. Familial essential thrombocythemia associated with a dominant-positive activating mutation of the c-MPL gene, which encodes for the receptor for thrombopoietin. Blood. 2004;103:4198–4200. doi: 10.1182/blood-2003-10-3471. [DOI] [PubMed] [Google Scholar]
- 24.Pikman Y, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tartaglia M, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 26.Kong-Beltran M, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 27.Nister M, Heldin CH, Wasteson A, Westermark B. A glioma-derived analog to platelet-derived growth factor: Demonstration of receptor competing activity and immunological crossreactivity. Proc Natl Acad Sci USA. 1984;81:926–930. doi: 10.1073/pnas.81.3.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver AM, Silva CM. Modulation of signal transducer and activator of transcription 5b activity in breast cancer cells by mutation of tyrosines within the transactivation domain. Mol Endocrinol. 2006;20:2392–2405. doi: 10.1210/me.2005-0418. [DOI] [PubMed] [Google Scholar]
- 29.Tyner JW, et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008;111:2238–2245. doi: 10.1182/blood-2007-06-097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baskar S, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14:396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 31.Shabani M, et al. Expression profile of orphan receptor tyrosine kinase (ROR1) and Wilms' tumor gene 1 (WT1) in different subsets of B-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2008;49:1360–1367. doi: 10.1080/10428190802124000. [DOI] [PubMed] [Google Scholar]
- 32.Sawai N, et al. Thrombopoietin augments stem cell factor-dependent growth of human mast cells from bone marrow multipotential hematopoietic progenitors. Blood. 1999;93:3703–3712. [PubMed] [Google Scholar]
- 33.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 34.Walters DK, et al. Phosphoproteomic analysis of AML cell lines identifies leukemic oncogenes. Leuk Res. 2006;30:1097–1104. doi: 10.1016/j.leukres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loriaux MM, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 38.Tomasson MH, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone DJ, et al. High-throughput screening by RNA interference: Control of two distinct types of variance. Cell Cycle. 2007;6:898–901. doi: 10.4161/cc.6.8.4184. [DOI] [PubMed] [Google Scholar]
- 40.Doherty L, et al. Pilot study of the combination of EGFR and mTOR inhibitors in recurrent malignant gliomas. Neurology. 2006;67:156–158. doi: 10.1212/01.wnl.0000223844.77636.29. [DOI] [PubMed] [Google Scholar]
- 41.Eberhard DA, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 42.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.