Abstract
Many studies have demonstrated that intracellular proteins, which are involved in carcinogenesis, can provoke autoantibody responses. Therefore, autoantibodies can be used clinically for cancer detection and for proteomic analysis in identification of tumor-associated antigens (TAAs) that are potentially involved in malignant transformation. Liver cancer, especially hepatocellular carcinoma (HCC), is one of the most common tumors in the world. The majority of people with HCC will die within 1 year of its detection. This high case fatality rate can partially be attributed to a lack of diagnostic methods that allow early detection. In the present study, sera from 20 patients with HCC, 30 patients with chronic hepatitis (CH), and 30 patients with liver cirrhosis (LC) as well as sera from 10 normal individuals were used in a proteomic approach to identify HCC-related TAAs. Thirty-four immunoreactive protein spots were excised from the two-dimensional gel electrophoresis (2DE), digested with trypsin, and subsequently analyzed by liquid chromatography-tandem mass spectrometry (LC–MS/MS). Of 34 immunoreactive protein spots, 28 were identified. Seventeen of them were not only reactive with serum antibodies in HCC but also with antibodies in pre-HCC conditions, and 11 were only reactive with serum antibodies in HCC but not with antibodies in pre-HCC conditions. In the subsequent analysis, two representative proteins, HSP60 and HSP70, were selected as examples for the validation purpose. The results from immunoassay were consistent with the data from proteomic analysis, supporting our hypothesis that proteins identified with autoantibodies that have been present in precancer conditions may be not appropriate to use as TAA markers in cancer detection.
Keywords: Hepatocellular carcinoma (HCC), autoantibody, tumor-associated antigen (TAA), biomarker, proteomic approach, cancer detection
Introduction
Liver cancer, especially hepatocellular carcinoma (HCC), is one of the most common malignant tumors worldwide and particularly prevalent in Africa and Asia. Many of the previous studies demonstrated that hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, dietary exposure to aflatoxin, and alcohol drinking were the principal etiological factors for HCC.1-3 Although HCC is not a common type of cancer in the United States, it affects the Hispanic population of this country at a rate double that of the Caucasian population. This statistic has been evident since the early 1970s and continues to hold throughout the diverse Hispanic–American population.4,5 The majority of people with HCC will die within 1 year after its detection. This high case-fatality rate can in part be attributed to lack of diagnostic methods that allow early detection. Although alpha fetoprotein (AFP) is the most effective serological marker available to detect HCC, its sensitivity and specificity are not optimal.6 Therefore, there is a need for the discovery of more sensitive and specific markers that supplement AFP in the early detection of this cancer.
Cancer has long been recognized as a multistep process, which involves not only genetic changes conferring growth advantage but also factors that disrupt regulation of growth and differentiation.7,8 It is possible that some of these factors could be identified and their functions evaluated with the aid of autoantibodies arising during tumorigenesis. Although the mechanisms leading to autoantibody production in cancer patients are not completely understood, emerging evidence indicates that most tumor-associated antigens (TAAs) are cellular proteins whose aberrant regulation of function could be linked to malignancy.9 Several approaches are currently available for the identification of TAAs in cancer. One of the approaches is the utilization of serum antibodies from cancer patients to immunoscreen cDNA expression library to identify TAAs in cancer, and some of these identified TAAs may have potential diagnostic values in cancer diagnosis. Using this approach, several novel TAAs such as p62 and p90 have been isolated in our previous studies.10,11 Another approach involves the use of a proteome-based methodology, which has been recently implemented for the identification of TAAs in cancer.12 Compared to the first approach, which we have previously used, the proteome-based methodology allows individual screening of a large number of sera, as well as determination of a large number of autoantigens. The proteome-based approach can also distinguish isoforms and the detection of autoantibodies directed against post-translational modifications (PTMs) of specific targets. It is well-known that mRNA levels do not necessarily correlate with corresponding protein abundance.13 Additional complexity of protein is conferred by PTMs including phosphorylation, acetylation, and glycosylation, as well as protein cleavage.14 These modifications may not reflect any change at the mRNA level but play important roles in protein stability, activity and functions. Intracellular proteins may also participate in the transformation of a healthy cell into a neoplastic cell. Therefore, protein levels may be more accessible and relevant to therapeutic targets than mRNA levels. Cancer proteomics is aimed at not only the identification of proteins, but also at the study of changes of protein expression, structure and PTMs as well as subcellular localization.15 Some of these changes may be associated with tumorgenesis, and therefore there is a substantial interest in the application of proteomics for the discovery of biomarkers in cancer detection. More recently, several studies have applied proteomic techniques to study differentially expressed proteins and to further identify liver toxicity markers in cell lines and liver tumor tissues.16-18 Also, a few studies have used this approach to identify TAAs in HCC.19-21 The practical utility of this approach remains to be established with the proviso that efforts should be made to identify tumor-associated from tumor-irrelevant antigens.
A feature of HCC is that antecedent liver cirrhosis and chronic hepatitis are common precursor conditions and during transition to malignancy some patients develop autoantibodies, which were not present during the preceding chronic liver disease phase.22-24 The hypothesis is that these antibody responses may be stimulated by cellular proteins that are involved in tumorigenesis. Therefore, these autoantibodies can be used as probes in a proteome-based approach to identify antigens that are potentially involved in malignant transformation. In the present study, the proteomic approach was used to immunoscreen sera from patients with HCC, liver cirrhosis, and chronic hepatitis, as well as sera from normal individuals to identify more HCC-specific TAAs as markers in cancer detection.
Materials and Methods
Serum Samples
In this study, sera from 30 patients with chronic hepatitis (CH), 30 patients with liver cirrhosis (LC), and 20 patients with HCC were derived from the US–Mexico border region of El Paso, Texas. In addition, 24 normal human sera from individuals who had no obvious evidence of malignancy were collected during annual health examinations in William Beaumont Army Medical Center, El Paso, Texas. All HCC patients were diagnosed according to the criteria described in a previous study.25 This study was approved by the Institutional Review Board of the University of Texas at El Paso (IRB-UTEP) and collaborating institutions.
Cell Culture and Cell Extracts
The hepatocellular carcinoma cell line (HepG2) was purchased from American type Culture Collection (ATCC, Manassas, VA) and cultured following the specific protocol as provided. Cells were seeded in 250 mL FALCON tissue culture flask (Becton Dickinson, Franklin Lakes, NJ) and allowed to grow to almost 100% confluence. Then, cells were rinsed once with Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) without fetal bovine serum (FBS) and removed from the flask by incubating them with a solution containing trypsin-EDTA (Gibco, Carlsbad, CA), and harvested in a 15 mL centrifuge tube for further study.
Two-Dimensional Gel Electrophoresis (2DE) Analysis
HepG2 cells were briefly sonicated with lysis buffer (10 mM Tris 5 mM EDTA, 50 mM NaCl, 30 mM Na4P2O7, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100, pH 7.6). Protein concentration was measured with Bradford assay (Bio-Rad, Hercules, CA). For the first dimensional gel electrophoresis (1DE) analysis, a total of 200 μg of protein was mixed with rehydration buffer containing 8 M Urea, 50 mM dithiothreitol (DTT), 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 0.2% carrier ampholytes, a trace bromophenol blue prepared in proteomics grade water and applied on a pH 3–10, 11 cm isoelectric focusing (IEF) strip (Bio-Rad, Hercules, CA). IEF was performed at a current of 50 mA per gel, 300 V for 30 min, followed by 8000 V for 2.5 h, and additional 8000 V for 5 h. Strips were immediately stored at −80 °C for the second dimensional gel electrophoresis analysis. For the second dimensional electrophoresis, 15% SDS-polyacrylamide gels (SDS-PAGE) were used. Proteins were transferred noto nitrocellulose membrane (Osmonics Inc., MA) for Western blotting analysis or stained with 0.1% Coomassie blue R-250 prepared in 45.4% methanol/4.6% acetic acid. The spots were visualized using PDQuest 2-DE analysis software as described in the manufacturer's manual (Bio-Rad, Hercules, CA).
Western Blotting
As described above, proteins were separated by first and second dimensional gels and subsequently transferred onto nitrocellulose membrane (Osmonics Inc., MA). After preblocking with 5% nonfat milk dissolved in phosphate-buffered saline (PBS) containing 0.05% Tween-20 (PBST) for 30 min at room temperature, the nitrocellulose membrane was incubated for 2 h at room temperature with 1:300 dilution of patient's serum. Horseradish peroxidase-conjugated goat antihuman IgG (Caltag Laboratories, San Francisco, CA) was used as secondary antibody (at 1: 3000 dilution) during 2 h of incubation at room temperature. Immunoreactive spots were detected using the enhanced chemiluminescence (ECL) kit (Amersham, Arlington Heights, IL) according to the manufacturer's instructions.
In-Gel Digestion
The proteins of interest were excised from a Coomassie blue-stained preparative gel and then washed with high performance liquid chromatograph (HPLC) grade water, destained with acetonitrile for 15 min to remove Coomassie blue staining, and dried in a vacuum centrifuge (Vacufuge, Eppendorf, Westbury, NY) as described previously.26 After the reduction of disulfide bounds with 5 mM dithiothreitol (DTT) and alkylation of cysteine residues with 10 mM iodoacetamide, digestion was performed by addition of 12.5 ng/μL of sequencing-grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate containing 5 mM CaCl2. Following the enzymatic digestion overnight at 37 °C, the peptides were extracted with 25 mM ammonium bicarbonate in 50% acetonitrile, followed by 5% formic acid in 50% acetonitrile solution.
After removal of acetonitrile in a vacuum centrifuge (Vacufuge, Eppendorf, Westbury, NY), the sample was desalted by C18 bead ziptips (POROS R2, Applied Biosystems, Framingham, MA)27 and dried out by a vacuum centrifuge before mass spectrometry analysis.
Liquid Chromatography—Tandem Mass Spectrometry (LC—MS/MS) Analysis
Samples were resuspended in 0.1% formic acid before the LC—MS/MS analysis. The LC—MS/MS analysis was performed using a nanoHPLC (Ultimate Plus, Dionex LC Packings, Sunnyvale, CA) equipped with a Famos autosampler (Dionex LC Packings, Sunnyvale, CA) and coupled to an ESI—QUAD-TOF-MS (Micromass Qtof-1, Waters, Framing-ham, MA). Peptides were separated in a capillary column (PepMap, C18 3 μm, 75 μm × 15 cm, LC Packings, Waters, Framingham, MA) with a flow rate of 300 nL/min and the following gradient: 100% solvent A for 5 min, 0—50% solvent B in 25 min, 50—90% solvent B in 1 min, and 90% solvent B for 5 min (solvent A, 5% acetonitrile/0.1% formic acid; solvent B, 80% acetonitrile/0.1% formic acid). The mass spectrometry (MS) spectra were collected for 2 s in the400—1800 m/z range. Each peptide with intensity higher than 10 counts was submitted only once to fragmentation, using ramp collision energy (22—60 eV). The MS/MS spectra were collected for 3 s in the 50—2050 m/z range.
Protein Identification
Tandem MS or MS/MS data were converted into peak lists (PKL format) using ProteinLynx 2.0 (Waters, Framingham, MA). MS peaks were smoothed twice with 5 channels using Savitzky-Golay's method, and centered with 4 channels in top 80%. For the MS/MS peaks, a threshold of 5% was set. After smoothing twice with 3 channels using Savitzky-Golay's method, the peaks were centered with 4 channels in the top 80% and converted into monoisotopic ions. Protein identification was performed using Mascot 2.1 software (Matrix Science, London, UK) by searching the peak lists against the International Protein Index (IPI) Human database (version 3.25, 67 250 sequences, http://www.ebi.ac.uk/IPI/IPIhuman.html). The following search parameters were used: trypsin was used as the digesting enzyme (allowing 1 missed cleavage site) and carbamidomethylation of cysteine residues and oxidation of methionine residues were chosen as fixed and variable modifications, respectively. Peptide mass tolerance for monoiso-topic ion was set to 500 ppm, and fragment mass tolerance was set to 0.8 Da. We validated only proteins with p < 0.05. An ion score cutoff of 20 was set to ensure the quality of valid peptides and to remove redundant protein hits. All valid proteins were submitted to a gene ontology (GO) analysis using GOblet tool, which is available at http://goblet.molgen.mpg.de. The analysis was performed by searching the identified protein sequences against the Translated European Molecular Biology Laboratory (TrEMBL) and Swiss-Prot databases, restricted to the human taxonomy. Only GO from proteins with e-value ≤ le−10 for database search was accepted.
Enzyme-Linked Immunosorbent Assay (ELISA)
Purified recombinant HSP60 and HSP70 proteins were commercially purchased (Abcam, Cambridge, MA). The protein purity was >95% by SDS-PAGE. Proteins were diluted in PBS to a final concentration of 0.5 μg/mL for coating polystyrene 96-well microtiter plates (Dynatech Laboratories, Alexandria, VA). A volume of 200 μL human serum samples at 1:200 dilution was added to the antigen-coated wells and incubated for 1.5 h at room temperature. Horseradish peroxidase-conjugated goat antihuman IgG (Caltag Laboratories, San Francisco, CA) at 1:5000 dilution and the substrate 2,2′-azinobis (3-ethylbenzthiazoline-6-sulfonic acid) (Boehringer Mannheim GmbH, Mannheim, Germany) were used as detecting reagents. Each sample was tested in duplicate, and the average OD at 405 nm was used for data analysis. The cutoff value designating positive reaction was the mean optical density (OD) of 24 normal human sera plus 3 standard deviations (SD). The detailed protocol of ELISA was used as described by Rubin.28
Slot Blotting
Slot blotting was performed in exactly the same way as the Western blotting except that the purified recombinant proteins were applied directly onto the nitrocellulose membrane at the concentration of 100 ng per well using a vacuum source. The different proteins were loaded into the different wells so that the immunoreactivity of each serum samples against different antigens could be tested in one blot simultaneously.
Statistical Analysis
To determine whether the frequencies of autoantibody to TAAs in each cohort of patients' sera were significantly higher than that in sera from normal individuals and other controls, the frequencies of antibody were compared using the Chi-squared (χ2) test with Yate's correction, and two significant levels (0.05 and 0.01) were used.
Results and Discussion
Identification of Immunoreactive Proteins in HCC
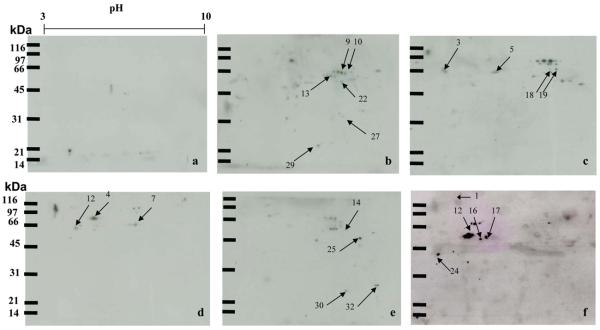
In the present study, a proteome-based approach was applied to identify potential TAAs as markers in HCC. In order to obtain a global view of the proteins in HepG2 cells, proteins extracted from HepG2 cells were separated by 2DE and subsequently transferred onto nitrocellulose membranes. As shown in Figure 1, a 15% SDS-PAGE was used as the second dimensional gel to separate proteins with relative molecular masses of 20—200 kDa. Sera from 20 patients with HCC were individually immunoscreened by Western blotting analysis (Figure 2). Pool of 10 normal human sera was used as control. The results showed that a number of protein spots were visualized on the membranes. By comparing and matching protein spots on each membrane to the equivalent protein spot on the original 2DE gels, it revealed that there were 34 immunoreactive spots which were detected with sera from patients with HCC (Figure 2, b—f) but not with sera from normal individuals (Figure 2, a). In the subsequent study, 34 immunoreactive protein spots were excised from the SDS-PAGE gels, digested with trypsin and further analyzed by LC—MS/MS. The resulting MS/MS spectra were searched with Mascot algorithm against the IPI human database, which is a comprehensive and so far the best annotated database for human protein sequences. As shown in Table 1, 28 of 34 protein spots were identified by LC—MS/MS. Despite repeated efforts, we could not identify 6 spots (spot #6, #8, #11, #15, #31, and #34). Of the 28 identified proteins, the molecular and cellular functions of 27 proteins have been documented in the literature, and several of these proteins were reported relating to cancer in previous studies. For example, the family of heat shock protein (HSP), including HSP60 and HSP70, and other proteins such as lamin A/C have been demonstrated to be associated with different types of cancer.29-31
Figure 1.
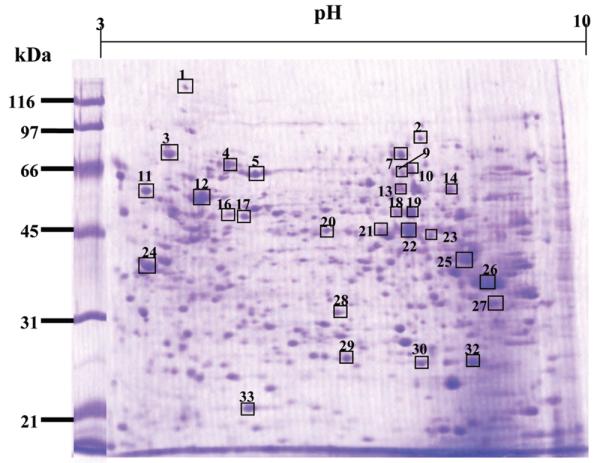
Total proteins from HepG2 cells were analyzed by 2DE. The proteins from HepG2 cells were separated by IEF (pH 3—10) and 15% SDS-PAGE, and subsequently processed with Coomassie blue staining. The MS/MS sequences for the protein spots numbered on the gel are listed in Table 1.
Figure 2.
Immunoreactive proteins were detected by autoantibodies in serum samples from HCC patients. The proteins extracted from the HepG2 cells were separated by 2DE and transferred onto nitrocellulose membranes for immunoblotting analysis. Immunoreactive spots from five representative HCC sera are shown (b—f). The pool of normal human sera (n = 10) was used as control (a), showing few reactive spots.
Table 1.
Proteins Identified by Mass Spectrometry
| spot no. |
accession no. | identified proteins | score | number of peptides |
protein functions |
|---|---|---|---|---|---|
| 1 | IPI00000877 | 150 kDa oxygen-regulated protein (ORP150) |
234 | 6 | Protein folding and secretion, suppression. |
| 2 | IPI00017855 | Aconitate dehydratase | 262 | 9 | Glycometabolism |
| 3 | IPI00003362 | 78 kDa glucose-regulated protein (GRP78) |
634 | 15 | Glycosylation, protein folding, assembly and stress-response |
| 4 | IPI00007765 | Heat shock protein 70 (HSP70) |
488 | 10 | Heat shock protein, stress-response |
| 5 | IPI00022367 | Lamin A/C | 120 | 3 | Cytoskeleton |
| 7 | IPI00022463 | Serotransferrin | 1118 | 22 | Iron binding, transport, and detoxificationates. |
| 9 | IPI00410693 | Splice Isoform 1 of Plasminogen activator inhibitor 1 RNA-binding protein (PAI-1) |
216 | 3 | Plasminogen activation regulator |
| 10 | IPI00027834 | Heterogeneous nuclear ribonucleoprotein L isoform a (hnRNP L) |
135 | 3 | Formation, packaging, processing, and function of mRNA. |
| 12 | IPI00472102 | heat shock protein 60 (HSP60) |
628 | 13 | Heat shock protein, protein folding and assembly. |
| 13 | IPI00015911 | Dihydrolipoyl dehydrogenase (DDH) |
183 | 5 | Glycometabolism. |
| 14 | IPI00220644 | Pyruvate kinase 3 isoform 2 |
103 | 3 | Glycometabolism. |
| 16 | IPI00025252 | Protein disulfide- isomerase A3 |
565 | 12 | Catalyzes the formation and rearrangement of disulphide bond. |
| 17 | IPI00022078 | N-myc downstream-regulated gene-1 protein (NDRG1) |
164 | 2 | Arresting cell growth and prompting cells to differentiate. |
| 18 | IPI00016801 | Glutamate dehydrogenase 1 (GLUD1) |
346 | 9 | Glutamate metabolism |
| 19 | IPI00028888 | Splice Isoform 1 of Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) |
82 | 1 | Regulation of mRNA stability |
| 20 | IPI00299000 | Proliferation-associated 28 protein 2G4 (PA2G4) |
132 | 4 | Cell regulation, differentiation and proliferation |
| 21 | IPI00014424 | Elongation factor 1 (EF-1) | 97 | 6 | Protein biosynthesis |
| 22 | IPI00465248 | Alpha-enolase | 511 | 13 | Glycometabolism. |
| 23 | IPI00296053 | Fumarate hydratase (FH) | 106 | 4 | Glycometabolism |
| 24 | IPI00220740 | Splice isoform 2 of nucleophosmin |
286 | 5 | Nucleolar phosphoprotein, RNA processing. |
| 25 | IPI00465439 | Fructose-bisphosphate aldolase A (ALDOA) |
315 | 9 | Glycometabolism |
| 26 | IPI00219018 | Glyceraldehyde-3-phosphate dehydrogenase, Liver (GAPDH) |
428 | 6 | Glycometabolism |
| 27 | IPI00216308 | 36 Voltage-dependent anion-selective channel protein 1 (VDAC1) |
362 | 8 | Membrane permeability. |
| 28 | IPI00219446 | Phosphatidylethanolamine- binding protein (PEBP) |
66 | 1 | Signal transduction. |
| 29 | IPI00418184 | WUGSC:H_NH0244E06.1 protein |
135 | 3 | Unknown |
| 30 | IPI00022314 | Superoxide dismutase (Mn) (Mn-SOD) |
81 | 1 | Detoxification |
| 32 | IPI00000874 | Peroxiredoxin | 127 | 5 | Detoxification |
| 33 | IPI00218733 | Superoxide dismutase (Cu-Zn) |
164 | 2 | Detoxification |
Functional Categorization and Cancer Association
To understand the function of these identified proteins, the gene ontology (GO) analysis using GOblet algorithm and searching sequences against TrEMBL and Swiss-Prot databases (http://www.expasy.ch/sprot/) was carried out. The 28 identified proteins were categorized in four groups including molecular function (93%, 26/28), biological process (79%, 22/28), physiological process (71%, 20/28), and cellular component (46%, 13/28). Proteins in each group were further categorized in different subgroups. For example, proteins in the group of molecular function were divided into 13 subgroups such as catalytic activity, oxidoreductase activity, nucleotide binding, and so on, and proteins in the group of biological process were divided into only two subgroups of cellular process and metabolism. For further exploring the cancer association of these identified proteins, a literature search was conducted at PubMed (http://www.ncbi.nlm.nih.gov/pubmed/). According to the literature search, 18 of the 28 identified proteins have been reported relating to cancer. For example, 3 proteins, namely, 150 kDa oxygen-regulated protein (ORP150), peroxiredoxin, and proliferation-associated protein 2G4 (PA2G4), were reported relating to breast cancer.32-34 In contrast, 4 proteins, namely, 78 kDa glucose-regulated protein (GRP78), heat shock protein 70 (HSP70), serotransferrin, and superoxide dismutase-Mn (Mn-SOD), were reported to be associated with HCC.35-37 The remaining 11 proteins were reported relating to other cancers such as colon, prostate, lung, and ovarian cancers.31,38-47
Distinction of HCC-Related and Non-HCC-Related Proteins
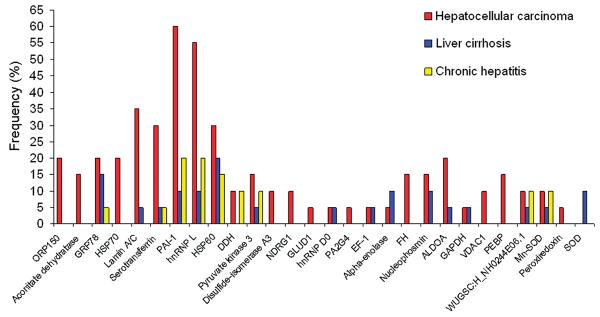
Chronic liver diseases such as chronic hepatitis and liver cirrhosis are clinically considered as early stages of HCC. Patients with chronic liver diseases are most likely to develop HCC after a period of ten or more years. It has been observed that some patients with the transition from chronic liver diseases to HCC may have an appearance of autoantibodies against cellular antigens.22,23 Because some autoantibodies may pre-exist in conditions that are not related to malignancy that arises later, it may be erroneous to include all cellular antigens identified by autoantibodies in cancer sera as TAAs. To distinguish HCC-related and non-HCC-related proteins, the frequency of identified proteins with sera from different conditions were analyzed. As shown in Figure 3, 17 proteins, including GRP78, serotransferrin, heterogeneous nuclear ribonucleoprotein L (hnRNP L), heat shock protein 60 (HSP60), elongation factor 1 (EF-1), α-enolase, and so on, were identified by antibodies in sera from patients with either chronic hepatitis, liver cirrhosis, or HCC, which suggests that antibodies against these proteins may have been already existed in precancer conditions and were not good candidates as cancer biomarkers. Whether or not autoantibodies against this group of proteins can be used as markers of molecular changes associated with early events leading to tumorigenesis remains to be investigated. Interestingly, one of our previous studies clearly demonstrated that antibodies against these two so-called “cancer antigens” elongation factor-1 (EF-1) and α-enolase are not unique to HCC and suggested that its detection in HCC might be attributed to the antibody already being present in the precursor liver diseases such as chronic hepatitis, liver cirrhosis, or primary biliary cirrhosis (PBC).48 Although there have been a steadily increasing number of studies describing and characterizing autoantibodies to “cancer-related antigens” in recent years, many of these antigen–antibody systems are not found to be useful in differentiating cancer from normal condition. This data also emphasizes the importance of a comprehensive analysis of antibody responses to putative cancer antigens in different precancer conditions such as chronic hepatitis and liver cirrhosis before conclusions can be made regarding their contribution to HCC.
Figure 3.
Frequency of immunoreactive proteins identified by autoantibodies in sera from patients with different conditions. Of 28 identified proteins, 17 proteins such as GRP78, sterotransferrin, HSP60, and so on were identified by antibodies in sera from patients with either HCC, liver cirrhosis, or chronic hepatitis, and 11 proteins such as ORP150, aconitate dehydratase, HSP70, and so on were only identified by antibodies in sera from patients with HCC.
In contrast, 11 proteins including ORP150, aconitate dehydratase, HSP70, protein disulfide-isomerase A3, N-myc downstream-regulated gene 1 (NDRG1), glutamate dehydrogenase 1 (GLUD1), PA2G4, fumarate hydratase, voltage-dependent anion-selective channel protein 1 (VDAC1), phosphatidylethanolamine-binding protein (PEBP), and peroxiredoxin were only identified with antibodies in sera from patients with HCC, indicating that this group of proteins may have great potential as cancer biomarkers. For example, ORP150 is a novel endoplasmic reticulum-associated polypeptide in the HSP family.32 This protein has been shown to be up-regulated in tumors, especially in breast tumors, and it is reported to be involved in tumor invasiveness.29 HSP70 plays a central role in the processing of cytosolic and secretory proteins, and it is involved in cell proliferation, differentiation, and tumorigenesis.49 A long line of evidence has proved that HSP70 is a cancer-relevant survival protein. The expression of HSP70 correlates with increased cell proliferation, poor differentiation, lymph node metastases, and poor therapeutic outcome in human breast cancer.50-52 Cancer cells depleted of HSP70 can display strikingly different morphologies (detached and round vs flat senescent-like), cell cycle distributions (G2/M vs G1 arrest), and gene expression profiles.29 More recently, a study showed that autoantibody against HSP70 might be used as diagnostic marker in esophageal carcinoma.53 The PA2G4 family of proteins was reported to be implicated in regulation of cell growth and differentiation.54 One of members in PA2G4 protein family, the ErbB-3 receptor binding protein (Ebp1) was known to interact with a number of proteins and RNAs involved in either transcription regulation or translation control.55 Several studies demonstrated that Ebp1 could be a potent tumor suppressor in breast and prostate adenocarcinomas.56,57 More recently, one study showed that ectopic expression of Ebp1 could mediate multiple antitumor activities against adenoid cystic carcinoma cells.34 Another study also indicated that Ebp1 was capable of eliciting CD8-mediated responses in vivo and in vitro, and it might be used as a possible target for anticancer immunotherapy.58 Peroxiredoxin is found in cytoplasmic and lysosomal organelles, and its function is to protect cells from reactive oxygen injury. Overexpression of peroxiredoxin in breast cancer tissues is suggested that this protein has a proliferative effect and may be related to cancer development or progression.33 PEBP has been known as Raf kinase inhibitor protein (RKIP) and contributed to many different physiologic activities including reproduction and neurophysiology.45 RKIP has been shown to contribute to several anticancer activities including induction of apoptosis and inhibition of metastasis.45 Of interesting notion was that four proteins, namely, HSP70, glyceraldehyde-3-phosphate dehydrogenase (GADPH), peroxiredoxin, and Mn-SOD, were also identified in HCC by Takashima's study.20 Both Takashima's study and our study have used a similar approach to identify HCC-associated antigens, and the difference was that, in Takashima's study, the tissue proteins were used in 2DE to assess the immunoreactivity with serum autoantibodies in HCC and controls, and in our study, the proteins extracted from HCC cells were used. Taken together, it could be considered that these proteins described above might be used as potential markers for monitoring tumor progression and prognosis as well as for tumor diagnosis. How these proteins can induce humoral immune response in HCC and whether autoantibodies to these proteins can be used as markers for in cancer detection remains to be further investigated.
Evaluation of Two Representative Proteins as Markers in HCC
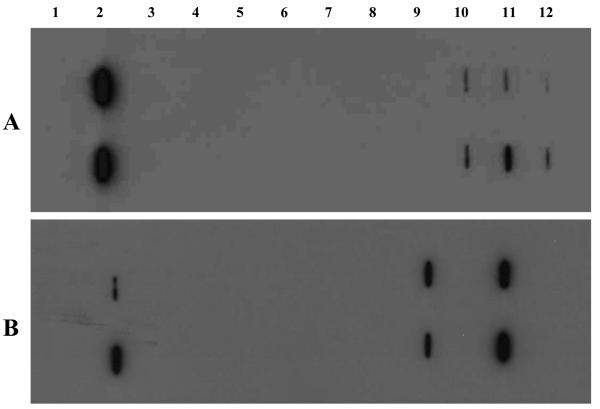
Interestingly, two identified proteins, namely HSP60 and HSP70, have been reported relating to cancer in several studies.29,31,35 In the current study, HSP70 was identified with sera only from patients with HCC, and HSP60 was identified with sera not only from patients with HCC but also with sera from patients with chronic hepatitis and liver cirrhosis. As described above, because some autoantibodies may have already existed in conditions such as chronic hepatitis and liver cirrhosis that are not related to malignancy that arises later, it will be imperative to evaluate whether or not the identified proteins can be used as biomarkers in cancer. HSP60 and HSP70 were selected as two representative examples for the purpose of this evaluation and validation study. These two proteins were commercially purchased and further used as coating antigens in ELISA for the detection of autoantibodies against these two proteins in sera from patients with HCC, liver cirrhosis and chronic hepatitis as well as in sera from normal individuals. Table 2 shows the frequency of antibodies to HSP60 and HSP70 in sera with different conditions. All positive sera were also confirmed by slot blot. A slot blot with several representative sera is shown in Figure 4. The cutoff value designating positive reaction was established as the mean OD of 24 normal human sera (NHS) plus 3 standard deviations (SD). Of the 20 sera with HCC analyzed, 5 (25%) were reactive with HSP70, only 1 (3.3%) sera in liver cirrhosis was positive, and no reactivity was found in chronic hepatitis and normal controls. Statistical analysis indicated that there were significant differences (p < 0.01) among these groups, suggesting that HSP70 may have potential possibility in use as marker in HCC. The result from Takashima's study, which was mentioned above, also demonstrated that the frequency of autoantibodies against HSP70 was 46.7% in HCC, which was significantly higher than that in normal controls (10.0%), indicating that serum anti-HSP70 antibody might be a patient-specific antibody in HCC and used as a candidate diagnostic biomarker for HCC.20 For antibodies to HSP60, 4 (20%) were positive in HCC, 11 (36.7%) in liver cirrhosis, 8 (26.7%) in chronic hepatitis, and there were no statistical differences among these three groups (p > 0.05). These results are consistent with our data above from proteomic analysis, supporting our hypothesis that proteins identified with autoantibodies that have appeared in precancer conditions might not be appropriate to use as TAA markers in cancer detection.
Table 2.
Frequency of Autoantibody Responses to Two Representative Proteins HSP60 and HSP70 in ELISA
| autoantibodies to |
|||
|---|---|---|---|
| liver diseases | number of sera tested |
HSP60 number of positive sera (%)a |
HSP70 number of positive sera (%) |
| HCC | 20 | 4 (20.0)b | 5 (25.0)b |
| liver cirrhosis | 30 | 11 (36.7)b | 1 (3.3) |
| chronic hepatitis | 30 | 8 (26.7)b | 0 (0.0) |
| normal controls | 24 | 1 (3.3) | 0 (0.0) |
Cutoff value, mean + 3 SD of 24 normal controls.
p-values relative to normal controls, p < 0.01.
Figure 4.
Slot blot analysis of representative sera. Each blot represents a duplicate test for autoantibodies against HSP70 (A) and HSP60 (B). Lane 1, phosphate-buffered saline (PBS) as a negative control; lane 2, monoclonal anti-HSP70 or anti-HSP60 antibody as a positive control; lanes 3–4, normal human sera; lanes 5–6, chronic hepatitis sera; lanes 7–8, liver cirrhosis sera; and lanes 9–12, HCC sera. Three HCC sera show reactivity with HSP70 (A, lanes 10–12), and two HCC sera show reactivity with HSP60 (B, lanes 9 and 11).
Conclusions
In the present study, we have applied a proteomic approach to immunoscreen sera from patients with HCC and pre-HCC conditions such as liver cirrhosis and chronic hepatitis as well as sera from normal individuals, and identified 28 HCC-associated tumor antigens. Of 28 identified proteins, seventeen were reactive not only to serum antibodies in HCC but also in pre-HCC conditions, indicating these proteins might not be appropriate TAA markers in HCC detection, and eleven were reactive with serum antibodies in HCC but not in pre-HCC condition, suggesting these proteins could be potential TAA markers in HCC. In the further analysis, two representative proteins, HSP60 and HSP70, were selected as examples for the validation purpose. The results from the immunoassay were consistent with the data from proteomic analysis, supporting our hypothesis that proteins identified with autoantibodies that have appeared in precancer conditions may be not appropriate to use as TAA markers in cancer detection. In cancer, the major task ahead is the continuing identification of TAAs, and the challenging problem is the separation of tumor-associated from nontumor-associated antigens, because autoantibodies to other cellular antigens can be present before appearance of new antibodies occurring with malignancy.59 In our experience, it has been necessary to validate a candidate TAA by testing not only with cancer sera but also with precancer sera when available and with sera from other autoimmune disorders. Our previous studies have demonstrated that a panel of recombinant TAAs could enhance the sensitivity and specificity of autoantibody detection in cancer.60 It will be expected that the further investigation on other identified proteins from this study may help us to define more proteins as candidate TAAs for the formulation of a more HCC-specific TAA array in immunodiagnosis of HCC. A mini-array protein chip of multiple TAAs can be also developed and evaluated as a novel noninvasive approach for early detection of HCC. The molecular identification and characterization of TAAs in HCC will also contribute to our understanding of their role in malignant transformation of the liver, thereby providing attractive candidates for early diagnosis and targeted therapies.
Acknowledgment
We thank Drs. Thomas M. Wertin and Daniel Kim at William Beaumont Army Medical Center, El Paso, Texas, for providing serum samples that were used in this study as well as Dr. Fabio Gozzo form National Laboratory of Synchrotron Light, Campinas, Brazil for open the access to his Mascot server. We also thank the Biomolecule Analysis Core Facility (BCAF) at University of Texas at El Paso, supported by NIH grant number 5G12RR008124, for LC–MS equipment access. This work was supported by NIH grants 2S06GM008012-37 and 5G12RR008124, as well as a grant from Paso del Norte Health Foundation.
References
- 1.Lopez LJ, Marrero JA. Hepatocellular carcinoma. Curr. Opin. Gastroenterol. 2004;20:248–253. doi: 10.1097/00001574-200405000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Ogunbiyi JO. Hepatocellular carcinoma in the developing world. Semin. Oncol. 2001;28:179–187. doi: 10.1016/s0093-7754(01)90090-9. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Trapido EJ, Buciaga Valdez R, Obeso JL, Strickman-Stein N, Rotger A, Pérez-Stable EJ. Epidemiology of cancer among Hispanics in the United States. J. Natl. Cancer Inst. Monogr. 1995;18:17–28. [PubMed] [Google Scholar]
- 5.Cooper SP, Sigurdson A, Labarthe D, Whitehead L, Downs T, Burau K, Vernon SW, Spitz M, New B. Assessing the burden of cancer in Texas using vital statistics data. South Med. J. 1998;91:173–181. doi: 10.1097/00007611-199802000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alphafetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127:S108–112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 8.Sarasin A. An overview of the mechanisms of mutagenesis and carcinogenesis. Mutat. Res. 2003;544:99–106. doi: 10.1016/j.mrrev.2003.06.024. [DOI] [PubMed] [Google Scholar]
- 9.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J. Clin. Invest. 2001;108:1411–1415. doi: 10.1172/JCI14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang JY, Chan EK, Peng XX, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J. Exp. Med. 1999;189:1101–1110. doi: 10.1084/jem.189.7.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soo Hoo L, Zhang JY, Chan EK. Cloning and characterization of a novel 90 kDa ‘companion’ auto-antigen of p62 overexpressed in cancer. Oncogene. 2002;21:5006–5015. doi: 10.1038/sj.onc.1205625. [DOI] [PubMed] [Google Scholar]
- 12.Casiano CA, Mediavilla-Varela M, Tan EM. Tumor-associated antigen arrays for the serological diagnosis of cancer. Mol. Cell. Proteomics. 2006;5:1745–1759. doi: 10.1074/mcp.R600010-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginestier C, Charafe-Jauffret E, Bertucci F, Eisinger F, Geneix J, Bechlian D, Conte N, Adelaide J, Toiron Y, Nguyen C, Viens P, Mozziconacci MJ, Houlgatte R, Birnbaum D, Jacquemier J. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am. J. Pathol. 2002;161:1223–1233. doi: 10.1016/S0002-9440(10)64399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 15.Chignard N, Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127:S120–125. doi: 10.1053/j.gastro.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Lim SO, Park SJ, Kim W, Park SG, Kim HJ, Kim YI, Sohn TS, Noh JH, Jung G. Proteome analysis of hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2002;291:1031–1037. doi: 10.1006/bbrc.2002.6547. [DOI] [PubMed] [Google Scholar]
- 17.Park KS, Cho SY, Kim H, Paik YK. Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int. J. Cancer. 2002;97:261–5. doi: 10.1002/ijc.1585. [DOI] [PubMed] [Google Scholar]
- 18.Thome-Kromer B, Bonk I, Klatt M, Nebrich G, Taufmann M, Bryant S, Wacker U, Kopke A. Toward the identification of liver toxicity markers: a proteome study in human cell culture and rats. Proteomics. 2003;3:1835–1862. doi: 10.1002/pmic.200300552. [DOI] [PubMed] [Google Scholar]
- 19.Naour F, B F, Misek DE, Brechot C, Hanash SM, Beretta L. A distinct repertoire of autoantibodies in hepatocellular carcinoma identified by proteomic analysis. Mol. Cell. Proteomics. 2002;1:197–203. doi: 10.1074/mcp.m100029-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Takashima M, Kuramitsu Y, Yokoyama Y, Iizuka N, Harada T, Fujimoto M, Sakaida I, Okita K, Oka M, Nakamura K. Proteomic analysis of autoantibodies in patients with hepatocellular carcinoma. Proteomics. 2006;6:3894–3900. doi: 10.1002/pmic.200500346. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Chen SH, Yu CH, Li YM, Wang SQ. Identification of hepatocellular-carcinoma-associated antigens and autoantibodies by serological proteome analysis combined with protein microarray. J. Proteome Res. 2008;7:611–620. doi: 10.1021/pr070525r. [DOI] [PubMed] [Google Scholar]
- 22.Zhang JY, Zhu W, Imai H, Kiyosawa K, Chan EK, Tan EM. De-novo humoral immune responses to cancer-associated autoantigens during transition from chronic liver disease to hepatocellular carcinoma. Clin. Exp. Immunol. 2001;125:3–9. doi: 10.1046/j.1365-2249.2001.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai H, Nakano Y, Kiyosawa K, Tan EM. Increasing titers and changing specificities of antinuclear antibodies in patients with chronic liver disease who develop hepatocellular carcinoma. Cancer. 1993;71:26–35. doi: 10.1002/1097-0142(19930101)71:1<26::aid-cncr2820710106>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JY, Wang X, Peng XX, Chan EK. Autoantibody responses in Chinese hepatocellular carcinoma. J. Clin. Immunol. 2002;22:98–105. doi: 10.1023/a:1014483803483. [DOI] [PubMed] [Google Scholar]
- 25.Johnson PJ, Leung N, Cheng P, Welby C, Leung WT, Lau WY, Yu S, Ho S. ‘Hepatoma-specific’ alphafetoprotein may permit preclinical diagnosis of malignant change in patients with chronic liver disease. Br. J. Cancer. 1997;75:236–240. doi: 10.1038/bjc.1997.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 27.Jurado JD, Rael ED, Lieb CS, Nakayasu E, Hayes WK, Bush SP, Ross JA. Complement inactivating proteins and intraspecies venom variation in Crotalus oreganus helleri. Toxicon. 2007;49:339–350. doi: 10.1016/j.toxicon.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Rubin RL. Enzyme-linked immunosorbent assays for antibodies to native DNA, histones, and (H2A-H2B)-DNA. In: Rose NR, de Macario EC, Folds JD, Lane HC, Nakamura RM, editors. Manual of Clinical Laboratory Immunology. 5th edition American Society for Microbiology; Washington DC: 1997. pp. 935–941. [Google Scholar]
- 29.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider J, Jiménez E, Marenbach K, Romero H, Marx D, Meden H. Immunohistochemical detection of HSP60-expression in human ovarian cancer. Correlation with survival in a series of 247 patients. Anticancer Res. 1999;19:2141–2146. [PubMed] [Google Scholar]
- 31.Gaedtke L, Thoenes L, Culmsee C, Mayer B, Wagner E. Proteomic analysis reveals differences in protein expression in spheroid versus monolayer cultures of low-passage colon carcinoma cells. J. Proteome Res. 2007;6:4111–4118. doi: 10.1021/pr0700596. [DOI] [PubMed] [Google Scholar]
- 32.Tsukamoto Y, Kuwabara K, Hirota S, Kawano K, Yoshikawa K, Ozawa K, Kobayashi T, Yanagi H, Stern DM, Tohyama M, Kitamura Y, Ogawa S. Expression of the 150-kd oxygen-regulated protein in human breast cancer. Lab. Invest. 1998;78:699–706. [PubMed] [Google Scholar]
- 33.Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21:2085–2090. [PubMed] [Google Scholar]
- 34.Yu Y, Chen W, Zhang Y, Hamburger AW, Pan H, Zhang Z. Suppression of salivary adenoid cystic carcinoma growth and metastasis by ErbB3 binding protein Ebp1 gene transfer. Int. J. Cancer. 2007;120:1909–1913. doi: 10.1002/ijc.22541. [DOI] [PubMed] [Google Scholar]
- 35.Lim SO, Park SG, Yoo JH, Park YM, Kim HJ, Jang KT, Cho JW, Yoo BC, Jung GH, Park CK. Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules. World J. Gastroenterol. 2005;11:2072–2079. doi: 10.3748/wjg.v11.i14.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellerbrand C, Poppl A, Hartmann A, Scholmerich J, Lock G. HFE C282Y heterozygosity in hepatocellular carcinoma: evidence for an increased prevalence. Clin. Gastroenterol. Hepatol. 2003;1:279–284. doi: 10.1016/s1542-3565(03)00132-0. [DOI] [PubMed] [Google Scholar]
- 37.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi F, Kusaba H, Asai A, Iwamoto Y, Yano K, Nakano H, Mizukami T, Saijo N, Kato H, Nishio K. hnRNP L enhances sensitivity of the cells to KW-2189. Int. J. Cancer. 2004;108:679–685. doi: 10.1002/ijc.11616. [DOI] [PubMed] [Google Scholar]
- 39.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 40.Huang LJ, Chen SX, Huang Y, Luo WJ, Jiang HH, Hu QH, Zhang PF, Yi H. Proteomics-based identification of secreted protein dihydrodiol dehydrogenase as a novel serum markers of non-small cell lung cancer. Lung Cancer. 2006;54:87–94. doi: 10.1016/j.lungcan.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, Lee JM. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 42.Koensgen D, Mustea A, Dahl E, Klaman I, Petschke B, Sun P, Lichtenegger W. Expression of the Plasminogen Activator Inhibitor- 1 RNA Binding Protein (PAI-RBP1) in epithelial ovarian cancer (OC) J. Clin. Oncol., ASCO Annual Meeting Proceedings. 2005;23:9628. [Google Scholar]
- 43.Bonafe N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, Rosen DG, Zhou Y, Feng L, Yang G, Liu J, Huang P. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: role in cell proliferation and response to oxidative stress. J. Biol. Chem. 2005;280:39485–39492. doi: 10.1074/jbc.M503296200. [DOI] [PubMed] [Google Scholar]
- 45.Keller ET, Fu Z, Brennan M. The biology of a prostate cancer metastasis suppressor protein: Raf kinase inhibitor protein. J. Cell Biochem. 2005;94:273–278. doi: 10.1002/jcb.20169. [DOI] [PubMed] [Google Scholar]
- 46.Caruso RP, Levinson B, Melamed J, Wieczorek R, Taneja S, Polsky D, Chang C, Zeleniuch-Jacquotte A, Salnikow K, Yee H, Costa M, Osman I. Altered N-myc downstream-regulated gene 1 protein expression in African-American compared with caucasian prostate cancer patients. Clin. Cancer Res. 2004;10:222–227. doi: 10.1158/1078-0432.ccr-0604-3. [DOI] [PubMed] [Google Scholar]
- 47.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 48.Tan EM, Zhang JY, Chan EK. Autoantibodies to insulin-like growth factor II mRNA-binding proteins in hepatocellular carcinoma. In: Manns MP, Paumgartner G, Leuschner U, editors. Immunology and Liver. Kluwer Academic Publishers; Lancaster, UK: 2000. pp. 8–15. [Google Scholar]
- 49.Cecconi D, Astner H, Donadelli M, Palmieri M, Missiaglia E, Hamdan M, Scarpa A, Righetti PG. Proteomic analysis of pancreatic ductal carcinoma cells treated with 5-aza-2′-deoxycytidine. Electrophoresis. 2003;24:4291–4303. doi: 10.1002/elps.200305724. [DOI] [PubMed] [Google Scholar]
- 50.Ciocca DR, Clark GM, Tandon AK, Fuqua SA, Welch WJ, McGuire WL. Heat shock protein hsp70 in patients with axillary lymph node-negative breast cancer: prognostic implications. J. Natl. Cancer Inst. 1993;85:570–574. doi: 10.1093/jnci/85.7.570. [DOI] [PubMed] [Google Scholar]
- 51.Lazaris A, Chatzigianni EB, Panoussopoulos D, Tzimas GN, Davaris PS, Golematis B. Proliferating cell nuclear antigen and heat shock protein 70 immunolocalization in invasive ductal breast cancer not otherwise specified. Breast Cancer Res. Treat. 1997;43:43–51. doi: 10.1023/a:1005706110275. [DOI] [PubMed] [Google Scholar]
- 52.Vargas-Roig LM, Gago FE, Tello O, Aznar JC, Ciocca DR. Heat shock protein expression and drug resistance in breast cancer patients treated with induction chemotherapy. Int. J. Cancer. 1998;79:468–475. doi: 10.1002/(sici)1097-0215(19981023)79:5<468::aid-ijc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 53.Fujita Y, Nakanishi T, Miyamoto Y, Hiramatsu M, Mabuchi H, Miyamoto A, Shimizu A, Takubo T, Tanigawa N. Proteomics-based identification of autoantibody against heat shock protein 70 as a diagnostic marker in esophageal squamous cell carcinoma. Cancer Lett. 2008;263:280–290. doi: 10.1016/j.canlet.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38–2G4, which varies with the cell cycle. Exp. Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 55.Kowalinski E, Bange G, Bradatsch B, Hurt E, Wild K, Sinning I. The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 2007;581:4450–4454. doi: 10.1016/j.febslet.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 56.Lessor TJ, Yoo JY, Xia X, Woodford N, Hamburger AW. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell Physiol. 2000;183:321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, Goloubeva O, Ross DD, Brodie A, Hamburger AW. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santegoets SJ, Schreurs MW, Reurs AW, Lindenberg JJ, Kueter EW, van den Eertwegh AJ, Hooijberg E, Brandwijk RJ, Hufton SE, Hoogenboom HR, Scheper RJ, Somers VA, de Gruijl TD. Identification and characterization of ErbB-3-binding protein-1 as a target for immunotherapy. J. Immunol. 2007;179:2005–2012. doi: 10.4049/jimmunol.179.3.2005. [DOI] [PubMed] [Google Scholar]
- 59.Tan EM, Zhang J. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol. Rev. 2008;222:328–340. doi: 10.1111/j.1600-065X.2008.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang JY, Megliorino R, Peng XX, Tan EM, Chen Y, Chan EK. Antibody detection using tumor-associated antigen mini-array in immunodiagnosing human hepatocellular carcinoma. J. Hepatol. 2007;46:107–114. doi: 10.1016/j.jhep.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]