Abstract
Since its activity was first reported in the mid-1960s, macrophage migration inhibitory factor (MIF) has gone from a cytokine activity modulating monocyte motility to a pleiotropic regulator of a vast array of cellular and biological processes. Studies in recent years suggest that MIF contributes to malignant disease progression on several different levels. Both circulating and intracellular MIF protein levels are elevated in cancer patients and MIF expression reportedly correlates with stage, metastatic spread and disease-free survival. Additionally, MIF expression positively correlates with angiogenic growth factor expression, microvessel density and tumor-associated neovascularization. Not coincidentally, MIF has recently been shown to contribute to tumoral hypoxic adaptation by promoting hypoxia-induced HIF-1α stabilization. Intriguingly, hypoxia is a strong regulator of MIF expression and secretion, suggesting that hypoxia-induced MIF acts as an amplifying factor for both hypoxia and normoxia-associated angiogenic growth factor expression in human malignancies. Combined, these findings suggest that MIF overexpression contributes to tumoral hypoxic adaptation and, by extension, therapeutic responsiveness and disease prognosis. This review summarizes recent literature on the contributions of MIF to tumor-associated angiogenic growth factor expression, neovascularization and hypoxic adaptation. We also will review recent efforts aimed at identifying and employing small-molecule antagonists of MIF as a novel approach to cancer therapeutics.
Keywords: angiogenesis, HIF, hypoxia, oxygen, proteasome, tumor, ubiquitin ligase
MIF, HIF-1α and Tumor-Associated Angiogenesis
MIF is over-expressed in a large variety of human neoplasms. Pancreatic, breast, prostate, colon, brain, skin and lung-derived tumors have all been shown to contain significantly higher levels of MIF message and protein than their non-cancerous cell counterparts (Bando et al., 2002; Kamimura et al., 2000; Markert et al., 2001; Meyer-Siegler et al., 1998; Shimizu et al., 1999; Takahashi et al., 1998; Winner et al., 2007). Several of these studies also report that MIF expression closely correlates with tumor aggressiveness and metastatic potential, suggesting an important contribution to disease severity and survival by MIF (del Vecchio et al., 2000; Han et al., 2008; Kamimura et al., 2000; Meyer-Siegler et al., 2002; Tomiyasu et al., 2002). Han and colleagues recently reported that immunohistochemical staining for MIF in pre-chemotherapy osteosarcoma biopsy specimens shows a strong and significant correlation between both disease- and metastases-free survival (Han et al., 2008). In a separate study, increased MIF expression in glioblastoma multiforma (GBM) tumor biopsies was found to localize predominantly to necrotic areas of GBM lesions and within tumor cells surrounding blood vessels (Bacher et al., 2003). Because necrotic regions within GBM and other neoplasms are commonly associated with the very low oxygen tensions (Louis, 2006) it is not unreasonable to speculate that MIF expression in GBM is positively regulated by hypoxia (Bacher et al., 2003).
MIF was first linked to tumor hypoxic responses when the Giaccia laboratory reported that MIF mRNA levels are induced by hypoxia in human squamous cell carcinoma cell lines (Koong et al., 2000). Subsequent studies confirmed these findings (Bacher et al., 2003; Baugh et al., 2006; Schmeisser et al., 2005; Takahashi et al., 2001) but it was not until the Giaccia lab demonstrated that loss of MIF phenocopies the loss of HIF-1α in inducing premature senescence that a functional role for hypoxia-induced MIF was first proposed (Welford et al., 2006). A subsequent study by this laboratory revealed that MIF is a direct transcriptional target of HIF-1α and, more importantly, loss of MIF results in inefficient HIF-1α stabilization induced by hypoxia and prolyl hydroxylase inhibitors (Winner et al., 2007). Consistent with these findings, MIF overexpression in human breast cancer cell lines was found to promote hypoxia-induced HIF-1α stabilization (Oda et al., 2008). Intriguingly, this study confirmed unpublished observations from this laboratory showing that the MIF receptor, CD74, is necessary for MIF-dependent HIF-1α stabilization (Oda et al., 2008).
Hypoxia-induced VEGF expression is significantly reduced in MIF-deficient cells and increased in MIF over-expressing cells consistent with its contribution to HIF-1α stabilization (Oda et al., 2008; Winner et al., 2007). Not coincidentally, numerous studies report that MIF intratumoral expression strongly correlates with VEGF expression, tumor vessel density and risk of recurrence after resection (Hagemann et al., 2007; Han et al., 2008; Hira et al., 2005; Ren et al., 2005; Shun et al., 2005; White et al., 2003; Xu et al., 2007). In mouse models, MIF-deficient mice crossed to adenomatous polyposis coli (ApcMin/+) “oncomice” exhibit significant reductions in both the number and size of adenomas that correspond to diminished tumor microvessel density (Wilson et al., 2005). Additionally, MIF-deficient mice show a 45% reduction in chronic ultraviolet B (UVB) irradiation induced epidermal tumorigenesis (Martin et al., 2008). Decreased tumor incidence and delayed tumor outgrowth in MIF-deficient mice exposed to UVB correlated with significantly less VEGF expression and intratumoral microvessel density. Thus, the single most consistent phenotype associated with loss of MIF in tumorigenesis is decreased angiogenic growth factor expression and microvascular density reminiscent of an impaired ability to adapt to hypoxia. While no studies to date have evaluated hypoxia either directly or indirectly with respect to intratumoral MIF, the invariability of this angiogenic phenotype suggests that MIF strongly influences tumoral hypoxic adaptation and associated neovascularization. Because low pO2-mediated induction of HIF-1α serves as more than just a vehicle by which angiogenic growth factors are generated, studies designed to elucidate the relative importance of MIF in hypoxia-induced metastatic spread and chemotherapeutic sensitivity are sorely needed.
Mechanism(s) of Action
Despite the aforementioned plethora of studies linking MIF to intratumoral angiogenesis, none has provided a clear mechanistic link between MIF, VEGF and tumor vascularization in normoxic tissues. In an effort to address this question, we recently reported that MIF, in addition to promoting hypoxia-induced VEGF expression (Winner et al., 2007), is also an important regulator of normoxic VEGF expression (Coleman et al., 2008). Specifically, we discovered that MIF cooperates with its only known homolog, D-dopachrome tautomerase (D-DT), in dictating the steady state expression of VEGF and IL-8 in non-small cell lung cancer (NSCLC) cell lines (Coleman et al., 2008). Angiogenic growth factor expression mediated by endogenous MIF family members was found to rely upon a c-Jun-N-terminal kinase (JNK)/AP-1-dependent signaling pathway. Importantly, MIF and D-DT-mediated activation of JNK leading to AP-1-dependent transcription of VEGF and IL-8 relied upon the presence of the cognate MIF cell surface receptor, CD74 (Coleman et al., 2008; Leng et al., 2003; Shi et al., 2006). Conditioned supernatants from one or both MIF family member siRNA transfected NSCLC cell lines were unable to induce endothelial cell migration or tube formation in vitro (Coleman et al., 2008). This effect could be reversed by adding back recombinant VEGF and/or IL-8 but not rMIF or rD-DT suggesting that decreased VEGF and IL-8 expression is responsible for defective endothelial cell migration and tube formation observed in MIF and/or D-DT-deficient cells.
As discussed above, Oda and colleagues recently recapitulated our findings showing that MIF functionally stabilizes HIF-1α in human cancer cell lines (Oda et al., 2008). Based on their observations that p53 null and p53 mutant cell lines were unresponsive to rMIF-induced HIF-1α stabilization, the authors concluded that MIF-dependent modulation of p53 was responsible for the effects of rMIF on HIF expression. Based on earlier reports that wildtype p53 acts to functionally stabilize HIF-1α in hypoxic and anoxic cells (Ravi et al., 2000; Sanchez-Puig et al., 2005) and coupled with the fact that p53 expression/activity is regulated by MIF (Hudson et al., 1999; Mitchell et al., 2002; Welford et al., 2006), this would seem to be a logical conclusion. However, other studies appear to contradict these findings as the pancreatic ductal adenocarcinoma cancer (PDAC) cell line used in earlier studies showing an important contributing role for MIF in HIF stabilization is p53 mutant (Cogoi et al., 2005; Sipos et al., 2003; Winner et al., 2007). Further studies from this laboratory reveal that several additional human PDAC cell lines that are also p53 mutant are similarly responsive to MIF-dependent HIF-1α stabilization (R.A.M., unpublished observations). One potential explanation that might help to resolve this apparent contradiction is the possibility that p53 mutant tumor suppressor proteins may similarly bind to and facilitate the degradation of HIF-1α. This explanation also could help to resolve why the HIF-1α/COP9 signalosome subunit 5 (CSN5) interaction is destabilized in MIF-deficient cells (Winner et al., 2007). CSN5, an important effector and interacting partner for MIF, also has been shown to functionally associate with p53 thus raising the possibility that MIF may influence CSN5/p53 interactions with HIF-1α and regulating its stability in hypoxic cells.
In order to better understand the contributions of MIF to HIF stability and hypoxic adaptation, it is first important to understand how HIF-1α stability is physiologically regulated by oxygen. HIF is a heterodimeric transcription factor in which both the α- and β-subunits are basic helix-loop-helix PAS (Per-ARNT-Sim) proteins (Kim and Kaelin, Jr., 2003). Isozymes of both HIF-1α and HIF-1β subunits have been identified with HIF-1α and HIF-2α being regulated by oxygen levels (Kim and Kaelin, Jr., 2003). The HIF-β subunit, identical to the aryl hydrocarbon receptor nuclear translocator (ARNT), is a constitutive nuclear protein involved in other transcriptional responses under normoxic conditions. In contrast, protein levels of the HIF-α subunit are virtually undetectable under normoxic conditions in most tissues, thus indicating that oxygen-dependent protein stability is a key mechanism regulating HIF function. Thus, under hypoxic conditions, protein levels of HIF-α rise, allowing HIF-α nuclear translocation, hetero-dimerization, and transcriptional activation.
Many studies have identified multiple molecular events involved in the degradation of HIF-α and have led to the identification of a family of Fe(II) and 2-oxoglutarate (2-OG) dependent dioxygenase enzymes acting as oxygen sensors (Kim and Kaelin, 2003). Under normoxic conditions, HIF-1α undergoes trans-4-hydroxylation at Pro-564 (CODD or C-terminal ODD) and Pro-402 (NODD or N-terminal ODD) which form part of highly conserved LXXLAP motifs in oxygen-dependent degradation domains (ODDs) (Chan et al., 2005; Kim and Kaelin, Jr., 2003). Hydroxylation allows recognition of HIF-1α by the von Hippel-Lindau tumor suppressor protein (pVHL) which serves as the recognition component of the ubiquitin E3 ligase complex consisting of VHL/Elongin C/Elongin B (VCB), Cullin 2, and the RING-H2 finger protein Rbx-1 (Hon et al., 2002). It is important to note that this HIF-degradation complex is distinct from SCF complexes which are made up of Cullin 1 (Cul1), Skp1 and F-box proteins. Importantly, structural analysis of the HIF-CODD and pVHL reveals that all five pVHL residues lining the 4-hydroxyproline-binding pocket are affected by mis-sense mutations in VHL disease (Kim and Kaelin, 2004), suggesting that failure to capture HIF-1α, and/or other hydroxylated targets, is important to the tumor-promoting mechanism associated with VHL disease. Subsequent ubiquitylation of HIF-α by the Cdc34/Ubc5 E2 ubiquitin conjugating complex targets HIF-α for transport to the proteasome and degradation.
CSN5 is a 38 kD protein and an essential component of the COP9 signalosome (CSN) which is composed of eight subunits designated CSN1-CSN8 (for review see Wolf et al., 2003). Until recently the function of the CSN was obscure although it appeared to control proteins that had high turnover rates. Mutational analysis in S. pombe revealed that disruption of CSN1 resulted in the accumulation of neddylated Cullins (Wolf et al., 2003). The conjugation of the small ubiquitin-like protein Nedd8 to Cullins is thought to be required for E2-recruitment and targeted ubiquitylation. CSN5 contains a JAB-1/MPN domain Metalloenzyme Motif (JAMM) that forms the catalytic region of the isopeptidase. In CSN5, the JAMM domain is responsible for the cleavage of Nedd8 from cullins. Cycles of cullin neddylation and de-neddylation are required for Cullin-dependent ubiquitin E3-ligase (Cul-Ub-E3) function. Thus, altering CSN function directly or indirectly has significant effects on the protein stability of Cul-Ub-E3 targets. This directly implicates the CSN in dynamically preventing ubiquitylation of certain proteins and subsequent 26S proteasome dependant degradation.
CSN5 binds both the CODD of HIF-1α and the pVHL tumor suppressor (Bemis et al., 2004). High CSN5 expression generates a pVHL-independent form of CSN5 that stabilizes HIF-1α aerobically by inhibiting HIF-1α prolyl-564 hydroxylation. Aerobic CSN5 association with HIF-1α occurs independently of the CSN holocomplex, leading to HIF-1α stabilization independent of Cullin 2 deneddylation. CSN5 also associates with HIF-1α under hypoxia and is required for optimal hypoxia-mediated HIF-1α stabilization (Bemis et al., 2004). Less clear from this study is whether the anaerobic binding of CSN5 to HIF-1α occurs independently of pVHL. Several studies have shown that CSN5 exists in small subunits or in monomeric form outside of the CSN in various species (Freilich et al., 1999; Kwok et al., 1998; Oron et al., 2002; Tomoda et al., 2002; Tomoda et al., 2004). Both monomeric and CSN-associated CSN5 has been found to functionally interact with a number of intracellular proteins and, in almost all cases, regulates their turnover (Richardson and Zundel, 2005). A notable exception to this is MIF (Kleemann et al., 2000). Jab1/CSN5 was initially identified by yeast 2 hybrid screening to interact with MIF (Kleemann et al., 2000). Interestingly, MIF was shown to modulate CSN5 function and subsequent CSN5-dependent effects on p27 degradation, JNK activation and AP-1-mediated transcription (Kleemann et al., 2000). Extracellular MIF is shown to functionally regulate the activities of intracellular CSN5 (Berndt et al., 2008; Kleemann et al., 2000; Kleemann et al., 2002; Lue et al., 2007; Meyer-Siegler et al., 2006). While the precise mechanism remains unclear, studies from this laboratory (Winner et al., 2007) and those of others (Kleemann et al., 2000; Kleemann et al., 2002; Leng et al., 2003) support the supposition that extracellular MIF may functionally regulate intracellular Jab1/CSN5 functions.
Consistent with the finding that MIF modulates CSN5-dependent p27 ubiquitylation and proteasomal degradation (Kleemann et al., 2000), MIF was recently found to be necessary for DNA damage checkpoint responses in developing lymphomas (Nemajerova et al., 2007). Specifically, MIF controls CSN5-dependent deneddylation and subsequent cullin (Cul1)-containing ubiquitin E3 (SCF) complex stability. Aberrant neddylation of the SCF complex in MIF-deficient B cell lymphomas results in defective checkpoint response protein (Chk1, Chk2, Cdc25A) accumulation resulting in defective DNA repair. As mentioned above, prior studies from this laboratory implicate CSN5/Jab1 in MIF-dependent HIF-1α stabilization (Winner et al., 2007). However, in contrast to the studies involving MIF regulation of the SCF complex resulting in enhanced degradation/decreased stability of p27 and Cdc25A proteins, MIF/CSN5 regulates HIF-1α turnover resulting in enhanced stability/decreased degradation of HIF-1α (Winner et al., 2007). These differences in p27 (the target of a Cullin-1Ub-E3), and HIF-1a (the target of a Cullin-2 Ub-E3), underscore the fact that CSN5 interacts with and affects its targets differently and may produce different effects depending on these interactions. Clearly more studies are needed to fully delineate how MIF influences HIF stabilization and what contributions, if any, p53 and CSN5 play in this process.
Irrespective of the mechanism by which MIF contributes to hypoxia-induced HIF-1α stability, another question that remains unanswered is that of the apparently divergent phenotypes of HIF-1α-deficient mice versus those associated with MIF-deficiency. HIF-1α homozygous null mice developmentally arrest and die by embryonic day 11 (Iyer et al., 1998; Ryan et al., 1998) while MIF-deficient mice are viable and develop relatively normally (Bozza et al., 1999). MIF-deficiency, however, does render these mice more resistant to several diseases including autoimmune, bacterial and parasitic infections, atherosclerosis and tumorigenesis (Bozza et al., 1999; de Jong et al., 2001; Fingerle-Rowson et al., 2003; Koebernick et al., 2002; McDevitt et al., 2006; Pan et al., 2004; Santos et al., 2008; Taylor, III et al., 2007; Wilson et al., 2005). Because existing data indicate that MIF contributes to, but is not required for, HIF-1α stabilization, it is more likely that MIF-deficiency more closely resembles HIF-1α heterozygous null mice than HIF-1α homozygous null mice. HIF-1α heterozygous null mice are viable and develop normally but do display specific phenotypes, some of which are similar to those of MIF−/− mice. For example, a recent study reveals that MIF-deficiency results in defective lung maturation and lethality in prematurely born pups (Kevill et al., 2008). Moreover, MIF is induced by heart ischaemia and contributes to AMPK activation leading to glucose uptake and cardiac repair following ischaemia-reperfusion (Miller et al., 2008). As these phenotypes are consistent with a role for MIF in hypoxic adaptation associated with HIF-1α activity (Cai et al., 2008; Compernolle et al., 2002; Land and Wilson, 2005; Loor and Schumacker, 2008), it is plausible that MIF acts to potentiate HIF-1α stabilization and function but is not absolutely essential for it. As such, when MIF is highly expressed in primary and/or metastatic malignant lesions, there is a correspondingly higher level of HIF-1α stabilization, angiogenic growth factor expression and intratumoral angiogenesis. As discussed above, high MIF expression within tumors is a negative predictor of disease survival and corresponds closely with enhanced VEGF expression and microvessel density (Hagemann et al., 2007; Han et al., 2008; Hira et al., 2005; Ren et al., 2005; Shun et al., 2005; White et al., 2003; Xu et al., 2007). Combining all of this information, it is not unreasonable to conclude that MIF is a critical determinant of hypoxic responses and plays an important role in maintaining microenvironmental adaptation within developing neoplasms.
MIF Structure, Function and Small Molecule Antagonists
Three-dimensional X-ray crystallographic studies have revealed that human MIF exists as a homotrimer structurally related to the bacterial enzymes 4-oxalocrotonate tautomerase and 5-carboxymethyl-2-hydroxymuconate isomerase (Sugimoto et al., 1996; Sun et al., 1996). MIF possesses the unusual ability to catalyze the tautomerization of the non-physiological substrates D-dopachrome and L-dopachrome methyl ester into their corresponding indole derivatives (Rosengren et al., 1996).
More recently, phenyl-pyruvic acid, p-hydroxy-phenylpyruvic acid (HPP), 3,4-dihydroxyphenylaminechrome, and norepinephrine-chrome also have been found to be MIF substrates. High Michaelis constant (Km) values, however, suggest that these also are unlikely to be natural substrates for MIF (Matsunaga et al., 1999; Rosengren et al., 1997). The N-terminal proline of MIF (Pro-1) appears to be a critical residue for enzymatic activity, as site-directed mutagenesis that substitutes a serine for this proline (P1S) is devoid of D-dopachrome tautomerase activity (Bendrat et al., 1997). Similarly, a proline to glycine (P1G) MIF mutant is also catalytically null for both D-dopachrome and HPP tautomerase activities (Lubetsky et al., 1999; Swope et al., 1998).
Structure-based drug design (SBDD) holds great promise but in itself is really the first step in one of many subsequent steps and disciplines that are needed for optimized drug discovery. The success of SBDD is well documented; it has contributed to the introduction of 50 compounds into clinical trials and to numerous drug approvals (Jorgensen, 2004). Virtual screening is a computational technique that can prescreen vast databases of small molecule structures against a three-dimensional structure to see which fit, or dock, into the chosen site. This can reduce the actual physical screening for lead compounds by many orders of magnitude.
Orita et al. first described virtual screening as a tool to identify novel small molecule antagonists of MIF enzymatic activity (Orita et al., 2001). From screening both ACD and ACDSC libraries (totaling nearly 1,000,000 compounds) the authors identified 524 potential inhibitors and tested them for MIF tautomerase inhibition. The top 14 candidate inhibitory compounds could be classified into four independent groups: 1) coumarin and derivatives; 2) 7-hydroxycoumarin and derivatives; 3) 7-hydroxy-chromen-4-one and derivatives; and, 4) 7-hydroxy-chroman-2,4-dione and derivatives. Co-crystallization studies revealed two hydrophobic binding regions that small molecule inhibitors require for MIF binding (Orita et al., 2001). This screen, like most studies attempting to identify MIF tautomerase inhibitors, targeted the N-terminal proline as the central target residue for inhibition. Despite excellent inhibitory activities reported for these compounds, it is unclear whether they also are capable of inhibiting the biological activities of MIF (Orita et al., 2001).
Of the MIF antagonists reported thus far (Dios et al., 2002; Lubetsky et al., 2002; Senter et al., 2002), ISO-1 (S,R-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester) is the best characterized (Al Abed et al., 2005; Lubetsky et al., 2002; Meyer-Siegler et al., 2006; Nicoletti et al., 2005; Rendon et al., 2007; West et al., 2008). Ours and others’ studies reveal that this compound is effective in blocking MIF-dependent malignant phenotypes nearly as well as MIF knockdown by siRNA (Meyer-Siegler et al., 2006; Rendon et al., 2007). Interestingly, ISO-1 was found to inhibit prostate cancer cell invasion, tumor volume and angiogenesis only in prostate cancer cells that express the cognate MIF receptor, CD74 (Meyer-Siegler et al., 2006). These findings indicate that small molecule inhibitors of MIF vestigial enzymatic activity may block MIF/CD74 interaction and/or signaling initiation leading to defective malignant cell growth, migration, invasion and tumor-associated angiogenesis (Meyer-Siegler et al., 2006; Rendon et al., 2007).
Using a novel virtual screening strategy we have discovered a novel suicide antagonist of MIF that is ~10x more potent than ISO-1 in blocking both MIF-dependent enzymatic and biologic functions (Winner et al., 2008). Unlike the prior virtual screening strategy described above this targeting strategy focused on identifying compounds that block methionine at position A2 (MetA2; the A2 refers to monomer A, position 2 from the crystal structure of MIF which is made up of A, B and C monomers to form a catalytically active trimer). This strategy was chosen for two reasons: 1) MetA2 resides at the base of the hydrophobic binding pocket adjacent to the N-terminal proline that actually resides on the side of the pocket (Sun et al., 1996); and 2) prior studies have shown that disrupting this hydrophobic substrate-binding pocket by insertion of a single amino acid residue adjacent to Met A2 leads to a complete loss of enzymatic and biologic activity (Lubetsky et al., 2002).
As mentioned above, this novel compound, 4-iodo-6-phenylpyrimidine (4-IPP), is unique compared to previously identified MIF tautomerase inhibitors in that it acts as a suicide substrate for MIF (Winner et al., 2008). Moreover, this compound is as effective as siRNA-mediated knockdown of MIF and ~5–10x more effective than ISO-1 in blocking migration and anchorage-independent growth of non-small cell lung cancer cells (Rendon et al., 2007; Winner et al., 2008). Importantly, 4-IPP delivered intraperitoneally effectively inhibits liver MIF enzymatic activity suggesting that 4-IPP is bio-available (Winner et al., 2008). Additionally, recent studies from our laboratory reveal that pancreatic ductal adenocarcinoma xenograft tumors in nude mice treated with 4-IPP are dramatically smaller and significantly less vascularized than vehicle control treated mice (unpublished observations). These findings are consistent with earlier studies showing ISO-1-mediated inhibition of tumor-associated angiogenesis of xenograft prostate adenocarcinoma tumors (Meyer-Siegler et al., 2006). These findings lend further support to a potential role for MIF antagonists in inhibiting HIF-1α stabilization in hypoxic tumors and the subsequent loss of hypoxia-induced VEGF expression and intratumoral angiogenesis.
Conclusions and Future Prospects
It is becoming more and more evident that MIF influences several important biological mechanisms and processes by which tumors thrive and spread. One of the most important of these is the modulation of hypoxic adaptation within the tumor microenvironment by directly influencing hypoxia-induced HIF-1α stabilization (Oda et al., 2008; Winner et al., 2007). Evidence of tumor microenvironmental modulation by MIF is found in numerous human and rodent studies demonstrating a clear and pronounced role for MIF in modulating tumor-associated neoangiogenesis.
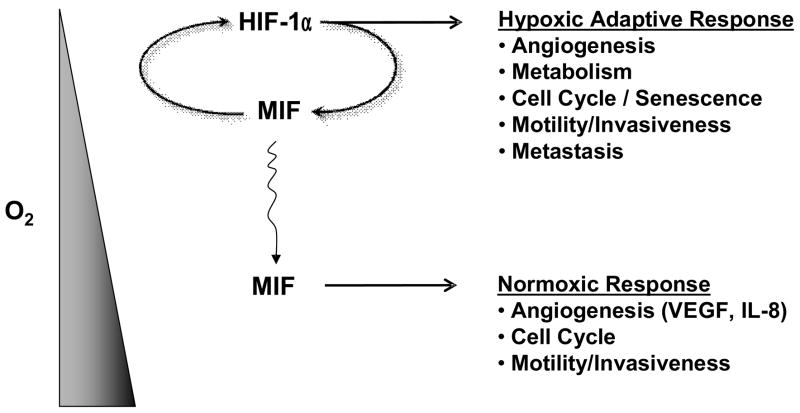
MIF influences both hypoxic and normoxic neovascular processes within a tumor’s microenvironment. We envision MIF transcription and secretion being induced in hypoxic areas of tumors where increased MIF then acts to further amplify HIF-1α expression and activity (Figure 1). Hypoxia-induced MIF can diffuse into normoxic regions of a tumor to further enhance VEGF and IL-8 angiogenic growth factor expression leading to neovascularization (Figure 1). The success of angiogenesis inhibitors such as Avastin (Omuro and Delattre, 2008; Sachdev and Jahanzeb, 2008), and the recent discovery of potent small molecule MIF antagonists, suggest that MIF targeting may represent a novel cancer chemotherapeutic strategy.
Figure 1.
Scheme demonstrating hypoxic/normoxic contributions to tumor microenvironmental adaptation by macrophage migration inhibitory factor (MIF). Hypoxia induces HIF-1α-dependent MIF transcription and secretion in human cancers. When over-expressed, MIF functionally enhances hypoxia-induced HIF-1α stabilization (Winner et al., 2007) and subsequent hypoxic adaptive responses thus contributing to angiogenic, metabolic, cell cycle and metastatic responses to low oxygen tension. Hypoxia-induced MIF diffuses into normoxic regions of the tumor where it can additionally modulate angiogenic growth factor expression, cell cycle progression and migration/invasion of normoxic malignant cells (Coleman et. al., 2008; Rendon et al., 2007).
Acknowledgments
This work has been supported by National Institutes of Health grants 5P20RR018733 and 5R01CA102285 (R.A.M.), and a grant from Philip Morris USA Inc. and Philip Morris International (R.A.M).
Abbreviations
- CSN5
COP9 signalosome subunit 5
- PDAC
Pancreatic ductal adenocarcinoma
- PHD
Prolyl hydroxylase
- VEGF
Vascular endothelial growth factor
- VHL
von Hippel-Lindau
Footnotes
Conflict of Interest. R. A. Mitchell is a co-inventor on patents and patent applications describing the therapeutic value of 4-IPP and 4-IPP analog MIF antagonists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz C, Pavlov VA, Miller EJ, Tracey KJ. ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- Bacher M, Schrader J, Thompson N, Kuschela K, Gemsa D, Waeber G, Schlegel J. Up-regulation of macrophage migration inhibitory factor gene and protein expression in glial tumor cells during hypoxic and hypoglycemic stress indicates a critical role for angiogenesis in glioblastoma multiforme. Am J Pathol. 2003;162:11–17. doi: 10.1016/S0002-9440(10)63793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando H, Matsumoto G, Bando M, Muta M, Ogawa T, Funata N, Nishihira J, Koike M, Toi M. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn J Cancer Res. 2002;93:389–396. doi: 10.1111/j.1349-7006.2002.tb01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun. 2006;347:895–903. doi: 10.1016/j.bbrc.2006.06.148. [DOI] [PubMed] [Google Scholar]
- Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, Chen X, Stenmark K, Giaccia AJ, Zundel W. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 2004;18:739–744. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendrat K, Al Abed Y, Callaway DJ, Peng T, Calandra T, Metz CN, Bucala R. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry. 1997;36:15356–15362. doi: 10.1021/bi971153a. [DOI] [PubMed] [Google Scholar]
- Berndt K, Kim M, Meinhardt A, Klug J. Macrophage migration inhibitory factor does not modulate co-activation of androgen receptor by Jab1/CSN5. Mol Cell Biochem. 2008;307:265–271. doi: 10.1007/s11010-007-9578-3. [DOI] [PubMed] [Google Scholar]
- Bozza M, Satoskar AR, Lin G, Lu B, Humbles AA, Gerard C, David JR. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J Exp Med. 1999;189:341–346. doi: 10.1084/jem.189.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Zhong H, Bosch-Marce M, Fox-Talbot K, Wang L, Wei C, Trush MA, Semenza GL. Complete loss of ischaemic preconditioning-induced cardioprotection in mice with partial deficiency of HIF-1 alpha. Cardiovasc Res. 2008;77:463–470. doi: 10.1093/cvr/cvm035. [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1 alpha. Mol Cell Biol. 2005;25:6415–6426. doi: 10.1128/MCB.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S, Codognotto A, Rapozzi V, Meeuwenoord N, van der MG, Xodo LE. Transcription inhibition of oncogenic KRAS by a mutation-selective peptide nucleic acid conjugated to the PKKKRKV nuclear localization signal peptide. Biochemistry. 2005;44:10510–10519. doi: 10.1021/bi0505215. [DOI] [PubMed] [Google Scholar]
- Coleman AM, Rendon BE, Zhao M, Qian MW, Bucala R, Xin D, Mitchell RA. Cooperative regulation of non-small cell lung carcinoma angiogenic potential by macrophage migration inhibitory factor and its homolog, D-dopachrome tautomerase. J Immunol. 2008;181:2330–2337. doi: 10.4049/jimmunol.181.4.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, Nemery B, Dewerchin M, Van VP, Plate K, Moons L, Collen D, Carmeliet P. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, Mizoguchi E, Metz CN, Alsahli M, ten Hove T, Keates AC, Lubetsky JB, Farrell RJ, Michetti P, van Deventer SJ, Lolis E, David JR, Bhan AK, Terhorst C, Sahli MA. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–1066. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- del Vecchio MT, Tripodi SA, Arcuri F, Pergola L, Hako L, Vatti R, Cintorino M. Macrophage migration inhibitory factor in prostatic adenocarcinoma: correlation with tumor grading and combination endocrine treatment-related changes. Prostate. 2000;45:51–57. doi: 10.1002/1097-0045(20000915)45:1<51::aid-pros6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Dios A, Mitchell RA, Aljabari B, Lubetsky J, O’Connor K, Liao H, Senter PD, Manogue KR, Lolis E, Metz C, Bucala R, Callaway DJ, Al Abed Y. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J Med Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- Fingerle-Rowson G, Petrenko O, Metz CN, Forsthuber TG, Mitchell R, Huss R, Moll U, Muller W, Bucala R. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci USA. 2003;100:9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Hagemann T, Robinson SC, Thompson RG, Charles K, Kulbe H, Balkwill FR. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol Cancer Ther. 2007;6:1993–2002. doi: 10.1158/1535-7163.MCT-07-0118. [DOI] [PubMed] [Google Scholar]
- Han I, Lee MR, Nam KW, Oh JH, Moon KC, Kim HS. Expression of macrophage migration inhibitory factor relates to survival in high-grade osteosarcoma. Clin Orthop Relat Res. 2008;466:2107–2113. doi: 10.1007/s11999-008-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira E, Ono T, Dhar DK, El Assal ON, Hishikawa Y, Yamanoi A, Nagasue N. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 2005;103:588–598. doi: 10.1002/cncr.20818. [DOI] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity [see comments] J Exp Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- Kamimura A, Kamachi M, Nishihira J, Ogura S, Isobe H, Dosaka-Akita H, Ogata A, Shindoh M, Ohbuchi T, Kawakami Y. Intracellular distribution of macrophage migration inhibitory factor predicts the prognosis of patients with adenocarcinoma of the lung. Cancer. 2000;89:334–341. [PubMed] [Google Scholar]
- Kevill KA, Bhandari V, Kettunen M, Leng L, Fan J, Mizue Y, Dzuira JD, Reyes-Mugica M, McDonald CL, Baugh JA, O’Connor CL, Aghai ZH, Donnelly SC, Bazzy-Asaad A, Bucala RJ. A role for macrophage migration inhibitory factor in the neonatal respiratory distress syndrome. J Immunol. 2008;180:601–608. doi: 10.4049/jimmunol.180.1.601. [DOI] [PubMed] [Google Scholar]
- Kim W, Kaelin WG., Jr The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr Opin Genet Dev. 2003;13:55–60. doi: 10.1016/s0959-437x(02)00010-2. [DOI] [PubMed] [Google Scholar]
- Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Kleemann R, Grell M, Mischke R, Zimmermann G, Bernhagen J. Receptor binding and cellular uptake studies of macrophage migration inhibitory factor (MIF): use of biologically active labeled MIF derivatives. J Interferon Cytokine Res. 2002;22:351–363. doi: 10.1089/107999002753675785. [DOI] [PubMed] [Google Scholar]
- Kleemann R, Hausser A, Geiger G, Mischke R, Burger-Kentischer A, Flieger O, Johannes FJ, Roger T, Calandra T, Kapurniotu A, Grell M, Finkelmeier D, Brunner H, Bernhagen J. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- Koebernick H, Grode L, David JR, Rohde W, Rolph MS, Mittrucker HW, Kaufmann SH. Macrophage migration inhibitory factor (MIF) plays a pivotal role in immunity against Salmonella typhimurium. Proc Natl Acad Sci USA. 2002;99:13681–13686. doi: 10.1073/pnas.212488699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koong AC, Denko NC, Hudson KM, Schindler C, Swiersz L, Koch C, Evans S, Ibrahim H, Le QT, Terris DJ, Giaccia AJ. Candidate genes for the hypoxic tumor phenotype. Cancer Res. 2000;60:883–887. [PubMed] [Google Scholar]
- Kwok SF, Solano R, Tsuge T, Chamovitz DA, Ecker JR, Matsui M, Deng XW. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land SC, Wilson SM. Redox regulation of lung development and perinatal lung epithelial function. Antioxid Redox Signal. 2005;7:92–107. doi: 10.1089/ars.2005.7.92. [DOI] [PubMed] [Google Scholar]
- Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loor G, Schumacker PT. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ. 2008;15:686–690. doi: 10.1038/cdd.2008.13. [DOI] [PubMed] [Google Scholar]
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- Lubetsky JB, Dios A, Han J, Aljabari B, Ruzsicska B, Mitchell R, Lolis E, Al Abed Y. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277:24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- Lubetsky JB, Swope M, Dealwis C, Blake P, Lolis E. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Luscher B, Bernhagen J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- Markert JM, Fuller CM, Gillespie GY, Bubien JK, McLean LA, Hong RL, Lee K, Gullans SR, Mapstone TB, Benos DJ. Differential gene expression profiling in human brain tumors. Physiol Genomics. 2001;5:21–33. doi: 10.1152/physiolgenomics.2001.5.1.21. [DOI] [PubMed] [Google Scholar]
- Martin J, Duncan FJ, Keiser T, Shin S, Kusewitt DF, Oberyszyn T, Satoskar AR, Vanbuskirk AM. Macrophage migration inhibitory factor (MIF) plays a critical role in pathogenesis of ultraviolet-B (UVB) -induced nonmelanoma skin cancer (NMSC) FASEB J. 2008 2008 Oct 24; doi: 10.1096/fj.08-119628. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Matsunaga J, Sinha D, Pannell L, Santis C, Solano F, Wistow GJ, Hearing VJ. Enzyme activity of macrophage migration inhibitory factor toward oxidized catecholamines. J Biol Chem. 1999;274:3268–3271. doi: 10.1074/jbc.274.6.3268. [DOI] [PubMed] [Google Scholar]
- McDevitt MA, Xie J, Shanmugasundaram G, Griffith J, Liu A, McDonald C, Thuma P, Gordeuk VR, Metz CN, Mitchell R, Keefer J, David J, Leng L, Bucala R. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J Exp Med. 2006;203:1185–1196. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Siegler K, Fattor RA, Hudson PB. Expression of macrophage migration inhibitory factor in the human prostate. Diagn Mol Pathol. 1998;7:44–50. doi: 10.1097/00019606-199802000-00008. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler KL, Bellino MA, Tannenbaum M. Macrophage migration inhibitory factor evaluation compared with prostate specific antigen as a biomarker in patients with prostate carcinoma. Cancer. 2002;94:1449–1456. doi: 10.1002/cncr.10354. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- Miller EJ, Li J, Leng L, McDonald C, Atsumi T, Bucala R, Young LH. Macrophage migration inhibitory factor stimulates AMP-activated protein kinase in the ischaemic heart. Nature. 2008;451:578–582. doi: 10.1038/nature06504. [DOI] [PubMed] [Google Scholar]
- Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, Bucala R. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci USA. 2002;99:345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemajerova A, Mena P, Fingerle-Rowson G, Moll UM, Petrenko O. Impaired DNA damage checkpoint response in MIF-deficient mice. EMBO J. 2007;26:987–997. doi: 10.1038/sj.emboj.7601564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti F, Creange A, Orlikowski D, Bolgert F, Mangano K, Metz C, Di Marco R, Al Abed Y. Macrophage migration inhibitory factor (MIF) seems crucially involved in Guillain-Barre syndrome and experimental allergic neuritis. J Neuroimmunol. 2005;168:168–174. doi: 10.1016/j.jneuroim.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Oda S, Oda T, Nishi K, Takabuchi S, Wakamatsu T, Tanaka T, Adachi T, Fukuda K, Semenza GL, Hirota K. Macrophage migration inhibitory factor activates hypoxia-inducible factor in a p53-dependent manner. PLoS ONE. 2008;3:e2215. doi: 10.1371/journal.pone.0002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro AM, Delattre JY. What is the place of bevacizumab and irinotecan in the treatment of glioblastoma and other malignant gliomas? Curr Opin Neurol. 2008;21:717–719. doi: 10.1097/WCO.0b013e3283184625. [DOI] [PubMed] [Google Scholar]
- Orita M, Yamamoto S, Katayama N, Aoki M, Takayama K, Yamagiwa Y, Seki N, Suzuki H, Kurihara H, Sakashita H, Takeuchi M, Fujita S, Yamada T, Tanaka A. Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography. J Med Chem. 2001;44:540–547. doi: 10.1021/jm000386o. [DOI] [PubMed] [Google Scholar]
- Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, Chamovitz DA. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129:4399–4409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- Pan JH, Sukhova GK, Yang JT, Wang B, Xie T, Fu H, Zhang Y, Satoskar AR, David JR, Metz CN, Bucala R, Fang K, Simon DI, Chapman HA, Libby P, Shi GP. Macrophage migration inhibitory factor deficiency impairs atherosclerosis in low-density lipoprotein receptor-deficient mice. Circulation. 2004;109:3149–3153. doi: 10.1161/01.CIR.0000134704.84454.D2. [DOI] [PubMed] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Law S, Huang X, Lee PY, Bacher M, Srivastava G, Wong J. Macrophage migration inhibitory factor stimulates angiogenic factor expression and correlates with differentiation and lymph node status in patients with esophageal squamous cell carcinoma. Ann Surg. 2005;242:55–63. doi: 10.1097/01.sla.0000168555.97710.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon BE, Roger T, Teneng I, Zhao M, Al Abed Y, Calandra T, Mitchell RA. Regulation of human lung adenocarcinoma cell migration and invasion by macrophage migration inhibitory factor. J Biol Chem. 2007;282:29910–29918. doi: 10.1074/jbc.M704898200. [DOI] [PubMed] [Google Scholar]
- Richardson KS, Zundel W. The emerging role of the COP9 signalosome in cancer. Mol Cancer Res. 2005;3:645–653. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren E, Aman P, Thelin S, Hansson C, Ahlfors S, Bjork P, Jacobsson L, Rorsman H. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 1997;417:85–88. doi: 10.1016/s0014-5793(97)01261-1. [DOI] [PubMed] [Google Scholar]
- Rosengren E, Bucala R, Aman P, Jacobsson L, Odh G, Metz CN, Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev JC, Jahanzeb M. Evolution of bevacizumab-based therapy in the management of breast cancer. Clin Breast Cancer. 2008;8:402–410. doi: 10.3816/CBC.2008.n.048. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1alpha ODD domain to p53. Mol Cell. 2005;17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Santos LL, Dacumos A, Yamana J, Sharma L, Morand EF. Reduced arthritis in MIF deficient mice is associated with reduced T cell activation: down-regulation of ERK MAP kinase phosphorylation. Clin Exp Immunol. 2008;152:372–380. doi: 10.1111/j.1365-2249.2008.03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser A, Marquetant R, Illmer T, Graffy C, Garlichs CD, Bockler D, Menschikowski D, Braun-Dullaeus R, Daniel WG, Strasser RH. The expression of macrophage migration inhibitory factor 1alpha (MIF 1alpha) in human atherosclerotic plaques is induced by different proatherogenic stimuli and associated with plaque instability. Atherosclerosis. 2005;178:83–94. doi: 10.1016/j.atherosclerosis.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Senter PD, Al Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc Natl Acad Sci USA. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Abe R, Nakamura H, Ohkawara A, Suzuki M, Nishihira J. High expression of macrophage migration inhibitory factor in human melanoma cells and its role in tumor cell growth and angiogenesis. Biochem Biophys Res Commun. 1999;264:751–758. doi: 10.1006/bbrc.1999.1584. [DOI] [PubMed] [Google Scholar]
- Shun CT, Lin JT, Huang SP, Lin MT, Wu MS. Expression of macrophage migration inhibitory factor is associated with enhanced angiogenesis and advanced stage in gastric carcinomas. World J Gastroenterol. 2005;11:3767–3771. doi: 10.3748/wjg.v11.i24.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch. 2003;442:444–452. doi: 10.1007/s00428-003-0784-4. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Suzuki M, Nakagawa A, Tanaka I, Nishihira J. Crystal structure of macrophage migration inhibitory factor from human lymphocyte at 2.1 A resolution. FEBS Lett. 1996;389:145–148. doi: 10.1016/0014-5793(96)00553-4. [DOI] [PubMed] [Google Scholar]
- Sun HW, Bernhagen J, Bucala R, Lolis E. Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc Natl Acad Sci USA. 1996;93:5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swope M, Sun HW, Blake PR, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Nishihira J, Shimpo M, Mizue Y, Ueno S, Mano H, Kobayashi E, Ikeda U, Shimada K. Macrophage migration inhibitory factor as a redox-sensitive cytokine in cardiac myocytes. Cardiovasc Res. 2001;52:438–445. doi: 10.1016/s0008-6363(01)00408-4. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nishihira J, Sato Y, Kondo M, Ogawa H, Ohshima T, Une Y, Todo S. Involvement of macrophage migration inhibitory factor (MIF) in the mechanism of tumor cell growth. Mol Med. 1998;4:707–714. [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, III, Kuchel GA, Hegde P, Voznesensky OS, Claffey K, Tsimikas J, Leng L, Bucala R, Pilbeam C. Null mutation for macrophage migration inhibitory factor (MIF) is associated with less aggressive bladder cancer in mice. BMC Cancer. 2007;7:135. doi: 10.1186/1471-2407-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyasu M, Yoshino I, Suemitsu R, Okamoto T, Sugimachi K. Quantification of macrophage migration inhibitory factor mRNA expression in non-small cell lung cancer tissues and its clinical significance. Clin Cancer Res. 2002;8:3755–3760. [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277:2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem. 2004;279:43013–43018. doi: 10.1074/jbc.M406559200. [DOI] [PubMed] [Google Scholar]
- Welford SM, Bedogni B, Gradin K, Poellinger L, Broome PM, Giaccia AJ. HIF1{alpha} delays premature senescence through the activation of MIF. Genes Dev. 2006;20:3366–3371. doi: 10.1101/gad.1471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West PW, Parker LC, Ward JR, Sabroe I. Differential and cell-type specific regulation of responses to Toll-like receptor agonists by ISO-1. Immunology. 2008;125:101–110. doi: 10.1111/j.1365-2567.2008.02825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White ES, Flaherty KR, Carskadon S, Brant A, Iannettoni MD, Yee J, Orringer MB, Arenberg DA. Macrophage migration inhibitory factor and CXC chemokine expression in non-small cell lung cancer: role in angiogenesis and prognosis. Clin Cancer Res. 2003;9:853–860. [PubMed] [Google Scholar]
- Wilson JM, Coletta PL, Cuthbert RJ, Scott N, MacLennan K, Hawcroft G, Leng L, Lubetsky JB, Jin KK, Lolis E, Medina F, Brieva JA, Poulsom R, Markham AF, Bucala R, Hull MA. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 2005;129:1485–1503. doi: 10.1053/j.gastro.2005.07.061. [DOI] [PubMed] [Google Scholar]
- Winner M, Koong AC, Rendon BE, Zundel W, Mitchell RA. Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res. 2007;67:186–193. doi: 10.1158/0008-5472.CAN-06-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner M, Meier J, Zierow S, Rendon BE, Crichlow GV, Riggs R, Bucala R, Leng L, Smith N, Lolis E, Trent JO, Mitchell RA. A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 2008;68:7253–7257. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- Xu X, Wang B, Ye C, Yao C, Lin Y, Huang X, Zhang Y, Wang S. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett. 2009;261:147–157. doi: 10.1016/j.canlet.2007.11.028. [DOI] [PubMed] [Google Scholar]