Abstract
Background
H3N2 is one of the main subtypes of influenza virus which circulates in human and swine population throughout the world.
Objectives
To investigate the genetic correlation between H3N2 human and swine influenza viruses from the same region during the same season.
Study design
Five H3N2 human and four H3N2 swine influenza viruses were isolated from Guangdong province of China in the winter of 2005. The molecular evolution of eight gene segments was analyzed.
Results
In the phylogenetic trees of gene segments, all H3N2 human isolates along with the 2000’s human isolates formed a cluster, and most of the H3N2 swine isolates along with the 1990’s human isolates formed another cluster except that the M and NS gene of A/Swine/Guangdong/01/2005 and the PA gene of A/Swine/Guangdong/02/2005 fell into the cluster of the classical swine influenza virus, indicating the reassortment between H3N2 human and H1N1 swine influenza viruses.
Conclusions
In this study, H3N2 swine influenza viruses in 2005 did not originate from the 2000’s H3N2 human influenza viruses, but from the 1990’s H3N2 human isolates. In addition, the reassortment of H3N2 human and H1N1 swine influenza virus in pigs was common in recent years.
Keywords: influenza virus, H3N2 subtype, human, swine, genetic correlation, reassortment
H3N2 subtype influenza viruses appeared in humans and caused a major pandemic in 1968. Subsequently, H3N2 influenza viruses were isolated regularly from pigs throughout the world 1–4. H1 and H3 subtype influenza viruses could transmit between humans and pigs 5–8. Pigs have been suggested as intermediate hosts and “mixing vessel” for gene segments reassortment among the influenza A viruses from human and/or avian and are of great significance for human health. There are instances of reassortment between swine, avian and human viruses occurring in pigs in nature 2, 9–11. So far, few reassortment cases in pigs of China have been reported 12–14.
To learn more information about the prevalence of H3N2 influenza virus and the genetic correlation between H3N2 human and swine isolates from the same region during the same season, 450 nasal swabs or lung tissues samples were collected from sick or dead pigs of some big farms, and 90 nasopharyngeal swab specimens were collected from influenza like cases of hospitals. These patients had no clear contact document. All these samples were collected from Guangdong province of China in the winter of 2005. Virus isolation and subtyping were done as previously reported15. As a result, five H3N2 human (A/Guangdong/03/2005, A/Guangdong/04/2005, A/Guangdong/05/2005, A/Guangdong/07/2005, A/Guangdong/09/2005) and four H3N2 swine (A/Swine/Guangdong/01/2005, A/Swine/Guangdong/02/2005, A/Swine/Guangdong/03/2005, A/Swine/Guangdong/04/2005) influenza viruses were isolated successfully.
RT-PCR was used to amplify the full-length protein coding regions of all eight viral RNA for sequencing analysis. Viral RNA was extracted from 200 μl of allantoic fluid using Trizol reagents (GIBCO-BRL), and reverse transcription was performed using influenza virus oligonucleotide universal primer: 5′-AGC AAA AGC AGG-3′. A series of primers were designed to amplify PB2, PB1, PA, HA, NP, NA, M and NS genes for sequencing. Sequencing of three independent clones of each PCR product was performed in order to eliminate errors resulting from the RT-PCR or cloning steps. The GenBank accession numbers of our isolates were EF455562-EF455569, EU604298-EU604305 and EU620723-EU620778. Sequence comparisons were made using the MegAlign software (DNAStar 5.01) and phylogenetic trees were drawn using the MEGA 3.1 program.
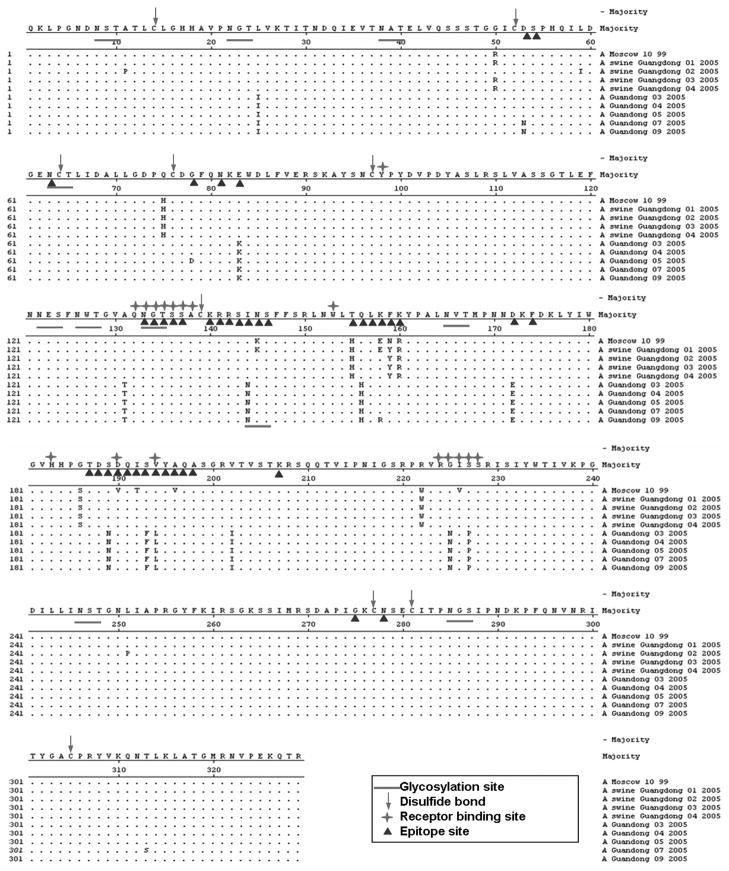
The results of molecular analysis of HA1 genes of these H3N2 influenza viruses were showed in Fig. 1. There were 10 glycosylation sites (8, 22, 38, 63, 122, 126, 133, 165, 246 and 285) in the HA1 genes of H3N2 swine influenza viruses. But in human isolates, except those 10 glycosylation sites, another glycosylation site at 144 was found. The disulfide bonds (14, 52, 64, 76, 97, 139, 277 and 281) in the HA genes of all these swine and human isolates were conservative. For these isolates, most of the receptor binding sites were conservative except three sites (G225N, S227P and V194L). The epitope sites (H155T, Q156H, Y159F, R160K, S189N, S193F and V194/L) were variable.
Fig. 1.
Alignment of HA1 amino acid sequences. Residues with triangles represent previously identified epitopes with H3 molecule. Residues with lines represent possible N-glycosylation sites. Residues with arrows represent disulfide bonds. Residues with red stars represent receptor binding sites.
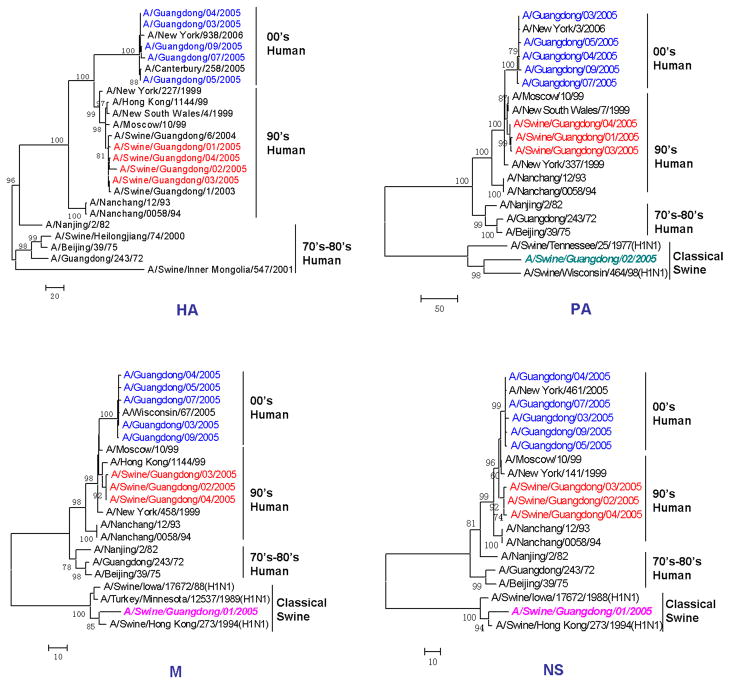
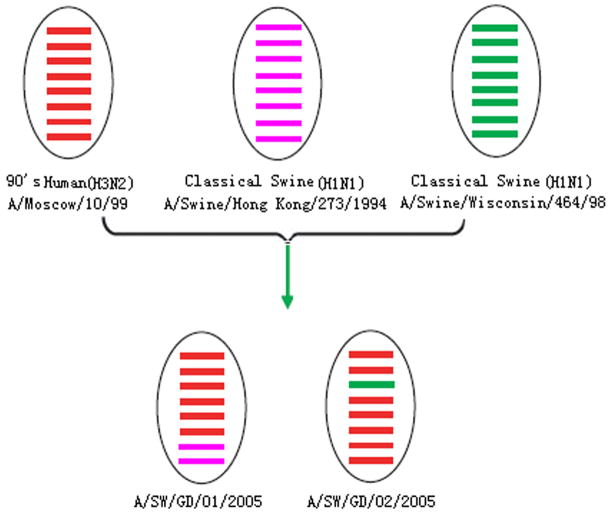
The sequence identities between human and swine isolates (94.4–95.5%) were lower than those among human (99.5–100%) or among swine isolates (98.4–99.8%). In the phylogenetic trees of HA (Fig. 2), NA, NP, PB1 and PB2 (data not shown) genes, five H3N2 human isolates along with the 2000’s human isolates formed a cluster (00′ human), four H3N2 swine isolates along with the 1990’s human isolates formed a different cluster (90′ human), and other selected H3N2 virus strains before 1990 formed another cluster (70′–80′ human). On the other hand, in the phylogenetic trees of M, NS and PA genes (Fig. 2), two swine isolates (A/Swine/Guangdong/01/2005 or A/Swine/Guangdong/02/2005) moved from 90’s human cluster to classical swine cluster. In general, reassortment is identified phylogenetically, when segments of a viral genome have inconsistent associations with distinct clades of viruses in segment-specific phylogenies. Therefore, in this study, two out of four swine influenza isolates were identified as reassortants. The M and NS gene of A/Swine/Guangdong/01/2005 fell into the cluster of the classical H1N1 swine influenza virus, while other genes originated from H3N2 human influenza virus. The reassortment was also found in A/Swine/Guangdong/02/2005 with its PA gene originated from the classical H1N1 swine influenza virus and other genes from H3N2 human influenza virus (Fig. 2). The reassortments of these two isolates were showed in Fig. 3.
Fig. 2.
Phylogenetic trees for the H3N2 HA, PA, M and NS genes of influenza A viruses. Analysis was based on the nucleotide sequences in open reading frames of the HA, PA, M and NS gene. Trees were generated with the MAGA program (version 3.1) by using neighbor-joining analysis.
Fig. 3.
Reassortments between H3N2 human and H1N1 swine influenza viruses. The eight gene segments in each schematic virus particle are represented in the order (top to bottom) PB2, PB1, PA, HA, NP, NA, M and NS.
The reassortment of human and swine influenza virus was common in recent years. Recent study showed that double and triple reassortants of human, avian and swine influenza viruses were detected in swine 12. In contrast, no avian fragment was detected in the four swine viruses. It had also been reported that 3 out of 32 H3N2 swine influenza isolates from pigs in Southern China during 1976–1982 were reassortants of human and swine influenza viruses 13. However, two out of 4 H3N2 swine isolates were reassortants in our studies. There were not enough isolates in this study, but it indicated that the reassortment of influenza virus was common in pigs in recent years to some extent.
In this study, H3N2 swine influenza viruses in 2005 did not originate from the 2000’s H3N2 human influenza viruses, but from the 1990’s H3N2 human isolates. It had been reported that the antigenic evolution of H3N2 swine viruses occurred at a rate approximately six times slower than the rate in human viruses 16. In our studies, prevailing H3N2 subtypes human viruses were not found in pigs from the same season during 2005 and swine influenza virus evolved at a lower rate than human influenza virus. The exact reason that these viruses fail to persist in pigs is still not clear. We speculated that if human influenza virus infected pigs, it needed a period of adaptation to the new host, five years or more years, before its prevalence in pig flocks. In addition, immune selection is not considered important in pigs, so strains with different antigenic characteristics may be disadvantaged compared to the ‘highly-adapted’ established viruses which continually circulate within a large susceptible population. On the other hand, previous studies showed that the old or prevailing H3N2 swine influenza viruses could infect human 6, 17.
In Guangdong province (located very close to Hong Kong), pigs are raised in increasing numbers. Some of the pig husbandry is carried out in large-scale farms and humans and pigs are housed closely in farming villages, providing the opportunity for interspecies transmission of influenza viruses. The frequent close contact between humans and pigs would facilitate the transmission of influenza virus between them and the generation of new reassortant genotypes of influenza viruses, which had potential threaten to human health.
Acknowledgments
This study was funded by the Ministry of Science and Technology of China program (2006BAD06A04, 2006BAD06A03), National Basic Research Program (973 Program) of China (2005CB523002), Chinese Academy of Sciences Innovation projects (KSCX2-YW-N-054), NIH-CDC (3 U19 AI051915-05S1) and Hundreds of Talents Program of Chinese Academy of Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nerome K, Ishida M, Nakayama M, Oya A, Kanai C, Suwicha K. Antigenic and genetic analysis of A/Hong Kong (H3N2) influenza viruses isolated from swine and man. J Gen Virol. 1981;56(Pt 2):441–5. doi: 10.1099/0022-1317-56-2-441. [DOI] [PubMed] [Google Scholar]
- 2.Ottis K, Sidoli L, Bachmann PA, Webster RG, Kaplan MM. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73(2):103–8. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 3.Shortridge KF, Cherry A, Kendal AP. Further studies of the antigenic properties of H3N2 strains of influenza A isolated from swine in South East Asia. J Gen Virol. 1979;44(1):251–4. doi: 10.1099/0022-1317-44-1-251. [DOI] [PubMed] [Google Scholar]
- 4.Tumova B, Mensik J, Stumpa A, Fedova D, Pospisil Z. Serological evidence and isolation of a virus closely related to the human A/Hong Kong/68 (H3N2) strain in swine populations in Czechoslovakia in 1969–1972. Zentralbl Veterinarmed B. 1976;23(7):590–63. [PubMed] [Google Scholar]
- 5.Smith TF, Burgert EOJ, Dowdle WR, Noble GR, Campbell RJ, Van Scoy RE. Isolation of swine influenza virus from autopsy lung tissue of man. N Engl J Med. 1976;294:708–10. doi: 10.1056/NEJM197603252941308. [DOI] [PubMed] [Google Scholar]
- 6.Gregory V, Lim W, Cameron K, Bennett M, Marozin S, Klimov A, et al. Infection of a child in Hong Kong by an influenza A H3N2 virus closely related to viruses circulating in European pigs. J Gen Virol. 2001;82(Pt 6):1397–406. doi: 10.1099/0022-1317-82-6-1397. [DOI] [PubMed] [Google Scholar]
- 7.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75(20):9679–86. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kida H, Shortridge KF, Webster RG. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 1988;162(1):160–6. doi: 10.1016/0042-6822(88)90405-9. [DOI] [PubMed] [Google Scholar]
- 9.Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol. 2003;41(7):3198–205. doi: 10.1128/JCM.41.7.3198-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73(10):8851–6. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, et al. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet Microbiol. 2000;74(1–2):47–58. doi: 10.1016/s0378-1135(00)00165-6. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Hua RH, Zhang Q, Liu TQ, Liu HL, Li GX, et al. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol. 2008;46(3):1067–75. doi: 10.1128/JCM.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu LL, Lin YP, Wright SM, Shortridge KF, Webster RG. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology. 1994;202(2):825–33. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 14.Qi X, Lu CP. Genetic characterization of novel reassortant H1N2 influenza A viruses isolated from pigs in southeastern China. Arch Virol. 2006;151(11):2289–99. doi: 10.1007/s00705-006-0796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309(5738):1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 16.de Jong JC, Smith DJ, Lapedes AS, Donatelli I, Campitelli L, Barigazzi G, et al. Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. Journal of Virology. 2007;81(8):4315–22. doi: 10.1128/JVI.02458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claas EC, Kawaoka Y, de Jong JC, Masurel N, Webster RG. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204(1):453–7. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]