Abstract
In addition to its central role in cellular stress signaling, the tumor suppressor p53 modulates mitochondrial respiration through its nuclear transcription factor activity and localizes to mitochondria where it enhances apoptosis and suppresses mitochondrial DNA (mtDNA) mutagenesis. Here we demonstrate a new conserved role for p53 in mtDNA copy number maintenance and mitochondrial reactive oxygen species (ROS) homeostasis. In mammals, mtDNA is present in thousands of copies per cell and is essential for normal development and cell function. We show that p53 null mouse and p53 knock-down human primary fibroblasts exhibit mtDNA depletion and decreased mitochondrial mass under normal culture growth conditions. This is accompanied by a reduction of the p53R2 subunit of ribonucleotide reductase mRNA and protein and of mitochondrial transcription factor A (mtTFA) at the protein level only. Finally, p53-depleted cells exhibit significant disruption of cellular ROS homeostasis, characterized by reduced mitochondrial and cellular superoxide levels and increased cellular hydrogen peroxide. Altogether, these results elucidate additional mitochondria-related functions for p53 and implicate mtDNA depletion and ROS alterations as potentially relevant to cellular transformation, cancer cell phenotypes, and the Warburg Effect.
Keywords: p53, mtDNA depletion, reactive oxygen species, mitochondrial transcription factor A, p53R2, cancer
1. Introduction
The human mitochondrial genome is a 16.5-kb circular molecule that is present in thousands of copies per cell in most tissues. Mitochondrial DNA (mtDNA) is maternally inherited and, because it encodes thirty-seven genes required for oxidative phosphorylation (OXPHOS), is essential for normal development and sustained function of cells and tissues throughout life [1]. Specifically, it encodes thirteen core subunits of four of the five OXPHOS complexes, two rRNA subunits of mitochondrial ribosomes, and twenty-two organelle-specific tRNAs. Mutations in mtDNA can affect the expression or function of individual OXPHOS subunits, if they occur in one of the protein-encoding genes, or can globally affect the OXPHOS system, if they occur in a tRNA, rRNA, or a non-coding control region for transcription or replication of the genome (e.g. the D-loop region). To date, hundreds of pathogenic mtDNA mutations have been identified that cause maternally inherited diseases, which, as a class, display unexpected tissue-specificity and complex genetics [2]. Somatic mutations in mtDNA also accumulate in tissues over time and have been implicated in aging and age-related pathology [3]. Finally, depletion of mtDNA is also pathogenic due to global disruption of mitochondrial gene expression and can result from inherited mutations in nuclear genes or represent a secondary consequence of other disease conditions and treatments [4–7].
Recent estimates indicate that the mammalian mitochondrial proteome comprises 1,130–1,500 proteins [8, 9]. Since mtDNA encodes only thirteen of these, the remaining majority are nuclear gene products that are translated in the cytoplasm and imported into the organelle. Consequently, mitochondrial dynamics is controlled by gene expression programs in the nucleus [10] and signaling pathways that relay information back and forth between these two organelles [11]. These two processes are essential to maintain cellular homeostasis and alter mitochondrial biogenesis and function in response to changing cellular needs or environmental conditions.
Mutations in nuclear genes that encode mitochondrial proteins cause mitochondrial diseases that are inherited in a Mendelian fashion. A subset of these involves factors that function directly in mtDNA expression and maintenance (e.g. mtDNA polymerase, Pol γ, and Twinkle helicase) [12]. In addition, nuclear mutations that affect factors that do not reside in mitochondria, but nonetheless perturb mitochondrial homeostasis or inter-organelle signaling pathways are likewise potentially pathogenic. A salient example of this type of mitochondrial genetic disruption is the identification of mutations in nuclear genes involved in deoxynucleotide metabolism as causative in mtDNA-depletion syndromes (i.e., diseases characterized by severely reduced mtDNA copy number in specific tissues) [13]. Recent efforts in this area have identified mutations in the p53-regulated subunit (p53R2) of ribonucleotide reductase (RNR) as causing a mtDNA-depletion syndrome [14]. This provided bona fide genetic proof in humans and mice for the previously proposed role for the RNR in mtDNA copy number regulation and stability in yeast and cultured human cells [15–19]. In this same vein, we recently reported that disruptions in RNR and associated mitochondrial perturbations may also play a role in the disease Ataxia-telangiectasia [4], representing one of the first examples of a disease that may involve altered nuclear-mitochondrial signaling.
The tumor suppressor p53 has documented roles in the response to DNA damage and cellular stress. In addition, p53 also triggers pro-oxidant genes and cell death pathways, presumably as a means to remove extensively damaged or genetically unstable cells that can contribute to cancer or other diseases. As a transcription factor, many of its effects are mediated through expression of target nuclear genes involved in the various processes it controls. However, effects of p53 that are independent of it transcription factor function have been uncovered, as have novel functions beyond its stereotypical safeguarding role [20]. For example, p53 has been implicated in regulating glucose metabolism and mitochondria respiration through expression of nuclear genes involved in glycolysis and cytochrome oxidase assembly [21]. In addition, p53 physically localizes to mitochondria where direct roles in apoptosis induction [22, 23], mtDNA stability and repair [24, 25], and mitochondrial transcription [26, 27] have been postulated. This includes reported physical interactions with Pol γ [24] and mitochondrial transcription factor A (mtTFA or Tfam) [28], proteins with documented roles in mtDNA replication/repair and mtDNA transcription/packaging, respectively. Therefore, the impetus for the current study was to examine the potential homeostatic role of p53 in maintaining mtDNA copy number in primary human and mouse cell lines in the absence of induced DNA damage.
2. Materials and Methods
2.1. Cell culture and retroviral shRNA
C57/B6 wild-type and p53 null mouse neonatal fibroblasts (MNFs) (provided by Douglas Brash and Patrick Rochette at Yale) were cultured in DMEM with 10% FCS. Primary human fibroblasts (GM07532) were purchased from Coriell Institute and grown in EMEM with 15% FCS. The scrambled and p53 shRNA retroviral constructs (provided by Kristin Yates and Daniel DiMaio at Yale) were used as described [29]. Primary human fibroblasts were infected with each retrovirus for 24 hours in the presence of 4 μg/ml polybrene, followed by a 4-day selection in puromycin (0.6 μg/ml).
2.2. Mitochondrial DNA copy number assay
Total cellular DNA was extracted from 1×105 cells as described [30] and resuspended in 20–50 μl of TE (pH 8.0). A quantitative, real-time PCR method was used to determine the relative abundance of mtDNA versus nuclear 18S rDNA using mitochondrial and nuclear primer sets in two parallel PCR reactions as described previously [4]. Each 25 μl PCR reaction contained 14 μl of SYBR Green Mix (for final concentrations in reaction: 5% PCR grade DMSO [Sigma], 1:10,000 dilution of SYBR Green I [Molecular Probes], 0.3% Tween 20, 20mM Tris pH 8.3, 0.04% gelatin [Sigma], 50 mM KCl, 3 mM MgCl2, 200 mM dNTPs, 10 nM fluorescein, 0.03 units Taq polymerase), 0.5 μl of each 25 μM primer, and 10 μl of a 1:20–1:100 dilution of DNA in water. The reactions were performed using a BioRad iCycler and analyzed using iCycler version 3.1 software. Relative mtDNA copy number was calculated as the ratio of the amount of amplification obtained with mtDNA versus nuclear 18S rDNA primer sets for each sample and plotted normalized to the control group.
2.3. Western blotting
Western blotting was performed as described previously [4]. Briefly, 50 μg of total cell protein were separated on 12% bis-acrylamide gels and transferred to nitrocellulose membrane, followed by overnight incubation with primary antibodies diluted in 5% milk in TBST. Commercially available antibodies used were obtained from the following sources: p53R2 (C-18), and p53 (FL-393) antibodies, Santa Cruz; R1 (H-300), Chemicon; VDAC1/porin, Abcam; and actin (20–33), Sigma. The rabbit polyclonal h-mtTFA antibody was a gift from David Clayton. Following incubation with HRP-conjugated secondary antibodies, Western Lightning ECL reagent (Perkin Elmer) was used to detect signal on extra-sensitive film (GE Healthcare). Exposures were adjusted to ensure the signals were in a linear range.
2.4 Mitochondrial membrane potential, mitochondrial mass and ROS detection
For FACS analysis, 1×105 cells were stained in PBS with the indicated fluorescent dye for 25 minutes, washed, and resuspended in 200 μl PBS with 1% FCS. Individual cellular fluorescence signals were then analyzed using a FACSCalibur (BD Biosciences). Mitochondrial membrane mass and mitochondrial potential were measured by staining with MitoTracker Green FM (60 nM; Invitrogen) and MitoTracker Red (80 nM; Invitrogen), respectively, for 25 minutes. Dihydrofluorescein diacetate (10 μg/ml) was used to stain for total cellular hydrogen peroxide, dihydroethidium (80 μM) for cellular superoxide, and MitoSox Red Red (5 μM; Invitrogen) for mitochondrial superoxide. Antimycin A treatment was performed by treating cells with vehicle (ethanol) or 20 μM antimycin A (250μM stock in ethanol) for 20 minutes followed by staining with MitoSox Red. Unstained cells were analyzed as controls and used to gate on live cells for final analysis using FlowJo software (TreeStar, Ashland, OR). Bar graphs show combined median fluorescence values of three independent cultures, with at least 3,000 cells analyzed for each. Histograms show the fluorescence-intensity distribution of cells as a percentage of total gated cells (% max).
2.5 Determination of p53R2 and h-mtTFA mRNA transcript levels
Cells were seeded at 1×105 cell per T150 flask and grown for 5 days to mid-confluence with one change of media. Cells were harvested and homogenized using QIAshredder (Qiagen). RNA was extracted using RNeasy Plus Mini Kit (Qiagen) and eluted with 30 μl of RNase-free water. 8 μg of total RNA were converted into gene-specific cDNA using M-MuLV Reverse Transcriptase (New England BioLabs) according to the manufacturer’s protocol and 1 μM of reverse primer for actin, p53R2 and mtTFA (sequence below). cDNA was purified using QIAquick PCR purification Kit (Qiagen), eluted with 10 mM Tris (pH 8.5), dialyzed, diluted 1:10 and 1:20 in water and analyzed by real-time PCR using the SYBR Green I protocol described above and primers listed below. Ct values for p53R2 and h-mtTFA were normalized to actin Ct values using the equation 2ΔCt and are reported relative to the mean of the scrambled group. Primers used for cDNA conversion and real-time PCR were: actin, forward: 5′-TGGCACCACACCTTCTACAATGAGC-3′; actin, reverse: 5′-GCACAGCTTCTCCTTAATGTCACGC-3′; p53R2, forward: 5′-GTTCCAGAGGCTCGCTGTTTCTATG-3′; p53R2, reverse: 5′-TGATCTCCCTGACCCTTTCTTCTG-3′; h-mtTFA, forward: 5′-AGGGCGGAGTGGCAGGTATATAAA-3′; h-mtTFA, reverse: 5′-CGACGTAGAAGATCCTTTCGTCCAAC-3′.
2.6 Data analysis and statistics
The bars in the graphs represent at least three biological replicates with error bars denoting the standard error of the mean. In all graphs, values for both the experimental (i.e. p53 null or p53-shRNA-expressing) and the control (i.e. wild-type or scrambled-shRNA-expressing) groups were normalized to the mean of the values obtained for the controls, so that the mean of the control group in each experiment is always set to 1. All experiments were repeated at least twice and representative experiments are shown. P-values were calculated based on un-paired Student’s t-tests, and are denoted in the graphs as follows: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
3. Results
3.1. p53 deficiency causes mtDNA depletion in primary mouse and human fibroblasts that is accompanied by reduced mitochondrial membrane potential and mass
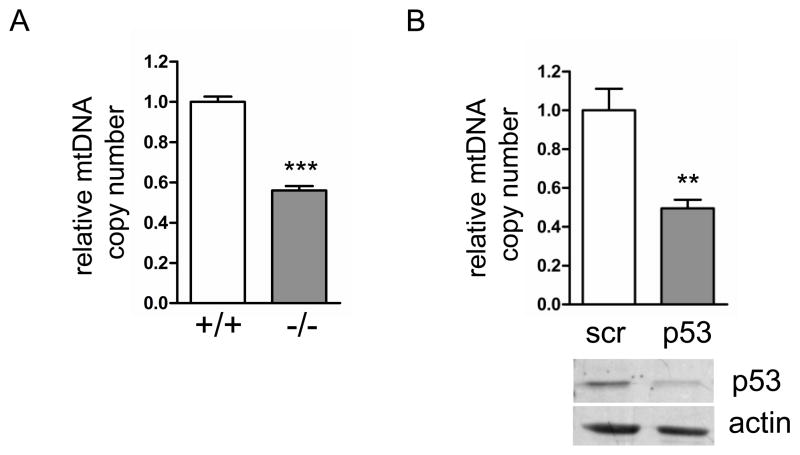
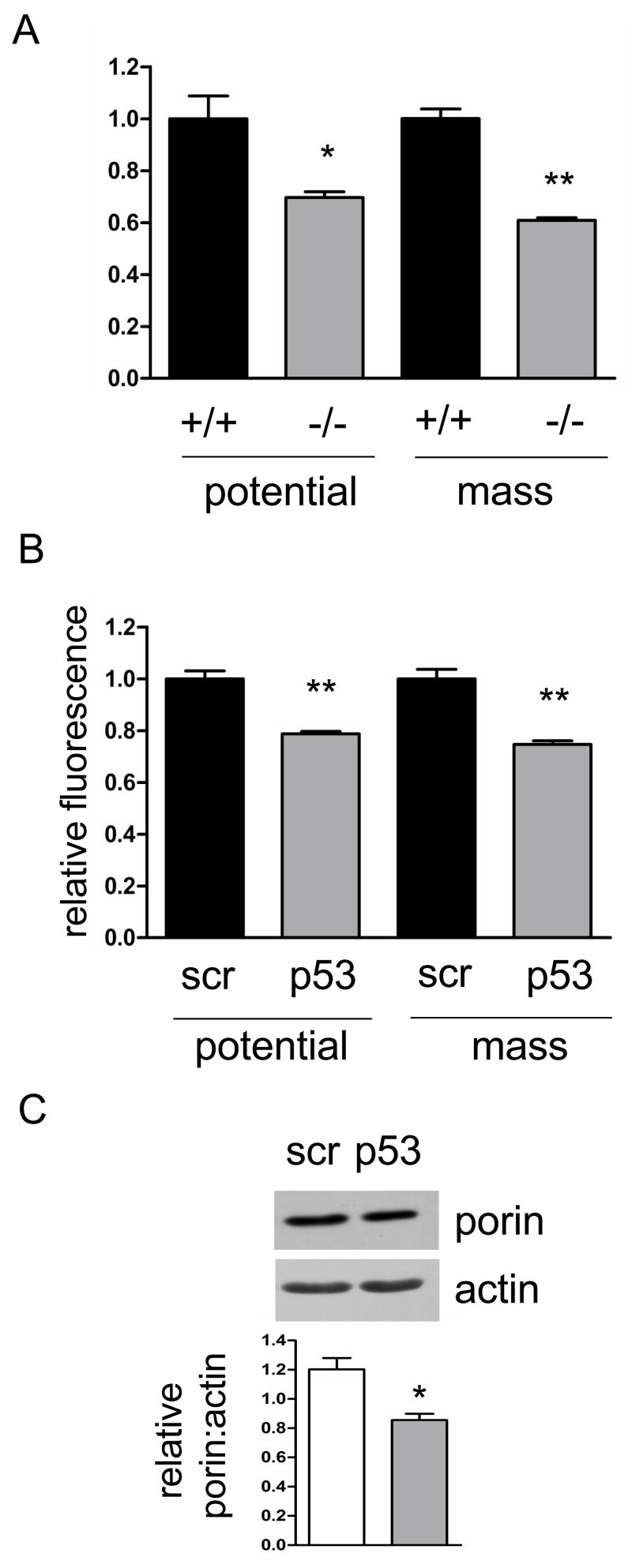
Based on our previous study [4] showing that the ATM pathway is essential for maintaining normal mtDNA copy number and mitochondrial homeostasis, we hypothesized that p53, one of the downstream targets of ATM kinase, may have a similar function. To test the potential role of p53 in mtDNA replication and stability, we first examined mtDNA copy number in mouse neonatal fibroblasts (MNFs) derived from p53 null and wild-type mice. The p53 null cells showed a 50% reduction in mtDNA copy number compared to isogenic wild-type cells, consistent with p53 playing a role in maintaining mtDNA (Fig. 1A). To address whether this effect on mtDNA copy number has a bearing on mitochondrial function and biogenesis in these cells, we measured mitochondrial membrane potential and mass using MitoTracker dyes and FACS analysis. Consistent with the mtDNA copy number results, p53 null cells showed a 30% reduction in total mitochondrial membrane potential and a 40% reduction in mitochondrial mass (Fig. 2A), again implicating p53 in promoting normal mitochondrial function.
Fig. 1.
Depletion of mtDNA in p53 null mouse neonatal fibroblasts and p53 knockdown primary human fibroblasts. A. Relative mtDNA copy number (determined by quantitative real-time PCR) in wild-type (+/+) and p53 null (−/−) MNFs. B. Primary human fibroblasts expressing scrambled control (scr) or p53 shRNA (p53) were analyzed for relative mtDNA copy number (graph) and primary p53 levels (Western blot).
Figure 2.
p53 null MNFs and human primary fibroblasts treated with p53 shRNA show mitochondrial membrane potential and mass defects. Mitochondrial membrane potential and mass measured by FACS in A. wild-type (+/+) and p53 null (−/−) MNFs; and B. primary human fibroblasts expressing scrambled control (scr) or p53 shRNA (p53). C. Western blot of porin as a mitochondrial marker with actin as a loading control. Quantification of porin signal normalized to actin is shown (graph) for scrambled control (scr) or p53 shRNA (p53) expressing primary human fibroblasts.
To test if this novel mitochondrial homeostasis function of p53 is conserved in human cells, and also to confirm that the mitochondrial defects observed in murine cells were a direct consequence of p53 loss, we depleted p53 in primary human fibroblasts using shRNA that was introduced via retroviral transduction. The levels of p53 were reduced by at least half in the p53-shRNA expressing cells, which also exhibited a 50% depletion of mtDNA compared to those transduced identically with a scrambled shRNA negative control (Fig. 1B). Mitochondrial membrane potential and mass were also decreased in the human p53 knock-down cells by 20–30% (Fig. 2B), indicating that the decrease in mitotracker red staining is most likely simply a reflection of less mitochondrial mass, rather than a down-regulation of membrane potential per se. The decrease in mitochondrial biogenesis observed via mitotracker staining was confirmed by a similar reduction in the mitochondrial protein marker porin in whole-cell lysates relative to actin (a cytoplamic marker)(Fig. 2C). These data largely confirm our findings in primary mouse cells (Fig 1A, 2A) and support a conserved role for p53 in maintaining mtDNA copy number and mitochondrial abundance.
3.2. Reduced p53 expression leads to down-regulation of h-mtTFA and the p53R2 subunit of ribonucleotide reductase
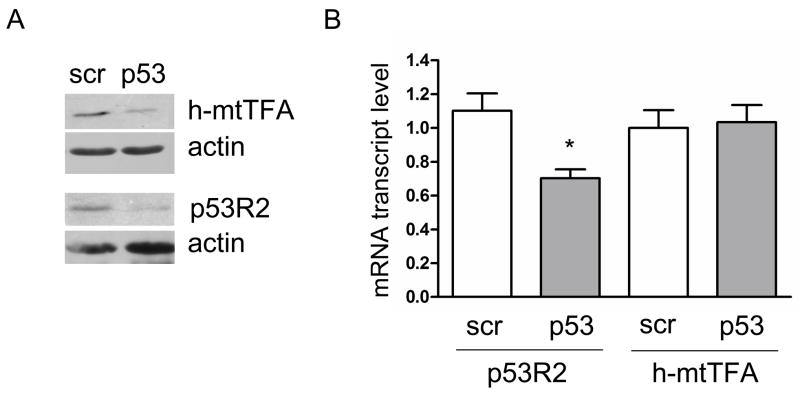
To begin to gain a mechanistic perspective on the mtDNA depletion observed in p53 deficient cells, we examined two factors previously implicated in mtDNA copy number homeostasis, the mitochondrial transcription and mtDNA-packaging factor, h-mtTFA/Tfam, and the p53-regulated subunit of RNR, p53R2. The steady-state level of both of these proteins was significantly reduced in the p53-shRNA cell lines compared to the negative control lines (Fig. 3A). The amounts of the R1 and R2 subunits in the p53-depleted cells were unchanged and modestly elevated, respectively (data not shown). The mRNA levels of p53R2 were reduced by ~35%, while h-mtTFA transcript levels were unchanged in p53 knockdown cells (Fig. 3B), suggesting that p53R2 is likely down-regulated at the transcriptional level, while h-mtTFA is down-regulated post-transcriptionally.
Figure 3.
p53 knockdown primary human fibroblasts have reduced levels of p53R2 and h-mtTFA. Cells expressing scrambled control (scr) or p53 shRNA (p53) analyzed for A. steady-state levels of p53R2 and h-mtTFA by Western blot (actin is shown as a loading control); and B. relative mRNA transcript levels of p53R2 and mtTFA normalized to actin message.
3.3. p53 has a pro-oxidant role in mitochondria and is required for cellular ROS homeostasis
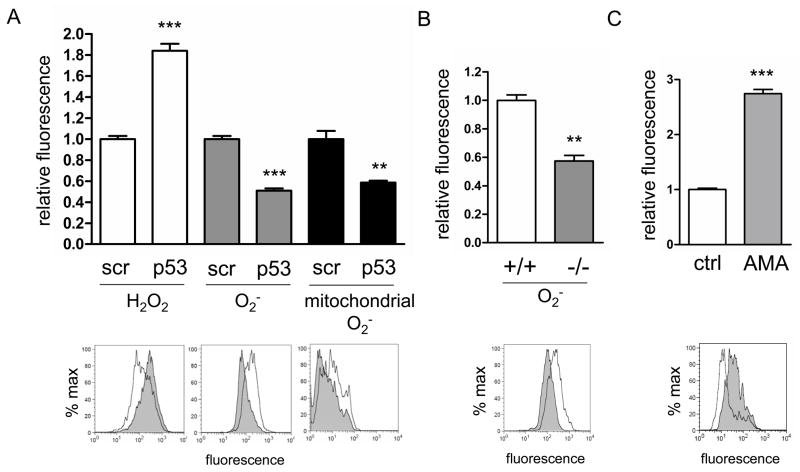
Certainly one of the predicted consequences of disrupted mitochondrial homeostasis is altered ROS production. In addition, p53 has been implicated previously in both anti- and pro-oxidant functions [20]. Therefore, we examined mitochondrial and cellular ROS levels in p53-shRNA primary human fibroblasts, using fluorescent ROS dyes MitoSox Red, dihydrofluorescein diacetate, and dihydroethidium and FACS analysis. In the p53-depleted cells, we observed a significant increase in cellular hydrogen peroxide levels, and reduced cellular and mitochondrial superoxide levels (Fig. 4A). We also observed reduced mitochondrial superoxide in p53 null MNFs (Fig. 4B). MitoSox Red is a hydroethidine derivative that is oxidized by superoxide to ethidium, which becomes highly fluorescent when intercalated into DNA. To rule out that reduced MitoSox Red fluorescence we observed in p53 deficient cells was due to lower mtDNA content in these cells, we treated p53 knockdown cells with antimycin A for 20 minutes to induce mitochondrial ROS [31] without increasing mtDNA copy number. Antimycin A treatment increased MitoSox Red signal in p53 knockdown cells (Fig 4C), confirming that mtDNA is not limiting for MitoSox Red staining in p53 deficient cells. These results indicate that in primary cells in culture p53 have a pro-oxidant role in mitochondria with regard to superoxide, but both pro-and anti-oxidant effects in the cell as a whole.
Figure 4.
Loss of p53 results in reduced mitochondrial superoxide and disrupted cellular ROS homeostasis. A. FACS analysis of cellular hydrogen peroxide (H2O2), cellular superoxide (O2−) and mitochondrial superoxide levels in primary human fibroblasts depleted of p53 by shRNA (p53) compared to those treated with the negative scrambled control shRNA (scr). Histograms of representative experiments for scrambled (black line) and p53 shRNA (gray shading) treated cells are shown underneath the corresponding bars in the graph. B. FACS analysis of cellular superoxide in wild-type (+/+) and p53 null (−/−) MNFs. The histogram shows a representative experiment with wild-type (black line) and p53 null (gray shading) MNFs. C. FACS analysis of p53 knockdown human primary fibroblasts treated with vehicle (ctrl) or antimycin A (AMA) and stained with MitoSox Red. The histogram shows a representative experiment with vehicle (black line) and antimycin A (gray shading) treated cells.
4. Discussion
There are three major outcomes of this study. First, primary mouse and human cells that lack or have reduced levels of the tumor suppressor p53, respectively, have depleted amounts of mtDNA that is accompanied by a parallel reduction in mitochondrial mass (Figs. 1 and 2). However, in terms of magnitude, there was not consistent one-to-one relationship between these parameters, suggesting that mtDNA copy number and mitochondrial biogenesis are not necessarily tightly linked. That is the mtDNA depletion was always greater in magnitude than the changes in mitochondrial mass indicating a larger role for p53 in mtDNA maintenance specifically. The observed decrease in mitochondrial mass without a compensatory increase in mitochondrial membrane potential (Fig. 2) indicates a reduced overall mitochondrial oxidative capacity in p53 deficient cells, consistent with reported decreases in respiration [32]. Second, p53-dependent mtDNA depletion was also accompanied by a reduction in mtTFA and p53R2, two factors that have been linked previously to mtDNA copy number maintenance [1]. And third, that there is a significant reduction of mitochondrial and total cellular superoxide levels when p53 is down-regulated, but an increase in cellular hydrogen peroxide, which points to complex regulation of cellular ROS homeostasis by p53. The fact that all of these changes are occurring in primary, non-transformed cells and in the absence of induced DNA-damage, implicates mtDNA depletion and perturbed mitochondrial ROS homeostasis as potential novel factors that promote p53-mediated cancer cells phenotypes. These results and conclusions are discussed further below.
The mechanism of mtDNA depletion accompanying the loss of p53 we report herein is worthy of discussion, as there are at least three plausible explanations that are not mutually exclusive (Fig. 5). First, the reduced expression of p53R2 is likely to be in part responsible for the mtDNA depletion (Fig. 3). This protein is a direct transcriptional target of p53 in the presence of DNA damage [33, 34] and inactivating mutations in its gene cause mtDNA depletion in humans and mice [14]. We observed that p53R2 was down-regulated at both the mRNA transcript and protein levels (Fig. 3), suggesting that p53R2 is a transcriptional target of p53 even in the absence of DNA damage. Reduced expression of p53R2 is expected to lead to diminished amounts of R1-p53R2 complexes of RNR (Fig. 5), the form of the enzyme that has been implicated specifically in providing dNTPs for mtDNA replication and repair [18, 19], and result in mtDNA depletion. Consistent with this premise is the fact that disruptions in RNR have also been implicated as a potential cause of mtDNA depletion in cultured Ataxia-telangiectasia patient fibroblasts [4]. A second possibility is that it is the down-regulation of mtTFA in the absence of p53 that we report here (Fig. 3A) that drives mtDNA depletion (Fig. 5). The loss of mtTFA expression is reported to cause mtDNA depletion [35, 36], but its protein levels also passively mirror those of mtDNA when mtDNA is depleted by treatment with ethidium bromide or inhibitors of Pol γ[37, 38]. Since we observed no change in the transcript levels of mtTFA (Fig. 3B), we conclude that its reduction at the protein level in p53 depleted cells is most likely due to its degradation in response to reduced mtDNA levels (i.e. that it is unstable when not bound to mtDNA) or some other form of post-transcriptional regulation. This instability may be driven by loss of its proposed physical interaction with p53 in mitochondria [28]. Given that p53 is physically located in mitochondria [39–41], it is also possible that loss of it in the organelle per se is the cause of the observed mtDNA depletion (Fig. 5). This could be due to abrogation of proposed interactions with mtTFA [28] and or mtDNA Pol γ[24] needed for efficient mtDNA replication or repair. It is also possible that p53 works as a transcription factor in mitochondria [26, 27] and hence its absence leads to reduced transcription-primed mtDNA replication [42]. However, this latter possibility seems the least likely given recent reports that p53 in mitochondria appears to act as a monomer, which is not its typical transcription mode [43]. Deciphering which of these pathways is primarily responsible for the mtDNA depletion in the absence of p53 is an intriguing area ripe for future investigation.
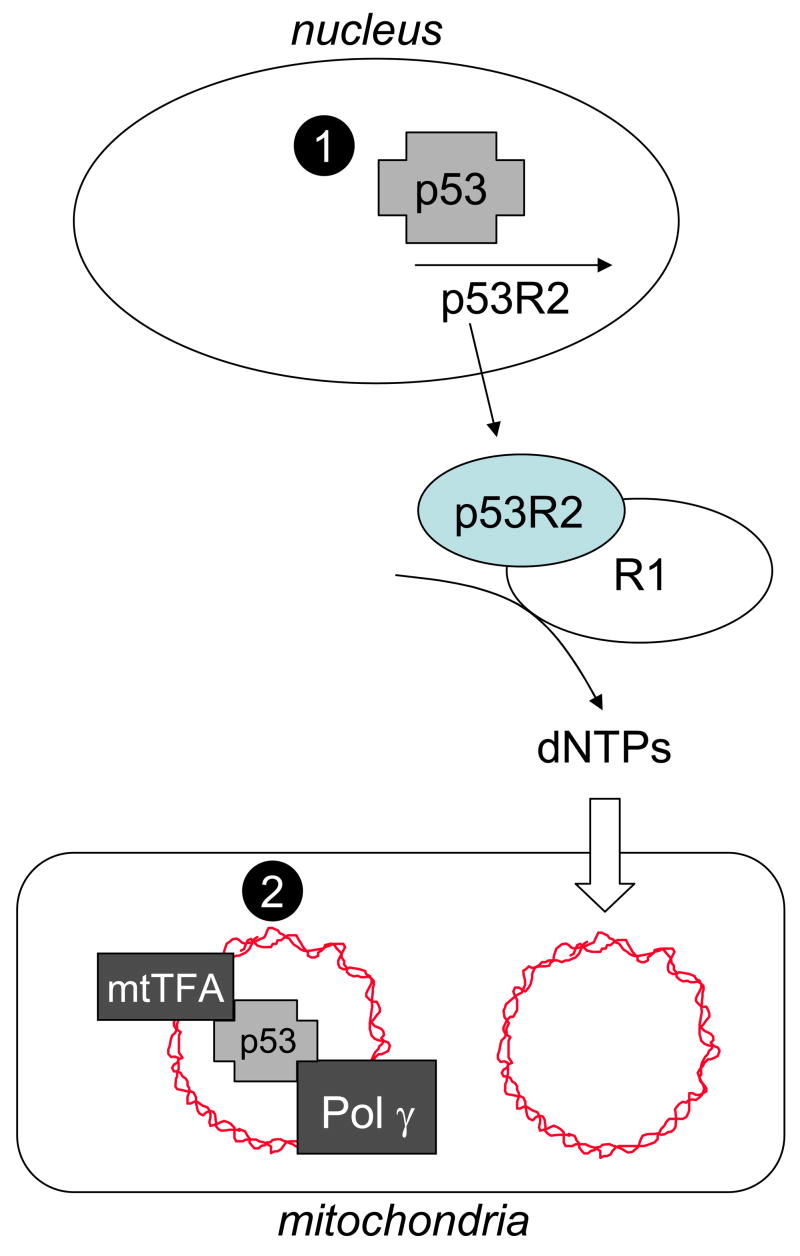
Figure 5.
Schematic representation of potential nuclear and mitochondrial functions of p53 in mtDNA copy number maintenance and how loss of such functions could lead to mtDNA depletion. At the top of the figure nuclear p53 is shown activating its known target gene p53R2, which we propose occurs at some level even in the absence of DNA damage. At the bottom of the figure, mitochondrial p53 is shown associated with mtDNA (ragged circle) and its putative binding partners, mtTFA and Pol γ, that are each required for mtDNA replication and maintenance. Loss of p53 is postulated to cause mtDNA depletion by one or more mechanisms (see text for details), that involve (1) its nuclear function as a transcription factor, or (2) its mitochondrial functions. Lack of p53 in the nucleus results in reduced expression of p53R2, dampened output of dNTPs by the p53R2-R1 form of RNR (ovals at center right) and compromised mtDNA replication or repair that causes mtDNA depletion. Reduced mtDNA levels could lead to reduced levels of mtTFA protein if DNA binding is required for its stability. Lack of p53 in mitochondria could alone cause mtDNA depletion if in fact p53 binds and stabilizes mtTFA or is needed for optimal activity of Pol γ in mtDNA replication or repair.
Our results also point to a novel role for p53 in promoting mitochondrial ROS. That is, loss of p53 results in decreased mitochondrial superoxide levels (Fig. 3A). This effect could be mediated through the rate of production of ROS by mitochondria or by disruption of normal pro- or anti-antioxidant pathways. The antioxidant function of p53 is well-established and involves both its ability to regulate the sestrins [44], anti-oxidant proteins responsible for regenerating oxidized peroxiredoxins, and its diversion of glucose to the pentose phosphate pathway to generate NADPH needed for enzymes involved in antioxidant defenses [45]. In addition, manganese superoxide dismutase (MnSOD), the mitochondrial enzyme responsible for the conversion of superoxide to hydrogen peroxide, is also affected by p53 status, although reports on the nature of this regulation are conflicting. One study shows that p53 positively regulates MnSOD in Li-Fraumeni patient fibroblasts and lymphoblasts [46], while others show that p53 represses the MnSOD promoter in a human carcinoma cell line [47] or reduces MnSOD levels when introduced into HeLa cells [48]. Consistent with p53 having an inhibitory effect on MnSOD, the physical binding of p53 to MnSOD in mitochondria has also been reported to decrease MnSOD activity [49]. Our results are most consistent with an inhibitory role for p53 on MnSOD, because, in this case, loss of p53 would result in increased MnSOD activity that would account for the lowered mitochondrial superoxide we observe in p53 deficient cells (Fig. 4A, B). In addition, increased MnSOD activity in the absence of p53 is predicted to generate increased levels of hydrogen peroxide that would add to that already culminating from the down-regulation of the sestrins. This scenario would explain the higher levels of cellular hydrogen peroxide we observe in p53-shRNA expressing primary human cells (Fig. 4A). In summary, our results point to the importance of p53 in maintaining ROS homeostasis, but show that its effects vary dramatically depending on the specific ROS species and where they are generated. Of course, in addition to imbalances of antioxidant systems described above, the decreased electron flux through the OXPHOS chain reported for p53 null cells [32] could certainly also contribute to the observed decrease in mitochondrial ROS we observe. In fact, it is probably a combination of both factors that is leading to the particular cellular ROS profiles we describe herein (Fig. 4).
Given the key role loss of p53 plays in cancer, our results are likely relevant in this regard. One key feature of many cancer cells is increased dependence on glycolysis for ATP production even in the presence of oxygen, when mitochondrial OXPHOS should more significantly contribute. This switch in metabolic behavior to “aerobic glycolysis” was postulated by Warburg [50] to be driven by mitochondrial dysfunction. While the precise mechanism of the “Warburg Effect” remains unknown, recent studies of novel p53 targets may have provided some insight. For example, the loss of p53 has been reported to result in decreased OXPHOS Complex IV activity and mitochondrial oxygen consumption, due to reduced expression of a cytochrome c oxidase assembly factor Sco2, that is a nuclear p53 target [32]. This was postulated by Hwang and colleagues to be a major mechanism driving the Warburg Effect [21] that could work in combination with up-regulation of glycolysis mediated by loss of expression of other p53 targets (e.g. TIGAR [45]). This study expands the list of specific mitochondrial defects that may be relevant to the mitochondrial dysfunction involved in the Warburg Effect or in other aspects of cancer progression to include mtDNA depletion and altered mitochondrial and cellular ROS. For example, mtDNA depletion could predispose mtDNA to instability and mutagenesis [24] that could subsequently alter mitochondrial function in unique ways that allow cellular transformation. Also, since ROS are increasingly implicated in signal transduction processes [51] alterations of mitochondrial ROS could impact signaling pathways that promote cancer development. Determining the full range of mitochondrial perturbations that contribute to cancer remains an important goal.
5. Conclusion
In this study we have expanded our understanding of how p53 impacts mitochondrial homeostasis. In addition to direct effects on respiration reported by others, we show here that p53 is required to maintain normal mtDNA copy number and biogenesis and both mitochondrial and cellular ROS homeostasis. These newly assigned functions for p53 are likely involved in the important “gate-keeping” function of p53 in cellular surveillance and relevant to how loss of its functions promotes alterations in metabolism and tumorigenesis.
Acknowledgments
This work was supported by NIH grant NS056206 from the National Institute of Neurological Disorders and Stroke awarded to G.S.S, and M.A.L. was supported by NIH training grant T32GM007223 from the National Institute of General Medical Sciences. The authors wish to thank Douglas Brash and Patrick Rochette for providing wild-type and p53 null MNFs, Daniel DiMaio and Kristin Yates for reagents and advice on retroviral shRNA, and David Clayton for the h-mtTFA antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. The American journal of pathology. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan KJ, Greaves LC, Reeve AK, Turnbull D. The ageing mitochondrial genome. Nucleic acids research. 2007;35:7399–7405. doi: 10.1093/nar/gkm635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. The Journal of Clinical Investigation. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis W. Cardiomyopathy, nucleoside reverse transcriptase inhibitors and mitochondria are linked through AIDS and its therapy. Mitochondrion. 2004;4:141–152. doi: 10.1016/j.mito.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Elpeleg O, Mandel H, Saada A. Depletion of the other genome-mitochondrial DNA depletion syndromes in humans. J Mol Med. 2002;80:389–396. doi: 10.1007/s00109-002-0343-5. [DOI] [PubMed] [Google Scholar]
- 7.Hakonen AH, Goffart S, Marjavaara S, Paetau A, Cooper H, Mattila K, Lampinen M, Sajantila A, Lonnqvist T, Spelbrink JN, Suomalainen A. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Human molecular genetics. 2008 doi: 10.1093/hmg/ddn280. [DOI] [PubMed] [Google Scholar]
- 8.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiological reviews. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 11.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Molecular cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 12.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annual review of medicine. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saada A. Deoxyribonucleotides and disorders of mitochondrial DNA integrity. DNA and cell biology. 2004;23:797–806. doi: 10.1089/dna.2004.23.797. [DOI] [PubMed] [Google Scholar]
- 14.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nature genetics. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 15.O’Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene. 2005;354:86–92. doi: 10.1016/j.gene.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 16.Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O’Rourke W, Siede TW, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Molecular biology of the cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebedeva MA, Shadel GS. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA Inheritance without compromising mitochondrial gene expression. Cell cycle (Georgetown, Tex. 2007;6:2048–2057. doi: 10.4161/cc.6.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. The Journal of biological chemistry. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 19.Pontarin G, Ferraro P, Hakansson P, Thelander L, Reichard P, Bianchi V. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. The Journal of biological chemistry. 2007;282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 20.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends in cell biology. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. Journal of bioenergetics and biomembranes. 2007;39:243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 22.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science (New York, NY) 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 23.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Molecular and cellular biology. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. The EMBO journal. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D, Yu Z, Zhu Z, Lopez CD. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer research. 2006;66:3485–3494. doi: 10.1158/0008-5472.CAN-05-4103. [DOI] [PubMed] [Google Scholar]
- 26.Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS letters. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 27.Donahue RJ, Razmara M, Hoek JB, Knudsen TB. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. Faseb J. 2001;15:635–644. doi: 10.1096/fj.00-0262com. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer research. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 29.Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging cell. 2008;7:609–621. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1989. [Google Scholar]
- 31.Turrens JF. Mitochondrial formation of reactive oxygen species. The Journal of physiology. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science (New York, NY. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 34.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 35.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature genetics. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 36.Goto A, Matsushima Y, Kadowaki T, Kitagawa Y. Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. The Biochemical journal. 2001;354:243–248. doi: 10.1042/0264-6021:3540243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic acids research. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulton J, Morten K, Freeman-Emmerson C, Potter C, Sewry C, Dubowitz V, Kidd H, Stephenson J, Whitehouse W, Hansen FJ, et al. Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Human molecular genetics. 1994;3:1763–1769. doi: 10.1093/hmg/3.10.1763. [DOI] [PubMed] [Google Scholar]
- 39.Merrick BA, He C, Witcher LL, Patterson RM, Reid JJ, Pence-Pawlowski PM, Selkirk JK. HSP binding and mitochondrial localization of p53 protein in human HT1080 and mouse C3H10T1/2 cell lines. Biochimica et biophysica acta. 1996;1297:57–68. doi: 10.1016/0167-4838(96)00089-1. [DOI] [PubMed] [Google Scholar]
- 40.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. The Journal of biological chemistry. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 41.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. The EMBO journal. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Molecular cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 43.Heyne K, Schmitt K, Mueller D, Armbruester V, Mestres P, Roemer K. Resistance of mitochondrial p53 to dominant inhibition. Molecular cancer. 2008;7:54. doi: 10.1186/1476-4598-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nature medicine. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 46.Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer research. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 47.Drane P, Bravard A, Bouvard V, May E. Reciprocal down-regulation of p53 and SOD2 gene expression-implication in p53 mediated apoptosis. Oncogene. 2001;20:430–439. doi: 10.1038/sj.onc.1204101. [DOI] [PubMed] [Google Scholar]
- 48.Pani G, Bedogni B, Anzevino R, Colavitti R, Palazzotti B, Borrello S, Galeotti T. Deregulated manganese superoxide dismutase expression and resistance to oxidative injury in p53-deficient cells. Cancer research. 2000;60:4654–4660. [PubMed] [Google Scholar]
- 49.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer research. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 50.Warburg O. On respiratory impairment in cancer cells. Science (New York, NY. 1956;124:269–270. [PubMed] [Google Scholar]
- 51.Finkel T. Oxidant signals and oxidative stress. Current opinion in cell biology. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]