Abstract
The human ZC3H14 gene encodes an evolutionarily conserved Cys3His zinc finger protein that binds specifically to polyadenosine RNA and is thus postulated to modulate post-transcriptional gene expression. Expressed sequence tag data predicts multiple splice variants of both human and mouse ZC3H14. Analysis of ZC3H14 expression in both human cell lines and mouse tissues confirms the presence of multiple alternatively spliced transcripts. Although all of these transcripts encode protein isoforms that contain the conserved C-terminal zinc finger domain, suggesting that they could all bind to polyadenosine RNA, they differ in other functionally important domains. Most of the alternative transcripts encode closely related proteins (termed isoform 1, 2, 3, and 3short) that differ primarily in the inclusion of three small exons, 9, 10, and 11, resulting in predicted protein isoforms ranging from 82 to 64 kDa. Each of these closely related isoforms contains predicted classical nuclear localization signals (cNLS) within exons 7 and 11. Consistent with the presence of these putative nuclear targeting signals, these ZC3H14 isoforms are all localized to the nucleus. In contrast, an additional transcript encodes a smaller protein (34 kDa) with an alternative first exon (isoform 4). Consistent with the absence of the predicted cNLS motifs located in exons 7 and 11, ZC3H14 isoform 4 is localized to the cytoplasm. Both EST data and experimental data suggest that this variant is enriched in testes and brain. Using an antibody that detects endogenous ZC3H14 isoforms 1-3 reveals localization of these isoforms to nuclear speckles. These speckles co-localize with the splicing factor, SC35, suggesting a role for nuclear ZC3H14 in mRNA processing. Taken together, these results demonstrate that multiple transcripts encoding several ZC3H14 isoforms exist in vivo. Both nuclear and cytoplasmic ZC3H14 isoforms could have distinct effects on gene expression mediated by the common Cys3His zinc finger polyadenosine RNA binding domain.
Keywords: mRNA processing/export, poly(A) binding proteins, S. cerevisiae Nab2, nuclear speckle, alternative splicing, post-transcriptional regulation of gene expression
1. Introduction
The function and fate of cells is dictated by the genes that are expressed at any given time. Thus control of gene expression is critical to cellular function and identity. Although transcriptional control is a key point for regulation of gene expression, there is an increasing appreciation of how extensively gene expression is modulated by post-transcriptional mechanisms including mRNA processing, export, stability, and translation (Moore, 2005). A large number of RNA binding proteins and associated factors contribute to these post-transcriptional regulatory mechanisms (Mangus et al., 2003; Moore, 2005; Lunde et al., 2007).
One family of proteins that are key post-transcriptional regulators of gene expression is composed of the poly(A) binding proteins (Pabs) that associate with the poly(A) tail of mRNA transcripts (Mangus et al., 2003). Functional studies in a wide variety of organisms ranging from yeast to humans have demonstrated that members of this evolutionarily conserved protein family (Mangus et al., 2003; Gorgoni and Gray, 2004; Kuhn and Wahle, 2004) directly contact the poly(A) tail of mRNA transcripts to regulate transcript polyadenylation (Amrani et al., 1997; Kerwitz et al., 2003), translation (Sachs and Davis, 1989; Tarun and Sachs, 1996; Le et al., 1997; Gallie, 1998; Kahvejian et al., 2005), stability (Caponigro and Parker, 1995; Caponigro and Parker, 1996), and possibly nuclear export (Brune et al., 2005; Dunn et al., 2005).
All conventional Pab family members specifically bind to poly(A) RNA via at least one RNA Recognition Motif (RRM) (Adam et al., 1986; Sachs et al., 1987). However, recent work reveals that evolutionarily conserved Cys3His zinc finger proteins can also mediate specific binding to polyadenosine RNA (Kelly et al., 2007) increasing the number of Pabs defined in eukaryotic cells. These zinc finger proteins are represented by the abundant nuclear poly(A) binding (Nab2) protein in budding yeast and the human Cys3His Zinc Finger Protein #14 (ZC3H14). Functional studies of Nab2 have revealed that this essential protein is required both for control of poly(A) tail length in the nucleus and proper mRNA export (Anderson et al., 1993; Green et al., 2002; Hector et al., 2002). In contrast, there has been a single in vitro study that characterized the RNA binding properties of ZC3H14 (Kelly et al., 2007). We have previously demonstrated that a ZC3H14-GFP fusion protein is localized to the nucleus and that the zinc finger domain of ZC3H14 binds specifically to polyadenosine RNA in vitro (Kelly et al., 2007). One additional study reported that ZC3H14, which was termed NY-Ren-37, was one of a number of antigens that were present at high levels in clear cell renal carcinoma as compared to normal tissues (Scanlan et al., 1999), but this was a large scale analysis and no follow-up on NY-Ren-37/ZC3H14 has been reported. Thus, preliminary studies suggest that, like Nab2 in budding yeast, ZC3H14 may contribute to control of gene expression in human cells through binding poly(A) RNA.
Examination of the NCBI database suggests that ZC3H14 is subject to alternative splicing to create multiple protein isoforms. Here we confirm the existence of multiple splice variants of ZC3H14 that encode distinct protein isoforms in both human cell lines and mouse tissues. We provide evidence for both nuclear and cytoplasmic isoforms of ZC3H14. Importantly, an antibody that recognizes the nuclear isoforms of ZC3H14 reveals that these isoforms are concentrated within nuclear speckles that co-localize with the splicing factor, SC35. Thus, results of this study provide the first characterization of ZC3H14 expression and identify both nuclear isoforms of ZC3H14 implicated in mRNA processing and, rather surprisingly, a cytoplasmic isoform of ZC3H14 that has the potential to modulate gene expression in the cytoplasm.
2. Materials and Methods
2.1. Cell culture and transfection
HEK293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. DNA plasmids were transfected into cultured cells using Lipofectamine2000 (Invitrogen) according to manufacturer's protocol.
2.2. Plasmid constructs
GFP fusion plasmids were generated by amplifying the ZC3H14 coding region (ATCC image clone 4298961 for ZC3H14 isoform 1 and image clone 4828241BS for ZC3H14 isoform 4) and subcloning into pEGFP-N1 (Clontech) to create a GFP fusion at the C-terminus of each protein isoform. FLAG fusion constructs were generated using PCR primers that included the FLAG sequence, creating N-terminally FLAG tagged protein isoforms. PCR products were then subcloned into the pcDNA3.1 vector (Invitrogen).
2.3 Phylogenetic analysis
psiBLAST searches were performed to identify amino acid sequences similar to the Nab2 protein across eukaryotic species. The putative homologous proteins resulting from the BLAST search were aligned with the S. cerevisiae sequence of Nab2 by using ClustalW (Thompson et al., 1994; Larkin et al., 2007) and corrections made by eye. The gap stripped alignment was 108 amino acids long. The program employed to perform phylogenetic inference was Mr. Bayes (Huelsenbeck and Ronquist, 2001). For the phylogenetic inference, we estimated from the data the amino acid frequencies necessary for estimating the rate of change and we took into account the heterogeneity of substitution rates across sites in the sequence (Huelsenbeck and Ronquist, 2001). In order to provide the support for the different clades of the inferred topology, we estimated the posterior probabilities supporting each node. Only nodes with support larger than 70% (posterior probability larger than 0.7) are shown. The sequences used in the analysis and their GenBank accession numbers are shown in Table I.
Table I. Accession numbers for protein sequences used in the phylogenetic analysis shown in Figure 1A.
| Species | Accession numbers |
|---|---|
| Anopheles gambiae | XP_313187 |
| Bos taurus | XP_588816 |
| Pongo abelii | NP_001125818 |
| Homo sapiens | AAH23641 |
| Mus musculus | EDL18926 |
| Rattus norvegicus | EDL81693 |
| Macaca fascicularis | BAB64440 |
| Xenopus laevis | Q08AZ1 |
| Danio rerio | Q5TYQ8 |
| Tetraodon nigroviridis | CAG09129 |
| Drosophila melanogaster | AAY33497 |
| Caenorhabditis elegans | NP_504239 |
| Plasmodium chabaudi | XP_744461 |
| Dictyostelium discoideum | XP_646803 |
| Candida glabrata | XP_446277 |
| Saccharomyces cerevisiae | NP_011393 |
| Kluyveromyces lactis | XP_455528 |
2.4. ZC3H14 homology modeling
Homology models of ZC3H14 were constructed using Swiss-Model [version 8.05; (Arnold et al., 2006)] starting with the known S. cerevisiae Nab2 crystal structure [PDB entry 2V75; (Grant et al., 2008)]. ClustalW sequence alignments (Chenna et al., 2003) between Nab2 and ZC3H14 were manually adjusted to account for correct alignment of Leu 18 (Figure 1D) due to their conserved functional importance (Nykamp, 2003). The model was energy minimized in CNS 1.2 to remove any steric strain introduced during modeling (Brunger et al., 1998; Brunger, 2007). This model was visualized and protein backbone torsion angles (phi and psi) were verified in Coot (Emsley and Cowtan, 2004).
Figure 1. ZC3H14 is a conserved tandem zinc finger protein.
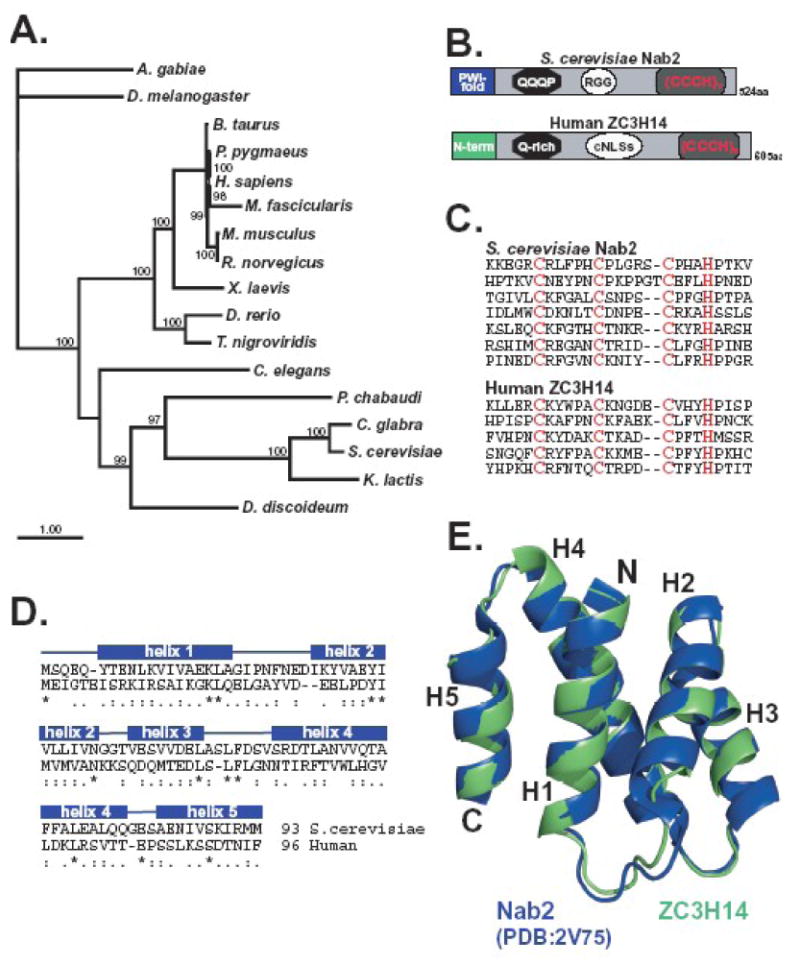
A. A Bayesian phylogenetic tree of proteins related to human ZC3H14 is shown. Nodes with posterior probabilities larger than 0.7 are indicated in the figure as percentages. The accession numbers corresponding to the sequences employed for the tree reconstruction are provided in Table I. B. The similarity in the arrangement of the domains within S. cerevisiae Nab2 and human ZC3H14 is illustrated. Both proteins contain N-terminal domains that assume (S. cerevisiae) or are predicted to assume (human) a proline-tryptophan-isoleucine (PWI-fold) followed by a glutamine-rich domain (QQQP) a nuclear targeting signal (RGG or cNLS) and finally tandem Cys3His (CCCH) zinc finger motifs. Human ZC3H14 has five Cys3His (CCCH) zinc fingers while Nab2 has seven. C. Alignment of the individual C-terminal zinc finger domains of S. cerevisiae Nab2 and human ZC3H14 illustrates the similarity in spacing between the cysteine and histidine residues. D. An alignment of the N-terminal domains of Nab2 (amino acids 1-93) and ZC3H14 (amino acids 1-96) used to generate the model in panel E is shown. Identical residues are indicated by the asterisks, very similar residues by the double dots, and similar residues by single dots. E. A homology model of the N-terminal domain (amino acids 1-96) of human ZC3H14 (green) overlaid with the N-terminal domain of S. cerevisiae Nab2 (PDB accession code 2V75; blue). Homology modeling was carried out as described in Materials and Methods. Molecular images were generated using PyMol (http://pymol.sourceforge.net).
2.5. RT-PCR
Total RNA was isolated from HEK293 cells or from male BALB/c mice tissues using TRIzol reagent (Invitrogen) as previously described (O'Connor et al., 2007). Briefly, the Expand High Fidelity PCR system (Roche) was used to amplify each of the ZC3H14 splice variants (see Figure 2A). The reverse transcriptase reaction was performed using 2.5 μg of total RNA in a 20 μl reaction and 1 μl of the resulting cDNA was used in the subsequent amplification step along with 2.5 μM of each primer. ZC3H14 primers were designed from GenBank accession number (NM_024824.3, NM_207660.2, NM_207661.2, NM_207662.2) to detect splice variants 1, 2, 3, and 4. To eliminate the possibility of genomic DNA contamination, ZC3H14 primers spanned exon-exon boundaries (see Figure 2A). Amplification consisted of 30 cycles (for isoforms 1, 2, and 3 as well as isoform 4 from HEK cells) or 60 cycles (for isoform 4 in mouse tissue) of 95°C for 30 s, 53°C for 30 s, 72°C for 30 s, and a final amplification step at 72°C for 5 min. As an internal control, either 18S cDNA was amplified with QuantumRNA 18S primers (Ambion) or GAPDH was amplified with primers that span exons 1 and 2. PCR products were resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
Figure 2. ZC3H14splice variants can be detectedin vivo.
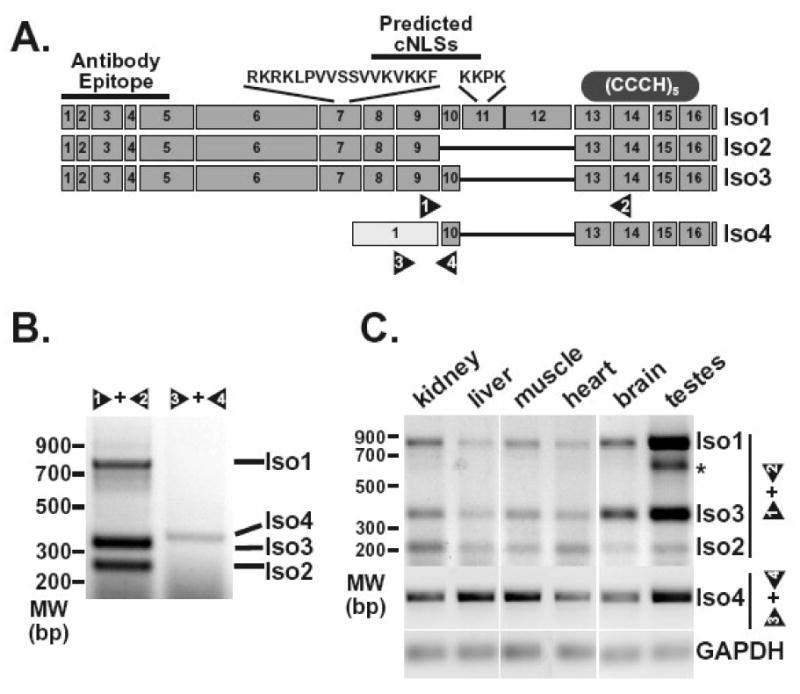
A. The diagram depicts exons predicted for the ZC3H14 splice variants encoding ZC3H14 isoform 1 (Iso1), isoform 2 (Iso2), isoform 3 (Iso3), and isoform 4 (Iso4). The sequences and approximate positions of predicted classical NLS motifs (cNLSs) are indicated at the top of the diagram. The location of the antibody epitope used to generate the polyclonal ZC3H14 antibody is also indicated as is the approximate location of the tandem Cys3His zinc finger domain (CCCH). Positions of primer pairs (1 and 2, 3 and 4) used to detect the splice variants are indicated at the bottom of the diagram. B. RT-PCR reactions (30 cycles) were performed on RNA isolated from HEK293 cells as described in Materials and Methods. Transcripts corresponding to ZC3H14 isoforms 1, 2, and 3 (Iso1, Iso2, Iso3) were detected using primer pair 1 and 2. Primer pair 3 and 4 was used to amplify the transcript corresponding to ZC3H14 isoform 4 (Iso4). Positions of the products corresponding to each splice variant are indicated. An additional band identified in the testes sample is indicated by the asterisk. This band likely reflects a ZC3H14 splice variant with additional alternative splicing of exons 10, 11, and 12. C. RT-PCR reactions were performed on RNA isolated from the mouse tissues indicated (kidney, liver, muscle, heart, brain, and testes). Isoform 1, 2 and 3 transcripts (Iso1, Iso2, Iso3) were detected using mouse primer pair 1 and 2 (see Figure 2A) with 30 cycles of amplification while the isoform 4 transcript (Iso4) was detected using mouse primer pair 3 and 4 with 60 cycles of amplification. The GAPDH transcript was amplified as a control for each tissue sample.
2.6. Generation of ZC3H14 polyclonal antibody
A DNA fragment encoding an N-terminal fragment of ZC3H14 was cloned into pGEX-4T to create an N-terminal GST-tagged fusion protein encoding the first 97 amino acids of isoform 1 (Amersham Biosciences). The GST fusion protein was purified by affinity chromatography on glutathione-Sepharose as previously described (Apponi et al., 2007). Cell lysate preparation and purification of recombinant proteins were performed as recommended by the resin manufacturer (Amersham Biosciences). The purified GST-ZC3H14 N-terminal domain (GST-ZC3H14 NT) was used for the immunization of rabbits to obtain polyclonal antibodies.
2.7. Microscopy
Cells were visualized by either direct fluorescence microscopy for GFP or indirect immunofluorescence microscopy. Prior to both direct fluorescence and indirect immunofluorescence, cells were fixed with 2% formaldehyde (EM Science) for 10 min, permeabilized with 0.1%Triton X-100 for 5 min, and incubated with Hoechst to mark nuclear DNA. The same protocol was employed to fix cells that had been treated with actinomycinD (5 μg/ml; Sigma). To localize endogenous ZC3H14 or SC35 by immunofluorescence, cells were probed with rabbit anti-ZC3H14 (1:5000) or mouse anti-SC35 (1:1000; BD Pharmigen) antibodies followed by staining with Texas Red or Fluorescein-conjugated secondary antibody (Jackson ImmunoResearch). Images were obtained using an Olympus IX81 microscope with a 0.3 NA 100X Zeiss Plan-Neofluor objective unless otherwise stated. Images were captured using a Hamamatsu digital camera with Slidebook software (version 1.63) and globally processed for brightness and contrast using Adobe Photoshop.
2.8. Immunoblotting
HeLa cells were harvested and washed in 1X PBS and then lysed on ice in RIPA-2 buffer (150 mM NaCl, 1% NP40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) containing protease inhibitors (PLAC: 3 μg/ml of pepstatin, leupeptin, aprotinin, and chymostatin and 0.5 mM PMSF). Immunoblots were performed using standard methods (Towbin et al., 1979). Briefly, 20 μg of total protein lysate per sample was resolved by SDS-PAGE and transferred onto a nitrocellulose membrane. For immunoblotting, a 1:10 000 dilution of rabbit anti-ZC3H14 antibody was used followed by a 1:4000 dilution of HRP-conjugated donkey anti-rabbit IgG secondary (Jackson ImmunoResearch).
3. Results
3.1. ZC3H14 is a member of an evolutionarily conserved family of zinc finger polyadenosine RNA binding proteins
Previous work identified the S. cerevisiae Nab2 protein as the founding member of a novel Cys3His zinc finger polyadenosine RNA binding protein family (Kelly et al., 2007). In order to assess the evolutionary relationship between Nab2 and putative Nab2 orthologues in higher eukaryotes, we used similarity searches to identify sequences that were potentially homologous to Nab2 (see Material and Methods). As shown in Figure 1A, putative orthologues of Nab2 including the human protein, ZC3H14, can be identified in all species examined. The topology obtained with this protein family is roughly consistent with the current relationship of these organisms (Kumar and Rzhetsky, 1996; Odronitz and Kollmar, 2007) and the high support inferred for the clades leads us to conclude that the observed similarity is the product of common ancestry rather than convergent evolution.
Comparison of the overall sequence similarity of the Nab2 protein with human ZC3H14 reveals a high level of similarity between the N-terminal and C-terminal domains of the two proteins with less conserved central domains (Figure 1B). The similarity in the N- and C-terminal domains is consistent with functional studies of Nab2 which reveal that the C-terminal zinc finger domain of Nab2 mediates RNA binding (Kelly et al., 2007) and the N-terminal domain assumes a proline-tryptophan-isoleucine (PWI) fold required to target Nab2 to the nuclear pore for export (Marfatia et al, 2004; Grant et al, 2008; Fasken et al, 2008). Thus, these domains which appear conserved mediate the critically important functions of the Nab2 protein.
Both Nab2 and ZC3H14 contain Cys3His (CCCH) zinc fingers with the spacing CX5CX4-6CX3H (Figure 1C). A BLAST search using this CCCH spacing motif revealed only a single zinc finger protein with spacing similar to Nab2 in the proteome of each species queried corresponding to those proteins depicted in the evolutionary tree shown in Figure 1A. While the Nab2 protein contains seven tandem Cys3His zinc fingers, the human protein and many other higher eukaryotic family members (not shown) contain only five tandem zinc fingers. Recent studies, however, reveal that the final three zinc fingers of the Nab2 protein are both necessary and sufficient for specific binding to polyadenosine RNA (Kelly et al., 2009). Thus, the five tandem zinc fingers found in higher eukaryotic members of this family should be more than sufficient to confer specific recognition of polyadenosine RNA. In fact, a previous in vitro study showed that recombinant ZC3H14 binds specifically to a polyadenosine RNA oligonucleotide (Kelly et al., 2007).
As mentioned above, the N-terminal domain of the yeast Nab2 protein is also functionally important. In fact, a deletion mutant of Nab2 where the N-terminal 97 amino acids are removed is only minimally functional and yeast cells that express this variant of Nab2 (Nab2-1/Nab2ΔN) as the sole copy of the essential Nab2 protein show defects in poly(A) RNA processing and export (Chekanova et al., 2001; Green et al., 2002). To assess the conservation of this domain between Nab2 and ZC3H14, we first aligned these domains (Figure 1D) and then determined whether the N-terminal domain of ZC3H14 could be modeled into the PWI-like fold that the N-terminal domain of Nab2 assumes (Grant et al., 2008). Figure 1E shows an overlay of the resolved structure of the N-terminal domain of yeast Nab2 (blue) with a model of the N-terminal domain of ZC3H14 (green). The Nab2 N-terminal domain assumes a five α-helical bundle (Figure 1E; labeled H1-5) with its N- and C-terminal helices juxtaposed (H1 and H5, respectively) (Grant et al., 2008). ZC3H14 could also be modeled as a five-helix bundle with one slight difference. The shortening of the loop between helix 1 and 2 by two amino acids possibly shortens helix 2 (labeled H2). Taken together, the phylogeny analysis, sequence comparison, and modeling reveal that ZC3H14 shares functionally important domains with Nab2 suggesting that ZC3H14, like Nab2, could play a role in the production of mature mRNA.
3.2. There are multiple isoforms of ZC3H14 in higher eukaryotes
Although there is a single transcript that encodes Nab2 in S. cerevisiae, where splicing is quite rare, genomic databases (NCBI, UCSC Genome Browser, ENSEMBL) predict multiple splice variants for ZC3H14 both in human (Figure 2A) and mouse. Most of the transcripts predicted contain the same translational start with differential inclusion of several central exons (exons 10, 11, 12) in the middle of the transcript (isoforms 1, 2, 3) that yield protein isoforms that all contain the evolutionarily conserved N-terminal PWI and C-terminal zinc finger domains. There is also evidence for a transcript corresponding to isoform 3 but containing an alternative translation start that encodes a slightly smaller isoform (isoform 3short). A fourth shorter splice variant is predicted by ESTs derived specifically from testes and brain (isoform 4). The protein isoform encoded by this splice variant would lack the conserved N-terminal PWI domain but retain an intact C-terminal zinc finger domain.
To confirm the existence of the alternatively spliced transcripts of ZC3H14, we first performed RT-PCR on RNA isolated from HEK293 cells using internal primers that can distinguish each of the splice variants (arrow heads, Figure 2A). Using a primer set specific for exon 9 and exon 14, which flank the predicted variant exons, we detected three PCR products that correspond to the predicted size of fragments derived from transcripts encoding ZC3H14 isoforms 1, 2, and 3/3short (Figure 2B). We also confirmed the presence of the transcript coding for ZC3H14 isoform 4 in HEK293 cells by using a primer within the predicted alternative first exon paired with a reverse primer located in exon 10 (Figure 2B). All reported ESTs corresponding to the isoform 4 transcript are derived from testes and brain. HEK293 cells possess some neuronal markers (Shaw et al., 2002) which could explain why this potentially tissue-specific transcript could be detected in these cells. Alternatively, the isoform 4 transcript may be present in other tissues, perhaps at lower abundance.
3.3. Multiple ZC3H14 transcripts can be detected in mouse tissues
To investigate the in vivo expression of the different splice variants of ZC3H14, we detected ZC3H14 transcripts in mouse tissues by RT-PCR. We isolated RNA from mouse kidney, liver, muscle, heart, brain and testes and performed RT-PCR on each RNA sample as described in Materials and Methods (Figure 2C). Using mouse primer sets similar to those described for human ZC3H14 transcripts, we identified multiple RT-PCR products corresponding to multiple transcripts. RT-PCR using mouse primers 1 and 2 confirmed at least two transcripts in mouse tissues, corresponding to ZC3H14 splice variants 1 and 2 (Figure 2C). Interestingly, we could also identify a transcript encoding a short mouse isoform corresponding to human ZC3H14 isoform 4 in the various tissue samples (Figure 2C). Consistent with the fact that most human isoform 4 ESTs are derived from testes, we found that the mouse splice variant corresponding to human isoform 4 could be detected with the fewest PCR amplification cycles in testes samples as compared to other tissues examined suggesting that this splice variant is most abundant in testes.
3.4. ZC3H14 isoforms localize both to the nucleus and the cytoplasm
Unlike S. cerevisiae Nab2, human ZC3H14 does not contain an arginine-glycine-glycine (RGG) domain, which mediates nuclear import of Nab2 (Aitchison et al., 1996); however, based on the predicted protein sequence, ZC3H14 isoforms 1, 2, and 3 all contain two putative classical nuclear localization signals (cNLS) in exon 7 and 11, that could target ZC3H14 into the nucleus. The first 9 exons are absent from the shortest splice variant encoding ZC3H14 isoform 4 and are replaced by an alternative first exon derived from the large intron between exons 8 and 9 (Figure 2A, isoform 4). Thus, exons 7 and 11 are both absent from ZC3H14 isoform 4 and this isoform lacks the predicted cNLS motifs (Figure 2A), possibly creating a differential cellular localization of ZC3H14 isoform 4 as compared to isoforms 1-3.
As an initial approach to examine the intracellular localization of distinct ZC3H14 isoforms, we fused the coding region of ZC3H14 isoform 1 and isoform 4 to the green fluorescent protein (GFP) as described in Material and Methods. We transfected each fusion construct into HeLa cells to examine the expression and steady-state localization of these fusion proteins. Probing immunoblots with an anti-GFP antibody revealed bands corresponding to the predicted size for each fusion protein (Figure 3A). We then examined the localization of the expressed GFP-tagged proteins using direct fluorescence microscopy. Similar to the Nab2 protein in S. cerevisiae, ZC3H14 isoform 1 is concentrated in the nucleus (Figure 3B). This localization is consistent with the presence of the predicted cNLS motifs within exons 7 and 11. Interestingly, as suggested by the lack of predicted cNLS motifs, the ZC3H14 isoform 4-GFP fusion protein is localized to the cytoplasm (Figure 3B).
Figure 3. The ZC3H14 gene encodes both nuclear and cytoplasmic isoforms.
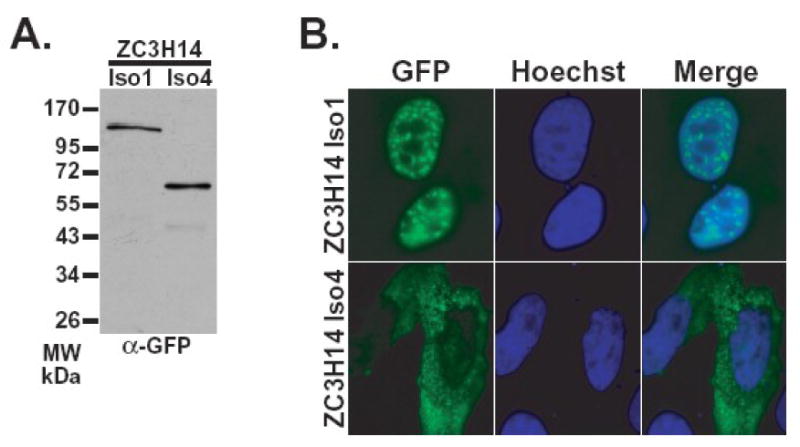
Plasmids encoding either ZC3H14 isoform 1-GFP or isoform 4-GFP were transiently transfected into HeLa cells. A. GFP fusion proteins were detected by immunoblotting cell lysates with anti-GFP antibody (Seedorf et al., 1999). B. Transfected HeLa cells were fixed and the localization of GFP fusion proteins was examined by directed GFP fluorescence microscopy (GFP) (green). Hoechst dye (blue) was used to stain DNA and indicate the position of the nucleus. A merged image is also shown.
3.4. Multiple endogenous ZC3H14 proteins are detected in cultured cells
To examine the localization of endogenous ZC3H14, we generated a polyclonal rabbit antibody directed against an N-terminal domain of ZC3H14, common to isoforms 1, 2, and 3 (see Figure 2A). To confirm that the antibody indeed recognizes ZC3H14, we probed lysates from HeLa cells expressing plasmid-encoded ZC3H14 isoform 1-GFP. The ZC3H14 antibody detects bands corresponding to the predicted size of endogenous ZC3H14 isoforms and, importantly, also recognizes a higher band corresponding to the predicted size of the ZC3H14 isoform 1-GFP fusion protein (Figure 4A). Immunoblotting with an anti-GFP antibody confirms that the highest band corresponds to ZC3H14 isoform 1-GFP. The ZC3H14 antibody recognizes two bands in lysate from untransfected HeLa cells. The higher band corresponds to the predicted size for ZC3H14 isoform 1. The lower band corresponds to the predicted size for ZC3H14 isoform 2 and/or isoform 3 of ZC3H14. As the difference in size between isoforms 2 and 3 is only two Daltons, we cannot presently confirm whether the lower band we detect is a combination of both isoforms 2 and 3 or whether it represents one or the other of the two isoforms.
Figure 4. Endogenous nuclear ZC3H14 colocalizes with nuclear speckles.
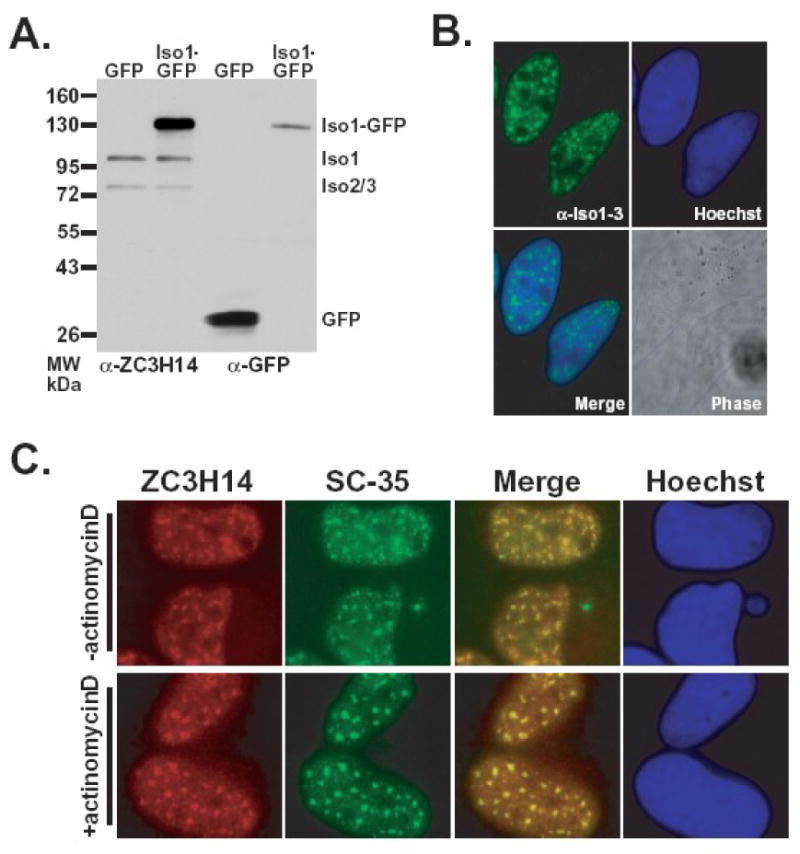
A. Lysates from HeLa cells expressing either GFP or ZC3H14 isoform 1-GFP (Iso1-GFP) fusion protein were probed with polyclonal anti-ZC3H14 (α-ZC3H14) or anti-GFP (α-GFP) antibody. B. The localization of endogenous ZC3H14 (isoforms 1-3) in HeLa cells was determined by indirect immunofluorescence using the polyclonal anti-ZC3H14 antibody. Hoechst dye was used to stain DNA and indicate the position of the nucleus. The merge reveals that endogenous ZC3H14 (isoforms 1-3) is located in foci within the nucleus. A corresponding Phase image is also shown. C. ZC3H14 localizes to nuclear speckles in HeLa cells. ZC3H14 was detected with anti-ZC3H14 antibody (green) and nuclear speckles were labeled with anti-SC-35 antibody (red). Colocalization (yellow) is indicated in the merge panel. To assess whether ZC3H14 shows dynamics typical for nuclear speckle proteins, ZC3H14 was localized either prior to (-actinomycinD) or following (+actinomycinD) treatment with actinomycinD for 4 hr. As a control, the –actinomycinD sample was treated with DMSO, which is the solvent for actinomycinD. The merge image shows that nuclear ZC3H14 colocalizes (yellow) with nuclear speckles under all conditions examined.
3.5. Endogenous nuclear ZC3H14 isoforms localize to nuclear speckles
To examine the endogenous localization of ZC3H14 isoforms 1 and 2/3, we performed indirect immunofluorescence on HeLa cells with the ZC3H14 antibody. Results of this analysis indicate that nuclear ZC3H14 localizes to distinct nuclear bodies (Figure 4B). Based on the function of Nab2 in mRNA processing, we hypothesized that these foci could correspond to mRNA processing bodies called nuclear speckles (Lamond and Spector, 2003). To test this hypothesis, we colocalized the ZC3H14 foci with the nuclear speckle marker SC35 (Figure 4C). The majority of the ZC3H14 foci overlap with SC35 as demonstrated by the yellow in the merge image (Figure 4C, merge).
To further examine ZC3H14 speckle localization, we treated HeLa cells with the transcription inhibitor actinomycinD. Nuclear speckles are altered in number and size in the presence of this inhibitor (Spector et al., 1983). Following a four hour treatment with actinomycinD, we examined the morphology of nuclear speckles (SC35) and the localization of nuclear ZC3H14 proteins (Figure 4C). We found the size and number of ZC3H14 foci altered in a similar fashion to nuclear speckles (SC35), suggesting that ZC3H14 localization to nuclear speckles, like that of other speckle components, is dependent on ongoing transcription.
4. Discussion
This study presents the first characterization of the human ZC3H14 gene which encodes a tandem Cys3His zinc finger protein with homology to the S. cerevisiae mRNA processing/export factor Nab2. We confirm that multiple splice variants of ZC3H14 encode distinct protein isoforms. Ubiquitously expressed nuclear ZC3H14 isoforms that are closely related to Nab2 localize to speckles within the nucleus suggesting a role in mRNA processing while a distinct isoform containing an alternative N-terminal domain is located in the cytoplasm. All isoforms characterized contain the C-terminal Cys3His zinc finger domain which has been defined as a novel polyadenosine RNA binding domain (Kelly et al., 2007) strongly suggesting that all isoforms could bind to and potentially regulate polyadenylated RNA transcripts.
ZC3H14 isoforms 1, 2, and 3 all contain an N-terminal sequence that has similarity to the N-terminal PWI-like domain of S. cerevisiae Nab2 (see Figure 1D and 1E). The N-terminal domain of Nab2 contributes to proper export of poly(A) RNA from the nucleus by targeting Nab2 to the nuclear pore complex via an interaction with nuclear pore associated Mlp proteins (Green et al., 2003; Fasken et al., 2008). One possibility is that the predicted PWI-like domain within isoforms 1, 2, and 3 of ZC3H14 could mediate interactions with Tpr, the human orthologue of the yeast Mlp proteins (Strambio-de-Castillia et al., 1999). In contrast to the evolutionarily conserved N-terminal domain of ZC3H14 isoforms 1, 2, and 3, the N-terminal domain of ZC3H14 isoform 4 does not contain a predicted PWI-like fold. Instead modeling of this domain suggests an extended alpha helix typically involved in protein/protein interactions (Phyre Protein Fold Recognition Server; http://www.sbg.bio.ic.ac.uk/∼phyre/). Further studies will be required to determine whether specific partners interact with the distinct N-terminal domains of the ZC3H14 isoforms.
Future studies will also examine functional interactions mediated by the Cys3His zinc finger domain of ZC3H14. A single in vitro binding study indicates that ZC3H14 isoform 1 binds to polyadenosine RNA (Kelly et al., 2007); however, this analysis carried out with a single protein isoform may not reflect the spectrum of RNA targets for various ZC3H14 isoforms in vivo. Furthermore, the zinc finger domain of S. cerevisiae Nab2 has also been implicated in interactions with an evolutionarily conserved mRNA binding protein, Pub1 (Apponi et al., 2007), which regulates mRNA stability (Vasudevan and Peltz, 2001). The function of Pub1 in yeast has been equated to the ubiquitous member of the Hu family of mRNA binding proteins, HuR, in higher eukaryotes (Hinman and Lou, 2008). Thus, ZC3H14 could interact with HuR to modulate mRNA stability.
In the studies presented here, we examined the steady-state localization of ZC3H14 isoforms. Consistent with the presence of predicted cNLS motifs, isoforms 1, 2, and 3 were found in intranuclear speckles. In contrast, ZC3H14 isoform 4, which lacks these predicted cNLS motifs, is localized to the cytoplasm and apparently excluded from the nucleus. However, since our studies examined only the steady-state localization of these protein isoforms, we can not rule out the possibility that both nuclear and cytoplasmic isoforms shuttle between the nucleus and cytoplasm as demonstrated for numerous other RNA binding proteins (Gama-Carvalho and Carmo-Fonseca, 2001).
The current study does not directly address the function of the ZC3H14 protein. The identification of multiple splice variants and the corresponding protein isoforms reveals the complexity of analyzing the cellular function/functions of ZC3H14. Clearly, the similarity to the S. cerevisiae Nab2 protein along with the localization of ZC3H14 isoforms 1, 2, and 3 to nuclear speckles strongly suggests that these isoforms play a role in mRNA biogenesis. Many potential functions for the cytoplasmic ZC3H14 isoform 4 can be postulated. As this isoform is enriched in both brain and testes, tissue-specific roles in mRNA transport, mRNA translation, and mRNA stability are all possible. In the future, specific knockdown of the distinct ZC3H14 isoforms will be required to delineate their functions in vivo.
Results of this analysis provide insight into the complexity of human ZC3H14. This tandem Cys3His zinc finger protein has the potential to play multiple roles in mRNA metabolism with both ubiquitous and tissue-specific functions. In addition, the protein may modulate mRNA processing and function both in the nucleus and in the cytoplasm. A major goal for the future will be to understand the interplay between ZC3H14 and the conventional nuclear and cytoplasmic poly(A) binding proteins. Although the data presented here focuses on a single Cys3His zinc finger gene which generates multiple protein isoforms, a recent genome-wide survey compiled the Cys3His zinc finger domain-containing proteins found in mice and revealed that this family consists of 58 members in mice and 55 in humans (Liang et al., 2008). Many of these proteins have not yet been characterized and thus their functions are largely unknown (Liang et al., 2008); however, a subset either bind RNA or are predicted to bind RNA. Thus, the Cys3His zinc finger family of proteins is likely to represent a large family of RNA regulatory factors that will each need to be studied in detail to understand their contributions to cell function and possibly regulation of gene expression.
Acknowledgments
We thank Dr. Roderick O'Connor for advice and contributions as well as Dr. Yue Feng, Dr. Maureen Powers, Dr. Songli Xu, and Marie Cross for experimental advice and reagents. We thank member of the Corbett laboratory for critical discussion and suggestions. This work was supported by a grant (GM58728) from the NIH to AHC.
Abbreviations
- Cys
cysteine
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- His
histidine
- NLS
nuclear localization signal
- PMSF
phenylmethylsulfonyl fluoride
- PWI domain
proline isoleucine tryptophan domain
- RT-PCR
reverse transcriptase polymerase chain reaction
- ZC3H14
Cys3His Zinc Finger Protein #14
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–43. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apponi LH, Kelly SM, Harreman MT, Lehner AN, Corbett AH, Valentini SR. An Interaction between Two RNA Binding Proteins, Nab2 and Pub1, Links mRNA Processing/Export and mRNA Stability. Mol Cell Biol. 2007 doi: 10.1128/MCB.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–31. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–32. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–49. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Belostotsky DA. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr Biol. 2001;11:1207–14. doi: 10.1016/s0960-9822(01)00371-2. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fasken MB, Stewart M, Corbett AH. Functional Significance of the Interaction Between the mRNA Binding Protein, Nab2, and the Nuclear Pore-Associated Protein, Mlp1, in mRNA Export. J Biol Chem. 2008 doi: 10.1074/jbc.M803649200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Carmo-Fonseca M. The rules and roles of nucleocytoplasmic shuttling proteins. FEBS Lett. 2001;498:157–63. doi: 10.1016/s0014-5793(01)02487-5. [DOI] [PubMed] [Google Scholar]
- Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic. 2004;3:125–41. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- Grant RP, Marshall NJ, Yang JC, Fasken MB, Kelly SM, Harreman MT, Neuhaus D, Corbett AH, Stewart M. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J Mol Biol. 2008;376:1048–59. doi: 10.1016/j.jmb.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci U S A. 2003;100:1010–5. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p Is Required for Poly(A) RNA Export in Saccharomyces cerevisiae and Is Regulated by Arginine Methylation via Hmt1p. J Biol Chem. 2002;277:7752–60. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH. Poly(A) RNA binding by Nab2 is required for correct mRNA 3′ end formation. 2009 doi: 10.1074/jbc.M110.141127. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM. Recognition of polyadenosine RNA by zinc finger proteins. Proc Natl Acad Sci U S A. 2007;104:12306–11. doi: 10.1073/pnas.0701244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–14. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kumar S, Rzhetsky A. Evolutionary relationships of eukaryotic kingdoms. J Mol Evol. 1996;42:183–93. doi: 10.1007/BF02198844. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–12. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–55. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nykamp KR. Coordination of mRNA 3′End Formation and Nuclear Export by a Nuclear Poly(A)-Binding Protein, Department of Molecular Genetics and Microbiology. University of Florida; Gainesville: 2003. [Google Scholar]
- O'Connor RS, Mills ST, Jones KA, Ho SN, Pavlath GK. A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J Cell Sci. 2007;120:149–59. doi: 10.1242/jcs.03307. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–67. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–76. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan MJ, Gordan JD, Williamson B, Stockert E, Bander NH, Jongeneel V, Gure AO, Jager D, Jager E, Knuth A, Chen YT, Old LJ. Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int J Cancer. 1999;83:456–64. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–55. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-eG. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedures and some application. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol Cell. 2001;7:1191–200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]


