Abstract
PDEF is an ETS transcription factor expressed in normal tissues with high epithelial cell content and non-invasive breast cancer cells. A putative tumor suppressor, PDEF protein expression is often lost during progression to a more invasive phenotype. Interestingly, PDEF mRNA has been found to retained or even over-expressed in the absence of protein; however, the mechanisms for this remain to be elucidated. This study identifies two microRNAs that directly act on and repress PDEF mRNA translation, leading to the loss of PDEF protein expression and the gain of phenotypes associated with invasive cells. In addition, we show that these microRNAs are elevated in human breast tumor samples. Together, these data describe a mechanism of regulation that explains for the first time the lack of correlation between PDEF mRNA and protein levels, providing insight into the under-explored role of post-transcriptional regulation and how this contributes to dysregulated protein expression in cancer. These observations have critical implications for therapeutically targeting microRNAs that contribute to cancer progression.
Keywords: microRNAs, PDEF, transcription factor, ets, breast cancer
INTRODUCTION
Cancer death is due in large part to metastases. One of the more interesting challenges is to understand the cellular changes that occur during progression towards invasive cancer. ETS proteins are a large family of transcription factors with diverse functions and activities that activate or repress the expression of genes that are involved in various biological processes, including cellular proliferation, apoptosis, differentiation, and transformation (1, 2). The ETS family gene, PDEF (prostate derived epithelial factor), is expressed in normal epithelial tissues including prostate, breast, and colon (3). In normal tissue and non-invasive cancers, mRNA and PDEF protein are easily detectable by northern and western blot. However, PDEF protein loss is correlated with prostate, breast, and colon cancer progression to an invasive phenotype both in vitro and in vivo (3–5). Interestingly, this loss of protein does not always correlate well with PDEF mRNA levels (5). Indeed, some invasive cancers retain or have elevated levels of PDEF mRNA in the absence of protein (3, 6, 7). PDEF re-expression in multiple invasive prostate, breast, and colon cancer cells results in reduced cell growth, migration and invasion (2, 3) (and unpublished data). Reciprocal siRNA-mediated knockdown experiments in PDEF expressing non-invasive cells results in increased migration and invasion together with an altered morphology consistent with a more invasive phenotype (2). Together, these and other data support the model that PDEF target genes control several aspects of the multi-step metastatic process and specifically, loss of PDEF regulatory networks is a key event in the development of invasive cancer (8). The processes involved in the loss of PDEF protein during cancer progression have not been elucidated. The goal of this study was to identify pathways involved in the post-transcriptional regulation of PDEF that ultimately results in protein loss, providing novel mechanistic insight into the cellular events leading to a more aggressive phenotype.
MicroRNAs (miRNAs) are endogenous 19–25 nucleotide non-coding RNAs that have recently emerged as a novel class of small, evolutionarily conserved important gene regulatory molecules involved in many critical developmental and cellular functions (9). Through specific base pairing with target mRNA sequences in the 3′ untranslated region (3′UTR), miRNAs induce mRNA degradation, translational repression, or both (10). Individual miRNAs can target numerous mRNAs, often in combination with other miRNAs, thereby providing a mechanism for controlling complex regulatory networks. It is estimated that there are over 600 miRNAs in mammalian cells, and that about 30% of all genes are regulated by miRNAs (11, 12). Over 3,000 identified mature miRNAs exist in species ranging from plants to humans. Their existence and conservation throughout species supports the concept that they perform critical functions in gene regulation (13). Indeed, the conserved evolution of both miRNAs and transcription factors highlights their importance in and the complexity of gene regulation (14). miRNAs have been implicated in the control of many fundamental cellular and physiological processes, including tissue development, cellular differentiation and proliferation, metabolic and signaling pathways, apoptosis and stem cell maintenance (15–17). Mounting evidence indicates that miRNAs may also play a significant role in cellular transformation and carcinogenesis acting either as oncogenes or tumor suppressors (18, 19). Hence, there are few cellular processes that are not affected by miRNAs. In addition, specific miRNA signatures have been identified for both solid cancers and hematologic malignancies (20–23), and mounting evidence suggest that the power of miRNAs lies in the ability to distinguish specific cancer subtypes based on their miRNA profile, including, and of direct relevance to the studies described herein, breast cancer (20, 24). Nonetheless, the identification and validation of specific targets has been limited. We report here PDEF as a novel target for miR-204 and miR-510 and describe for the first time a mechanism for the loss of PDEF protein expression during breast cancer progression.
EXPERIMENTAL PROCEDURES
Cell Culture
Human breast cancer cell lines (MCF7, BT474, CAMA-1, HBL100, HCC202, Hs578t, MDA-MB-157, MDA-MB-175 VII, MDA-MB-231, MDA-MB-361, MDA-MB-415, MDA-MB-436 and MDA-MB-453) were cultured according to the ATCC website. The breast cancer cell lines CAMA-1, HBL100, HCC202, MDA-MB-415 and MDA-MB-436 were a kind gift of R. Neve (University of California, CA). All other lines were obtained from ATCC. For the generation of stable MCF7 cells overexpressing miR-204, pSuppressor-neo vector (Imgenex; San Diego, CA) expressing miR-204 was transfected into MCF7 cells and stable cells were selected in medium containing G418 (Invitrogen; Carlsbad, CA). This vector system was also used for transient expression of miR-204 and miR-510 into MCF7 cells.
Tumor samples
Matched tumor and non-tumor breast samples were obtained from the Hollings Cancer Center tumor bank at MUSC. Prior to surgery at the Center, all patients provided written informed consent to allow any excess tissue to be used for research studies. Samples were snap frozen in OCT and stored at −80°C until use. The pathological status of the specimens was confirmed by histological examination of 10uM sections taken at the start, middle and end of the 5 × 20μM sections taken for RNA analysis. Each tumor section contained between 65–80% malignant epithelial cells and 0–5% non-malignant epithelial cells. Non-tumor sections contained 100% benign epithelial cells. To determine PDEF expression, human breast cancer paraffin blocks of tissues available from the same patients were obtained from the HCC Tumor Bank (MUSC).
Immunohistochemistry
Antigen retrieval was done by heating in a microwave oven for 2 × 5 min on half power in 10 mmol/L citrate (pH 6.0). Sections were washed, treated with 1% H2O2 for 15 min and non-specific binding was blocked with 2.5% horse serum (ImmPRESS Vector staining kit; Vector Laboratories, Burlington, CA) for 1h and then incubated overnight at 4°C with PDEF primary antibody at a 1:100 dilution in 2% BSA in PBS. Overnight incubation at 4°C was followed by 3 × 10 min washes in PBS, Immpress anti-rabbit secondary antibody was incubated (Vector Laboratories) for 2h at room temperature. After washing with H2O, 3,3′-diaminobenzidine substrate (Sigma, St Louis, MO) was added for 2 min followed by washing in H2O. Slides were counterstained with hematoxylin.
Quantitative reverse transcription PCR
Total RNA from cancer cell lines was extracted using the RNeasy Plus Mini Kit (Qiagen; Valencia, CA). Total RNA from breast tumor and non-tumor samples was extracted using Trizol as per the manufacturer’s instructions (Invitrogen, Carlsbad, CA). One microgram total RNA was reverse transcribed in a 20μl reaction using Superscript III reverse transcriptase (Invitrogen) for microRNA analyses and iScript (Bio-Rad; Hercules, CA) for all other studies. Real time PCR was performed with 1μl of a 1:10 dilution of reverse transcribed cDNA using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) in a LightCycler (Roche, Nutley, NJ). The cycling conditions for all genes were: pre-incubation at 50°C for 2 minutes, 95°C for 2 minutes, followed by 30–50 cycles of denaturation at 94°C for 10 seconds, annealing at one degree below the lowest Tm for each gene-specific primer pair (Supplemental Table 1) for 10 seconds and extension for 30 seconds at 72°C, with a single data acquisition at the end of each extension. All ramping was done at 20°C per second. Triplicate reactions were run for each cDNA sample. The relative expression of each gene was quantified on the basis of Ct value measured against an internal standard curve for each specific set of primers (Supplemental Table 1) using the software provided by the instrument manufacturer (Roche). These data were normalized to S26, GAPDH or U6 (see individual experiments). The size and purity of the PCR products were also analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining.
Plasmid construction
To over-express miR-204 and miR-510, the genomic region surrounding the pri-microRNA sequence of miR-204 and miR-510 were amplified with ThermalAce (Invitrogen). The cycling conditions were: pre-incubation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 1 minute, annealing at 65°C for 1 minute and extension for 1 minute at 72°C. The PCR products (~500bp) were directionally cloned into the pSuppressor-neo vector (Imgenex) using XhoI and XbaI. The 5′UTR, ORF, 3′UTR and full length sequences of PDEF were amplified from human genomic DNA of MCF7 cells and directionally cloned into pcDNA3 (Invitrogen). The wild type 3′UTR of PDEF was cloned into the XbaI site of the pGL3-promoter vector (Promega; Madison, WI). The sequences complementary to the seed of the miR-204 and miR-510 were deleted using a QuikChange Site-Directed Mutagenesis Kit (Stratagene; La Jolla, CA). For primer sequences and Ta’s, see Supplementary Table 2. All constructs were validated by sequencing at the MUSC sequencing facility.
Oligonucleotide Transfection
The miRNA inhibitors (Ambion; Austin, TX) are single-stranded chemically enhanced oligoribonucleotides designed to inhibit the endogenous miRNAs. Cells were transfected with 100nM of the indicated oligoribonucleotide using the Oligofectamine reagent as per the manufacturer’s instructions (Invitrogen). 48 hours after transfection, cells were harvested for protein or RNA extraction.
Luciferase assays
Cells were plated at 200,000 cells per well in a 6-well plate. The pGL3 reporter constructs (0.5μg, firefly luciferase) were co-transfected with pRL–TK (0.05μg, Renilla luciferase) using Lipofectamine 2000 as per the manufacturer’s instructions (Invitrogen). The media was changed the next day, and luciferase activity measured after 48h using the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well.
Western blot analysis
Cell lysate preparation and western blot analysis using enhanced chemiluminescence were performed as described previously (2). Experimental antibodies include human PDEF (prepared as described previously (3)) and E-Cadherin (BD BioSciences, San Jose, CA). GAPDH and beta-actin (Abcam, Cambridge, MA) were used as loading controls.
Transwell migration and invasion assay
Stably transfected MCF7 cells were seeded into the upper chamber of a Transwell insert pre-coated with 5μg/ml fibronectin for migration or a BD™ Matrigel invasion chamber for invasion, in serum-free medium at a density of 50,000 cells per well (24-well insert; pore size, 8μM; BD Biosciences). Medium containing 10% serum was placed in the lower chamber to act as a chemoattractant, and cells were further incubated for 24h. Non-migratory cells were removed from the upper chamber by scraping with a cotton bud. The cells remaining on the lower surface of the insert were stained using Diff-Quick (Dade Behring, Inc., Newark, DE). Cells were quantified as the number of cells found in 10 random microscope fields in two independent inserts. Error bars represent the SD from three separate experiments.
Immunofluorescence
Cells were seeded onto sterile cover slides (18mm diameter) coated with 5 g/ml fibronectin and allowed to attach overnight. Cells were then fixed with 2% formaldehyde, permeabilized with 0.1% Triton X-100 and blocked in 2% BSA for 1h at room temperature. E-Cadherin expression was examined using the antibody detailed above and visualized using Alexa Fluor secondary antibody (Invitrogen). Immunofluorescence was examined using an Olympus IX70 confocal microscope.
Colony formation Assay
Wild type and miR-204 stably transformed MCF7 cells were seeded at a cell density of ~4 cells/mm2 in normal growth media. Cells were incubated as normal and colonies were counted after 7–10 days.
Soft-agar assay
2ml of 0.6% agarose in 2 X DMEM was plated in each well of a 6-well plate and left to set for 20min. This layer was overlaid with 1.0 × 104 wild type and miR-204 stably transfected MCF7 cells in 3ml of 0.4% agarose diluted in 2x DMEM. Cells were incubated as normal for ~14 days and the colonies counted.
Statistical analysis
For statistical testing, two-sided paired Student’s t-tests were done using Excel spreadsheet. p values are given for each individual experiment, but in general, p < 0.05 was considered statistically significant. Error bars represent standard deviations of three independent experiments unless indicated otherwise.
RESULTS
PDEF is post-transcriptionally regulated during breast cancer progression
The discordance between PDEF mRNA and protein levels supported the model that PDEF may be post-transcriptionally regulated during breast cancer progression. To identify appropriate cell line systems to examine this model, we evaluated PDEF mRNA and protein levels in multiple breast cancer cell lines by real-time PCR and western analyses, respectively. MCF7, MDA-MB-361 and BT474 cell lines expressed detectable levels of both PDEF mRNA and protein, and MDA-MB-175 VII, MDA-MB-415, MDA-MB-453, CAMA-1 and HCC202 cell lines had PDEF mRNA, but little or no detectable levels of protein (Figure 1A & B). These cell lines are derived from luminal cells and appear more differentiated, form tight cell-cell junctions and are non- or weakly invasive (25). In contrast, MDA-MB-157, MDA-MB-436, MDA-MB-231, HBL100 and Hs578t cells had neither PDEF mRNA or protein (Figure 1A & B) and are derived from basal B cells, appear less differentiated, have a more mesenchymal-like appearance and are highly invasive (25). Therefore, cells lose PDEF protein during progression to a more invasive cancer.
Figure 1. PDEF is regulated post-transcriptionally through sequences present in the 3′UTR.
(A) Quantitative real-time PCR of PDEF mRNA normalized to S26. (B) Western blot analysis of breast cancer cell lines probed with primary antibodies for PDEF and GAPDH. Semi-quantitative analysis is shown in the lower panel. (C) Schematic representation of full length, open reading frame (ORF) alone, ORF + 5′UTR (5′UTR) and ORF + 3′UTR (3′UTR) constructs and western blot (WB) and RT-PCR analysis of PDEF expression in MDA-MB-157 cells transiently transfected with the constructs illustrated above. (D) Luciferase activity of MDA-MB-157 cells transfected with a reporter luciferase gene (pGL3) and pGL3 fused to the PDEF 3′UTR (3′UTR) normalized to Renilla luciferase activity. The data are expressed as the mean ± SD for 3 experiments conducted in triplicate.
Our previous gain-of-function studies utilized a construct expressing only the open reading frame (ORF) of PDEF to over-express the protein in multiple breast cancer cell lines (2, 3). These observations supported the notion that PDEF may be regulated post-transcriptionally through sequences in its untranslated regions (UTR). To study UTR-dependent translational control of PDEF transcripts, the level of PDEF mRNA and protein expression following transfection of MDA-MB-157 cells with constructs lacking 5′ and/or 3′UTR sequences were compared with that allowing expression of the full-length PDEF transcript (Figure 1C). RT-PCR analysis demonstrated that PDEF mRNA is expressed following transfection of each construct (Figure 1C). In contrast, PDEF protein was not detectable in cells transfected with PDEF constructs containing both or either UTR (Figure 1C), supporting the model that elements present within the 3′UTR of PDEF negatively regulate its mRNA translation in breast cancer. One mechanism of post-transcriptional regulation is through miRNA-mediated repression of elements present in the 3′UTR of genes. To investigate whether PDEF mRNA translation was regulated by factors binding to its 3′UTR, we fused the 3’UTR of PDEF mRNA to a luciferase reporter. The presence of the 3′UTR of PDEF resulted in a significant reduction in luciferase activity compared with the unmodified control luciferase reporter (Figure 1D).
microRNAs regulate PDEF expression through its 3′UTR
To determine whether miRNAs played a role in the post-transcriptional regulation of PDEF expression, we conducted a bioinformatics analyses and identified twelve potential miRNA recognition sequences within the 3′UTR of the PDEF transcript. Primers were designed for the top ten scoring miRNAs. Real-time PCR analysis on a series of breast cancer cell lines revealed that, compared to 8 other miRNAs, the expression of two miRNAs, miRNA 204 (miR-204) and miRNA 510 (miR-510), were most correlated with breast cancer cell lines having high PDEF RNA and low PDEF protein levels (Figure 2A and data not shown). Specifically, miR-204 and miR-510 are expressed at elevated levels relative to MCF7 in 4/5 and 5/5 of the breast cancer cell lines with high PDEF RNA and low PDEF protein (MDA-MB-175 VII, MDA-MB-415, MDA-MB-453, CAMA-1 and HCC202), while miR-28 is elevated in only 2/5 lines (MDA-MB-175 VII, MDA-MB-415). Although some elevated expression of miR-204 & -510 was also observed in other breast cancer cell lines when compared to MCF7, we chose these two miRNAs as the best candidates of the ten initially characterized for further investigation in the negative regulation of PDEF expression. The predicted sites for miRNAs within a 3′UTR can overlap or even be identical; however, miR-204 and miR-510 bind to separate sites within the 3′UTR of PDEF (Figure 2A). To examine whether PDEF expression is repressed by miR-204 and/or miR-510 through these elements, the luciferase reporter construct containing the 3′UTR of PDEF was co-transfected into HeLa cells together with a plasmid construct designed to over-express either miR-204 or miR-510. Co-transfection with either miR-204 or miR-510 further reduced the luciferase activity, to 55% and 35%, respectively (Figure 2B). The most important criteria for target recognition are the 5′ five to eight nucleotide core sequence of a miRNA, known as the ‘seed sequence’. To further validate that miR-204 and miR-510 are direct repressors of PDEF via binding to the identified sites within the 3′UTR, we mutated the seed sequences of miR-204 and miR-510, respectively, in the luciferase reporter construct. Transfection of HeLa cells with these mutated luciferase reporters resulted in ~25% and 40% increase in luciferase activity, respectively (Figure 2C). Furthermore, miR-204 and miR-510 had no significant repressive effect on luciferase activity when over-expressed in cells transfected with luciferase-UTR constructs containing the respective mutated seed sequences (Figure 2D).
Figure 2. miR-204 & miR-510 regulate PDEF through its 3′UTR.
(A, upper panel) Quantitative real-time PCR of miR-28 (black bars), miR-204 (white bars) & miR-510 (grey bars) normalized relative to the amount of U6 target (ΔCt). The relative levels of miRNA expression were measured by determining the ΔΔCt values of the indicated cell lines versus MCF7. (A, lower panel) Schematic representation of the predicted target sites of miR-204 & miR-510 in the 3′UTR of PDEF mRNA. The numbers (1–469) represent base pairs in the 3′UTR of PDEF and the numbers in parentheses (1425–1894) represent base pairs in the full length PDEF gene (GenBank Accession number AF071538 (42)). Complementary base sequences are highlighted in upper case letters. (B–D) Luciferase activity of HeLa cells transfected with (B) pGL3 PDEF 3’UTR reporter construct (3′UTR) and pSuppressor vector alone (pSupp), pSupp/miR-204 (miR204) or pSupp/miR-510 (miR510); (C) pGL3 reporter construct (pGL3), 3′UTR, 3′UTR reporter construct mutated in the miR204 seed sequence binding site (mut204) or 3′UTR reporter construct mutated in the miR510 seed sequence binding site (mut510); (D) or co-transfected with 3′UTR, mut204 or mut510 and pSupp (grey bars), miR204 (white bars) or miR510 (black bars). All luciferase assays were normalized to Renilla luciferase activity. The data are expressed as the mean ± SD for 3 experiments conducted in triplicate.
Regulation by miRNAs of protein translation can be due to translational repression and/or mRNA degradation. To assess whether miR-204 and/or miR-510 have a functional role in the down-regulation of endogenous PDEF expression by either of these mechanisms, two cell lines were selected as model systems: CAMA-1 (elevated PDEF mRNA, low PDEF protein) and MCF7 (PDEF mRNA and protein). Transfection of antisense oligoribonucleotides (ASO) targeted against miR-204 or miR-510 in CAMA-1 cells resulted in an increase in PDEF protein levels, while the PDEF mRNA levels remained unchanged (Figure 3A). Reciprocal over-expression of either miR-204 or miR-510 in MCF7 cells results in a loss of PDEF protein, without significantly affecting the levels of PDEF mRNA (Figure 3B & C). PDEF protein is present at very low (miR-204) or undetectable levels (miR-510) in cells transfected with 1 g plasmid. These results demonstrate that miR-204 and miR-510 posttranslationally regulate endogenous PDEF mRNA, most likely through a mechanism of translation inhibition.
Figure 3. miR-204 & miR-510 regulate endogenous PDEF protein expression.
(A) Western blot analysis of endogenous PDEF expression in CAMA-1 cells transfected with scrambled ASO (control) or ASO against miR-204 (204) or miR-510 (510). MCF7 and MDA-MB-231 cells are control cells positive and negative for PDEF protein expression, respectively. (B & C) Western blot analysis of MCF7 cells transfected with increasing concentrations of (B) miR-204 or (C) miR-510. Quantitative real-time PCR analysis of PDEF mRNA levels from the treatments in A, B & C normalized to GAPDH are shown to the right of the western blots.
Functional consequences of the regulation of PDEF by miR-204
Studies in our laboratory have shown that the over-expression of PDEF in an invasive breast cancer cell line leads to an altered cell morphology (2, 3). MCF7 cells typically have an epithelial morphology described as cobblestone in appearance. However, MCF7 cells that were stably transfected with miR-204 and showed concomitant lower levels of PDEF protein expression (Figure 4A) had an altered cell morphology (Figure 4C). These cells also had a more spindle-like morphology and appeared more mesenchymal, a change similar to that observed in cells that have undergone an EMT (epithelial-to-mesenchymal transition) (26). PDEF is a transcription factor involved in the negative regulation of genes involved in metastatic progression. Studies in our laboratory have identified uPA and slug as negative PDEF response genes, as their mRNA expression levels decrease when PDEF is over-expressed (3) (and unpublished data), and as direct targets as shown by chromatin immunoprecipitation (Turner et al, manuscript in preparation). uPA and slug are known to be involved in metastatic progression and slug up-regulation plays distinct roles during EMT (27, 28) and therefore these genes were selected for examination in our miR-204 stable over-expressing cells. Consistent with negative regulation by PDEF, miRNA-mediated reduction in PDEF protein expression resulted in an increase in the levels of both uPA and slug by quantitative real-time PCR analysis (Figure 4B). The adhesion protein E-cadherin plays critical roles during epithelial morphogenesis (29). Expression of this protein is down-regulated during the acquisition of invasive and metastatic phenotypes at late stages of epithelial tumor progression. Slug is established as a transcriptional repressor of E-cadherin gene expression in this process (28). Therefore, we performed quantitative real-time PCR analysis and confirmed that E-Cadherin mRNA levels were reduced in miR-204 over-expressing cells (Figure 4B). Concurrently, we observed a decrease in total and surface staining of E-Cadherin by immunofluorescence (Figure 4D) and total protein levels by western blot analysis (Figure 4A).
Figure 4. Functional consequences of miR-204 over-expression in MCF7 cells.
(A) Western blot analysis of PDEF and E-Cadherin (E-Cad) expression in MCF7 control cells (MCF7) and a miR-204 over-expressing stable clone (miR204). (B) Quantitative real-time PCR of E-cadherin, uPA and SLUG mRNA expression in MCF7 control (black bars) or miR-204 over-expressing cells (grey bars) normalized to GAPDH. (C) Bright field microscopy and (D) E-Cadherin immunofluorescence of MCF7 control cells (MCF7) and a miR-204 over-expressing MCF7 stable clone (miR204).
Cells that have undergone an EMT have higher migratory and invasive properties. To assess whether migration and/or invasion are altered in miR-204 over-expressing cells, we performed transwell migration assays across a chemokine gradient and invasion assays through matrigel. The number of cells found to migrate or invade in miR-204 over-expressing cells was significantly increased compared to the parental control (Figure 5A & B). To further explore the possible role of miR-204 in cancer progression, we investigated the ability of the stably transfected MCF7 cells to proliferate when seeded at low density in vitro using a clonogenic assay and observed an increase in the total number of colonies formed when compared to the parental control (Figure 5C). A hallmark of transformed cells is their ability to grow independent of anchorage; therefore, to determine whether the over-expression of miR-204 resulted in enhanced transformation, we performed a soft agar assay and observed an increase in the total number of colonies that were able to form in the cells overexpressing miR-204 (Figure 5D). Each of these molecular and cellular phenotypes were confirmed with multiple miR-204 stable clones (data not shown). Similar findings were obtained for cells following transient expression of miR-510 in MCF7 cells (Supplementary Figure 1).
Figure 5. miRNA over-expression increases migration, invasion and transformed phenotypes in vitro.
(A) Transwell migration (left panel) and matrigel invasion (right panel) of MCF7 control (black bars) or miR-204 over-expressing MCF7 (grey bars) cells. (B) Colony formation (left panel) and soft agar (right panel) assays of MCF7 control (black bars) or miR-204 over-expressing MCF7 (grey bars) cells. (C) Quantitative real-time PCR of uPA (left panel) and SLUG (right panel) mRNA expression of miR-204 over-expressing cells transiently transfected with PDEF (dark grey bars) or vector alone (black bars). (D) Transwell migration (left panel) and matrigel invasion (right panel) of miR-204 over-expressing cells transiently transfected with PDEF (dark grey bars) or vector alone (black bars).
PDEF expression inhibits the miR-204 over-expressing phenotype
MicroRNAs have multiple targets and therefore the effects observed following miR-204 expression may be the result of the increased PDEF protein as well as non-PDEF related miR-204 affects. One way to evaluate these possibilities is to examine the phenotypes in cells in which a non-targeted PDEF is expressed. To do this, the ORF of PDEF was transfected into the miR-204 over-expressing cells and molecular and cellular phenotypic effects were measured. Quantitative real-time PCR analysis showed that exogenous PDEF expression in miR-204 over-expressing cells resulted in a decrease in both uPA and SLUG mRNA levels when compared to the vector alone (Figure 6A & B). In addition, these cells were less migratory and less invasive (Figure 6C & D). Thus, the miRNA mediated loss of PDEF was responsible for the observed changes. The collective data suggest that during breast cancer progression, elevated miR-204 reduces PDEF protein expression, and the resultant change in expression of PDEF-regulated genes and their downstream targets contributes to a more invasive phenotype.
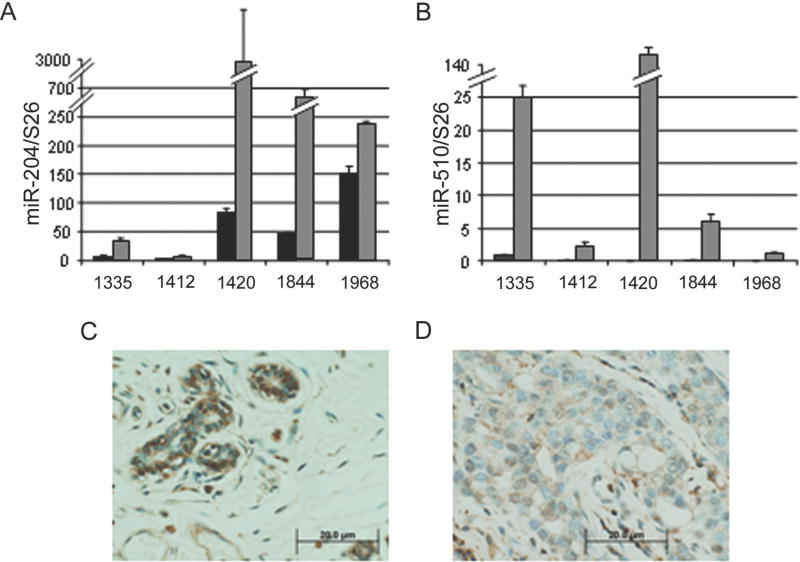
Figure 6. miR-204 & -510 levels are elevated in human breast cancer.
Quantitative real-time PCR analysis of (A) miR-204 and (B) miR-510 levels in human breast tumor (grey bars) and matched non-tumor (black bars) samples (For p values see Supplementary Table 3). Representative immunohistochemical staining of PDEF in human (C) non-tumor and (D) tumor breast tissue (Case 1335; magnification X400).
miR-204 and miR-510 levels are elevated in breast tumor samples
Based upon the impact of miR-204 and miR-510 on PDEF protein expression and PDEF dependent phenotypes, we evaluated the levels of miR-204 and miR-510 in RNA prepared from human breast tumor and matched non-tumor samples by quantitative real time PCR. Relative to that found in non-tumor tissue samples, the levels of miR-204 and miR-510 were found to be significantly elevated in all tumor samples (Figure 6A & B; Supplementary Table 3). These data suggest that miR-204 and miR-510 function as ‘oncomiRs’ and further support the model that elevated expression of miRNAs contribute to the loss of PDEF protein expression during breast cancer progression. We have previously demonstrated that PDEF protein expression is reduced in invasive breast cancer specimens (3). PDEF expression was evaluated in 4 of the 5 samples evaluated for miRNA expression as they were available as formalin-fixed paraffin embedded blocks from the same patients (1335, 1412, 1420 & 1844). As shown in figure 6C and 6D, PDEF protein is found predominantly in the nucleus of non-tumor epithelial cells. However, consistent with our previous findings, epithelial cells present in invasive ductal carcinomas show decreased protein expression. Taken together, these data suggest that miR-204 & -510 levels are elevated in invasive breast cancer and that there levels are inversely correlated to PDEF protein expression.
DISCUSSION
The rapidly evolving field of miRNAs has established close correlations between their altered expression and many types of cancer (30–32). However, the identification and validation of specific targets has been limited. This study identifies two miRNAs (miR-204 and miR-510) that are involved in the negative regulation of PDEF mRNA translation and describes for the first time a mechanism that contributes to PDEF protein loss during breast cancer progression. Specifically, over-expression of miR-204 and -510 in MCF7 cells results in a dose-dependent loss of PDEF protein expression. Although ~60% transfection efficiency was achieved in MCF7 cells, an apparently greater loss of PDEF protein expression was observed, perhaps due to a threshold level required for detection of protein by western blot analysis. A potential role for PDEF as a tumor suppressor has been demonstrated by studies showing reduced PDEF protein expression and/or loss by immunohistochemistry in invasive cancer (3), as well as the repression of survivin (33), uPA (3) and slug (Turner et al, manuscript in prep). Furthermore, recent studies showed PDEF protein expression inhibited xenograft tumor formation in vivo (33). Identifying the mechanisms of PDEF protein loss provides novel insight into the transcriptional networks that are active during metastatic progression. It is generally accepted that multiple mechanisms of regulation of any protein are active in normal cells and each of these represent pathways that can be altered during pathogenesis. Indeed, we show that regulatory elements are also present within the 5′UTR of PDEF, supporting a previous model that the 5′UTR of PDEF is involved in its post-transcriptional regulation in prostate cancer cells through inhibition of translation initiation (5). Interestingly, a recent study found that mRNA are repressed by engineering miRNA-binding sites in the 5′UTR as efficiently as in the 3′UTR, although no endogenous targets regulated in this manner have been found (34). In addition, both PDEF mRNA (4) and protein expression (Findlay et al., unpublished observations) are lost upon stimulation of cells with TGF-beta, a soluble growth factor able to induce EMT (35). The mechanism of TGF-beta dependent loss of PDEF mRNA and protein expression may include activation of inhibitory miRNAs, as a recent study illustrated a TGF-beta-induced EMT miRNA signature in human keratinocytes (36). Indeed, the EMT-like phenotype observed in MCF7 cells stably transfected with miR-204 or following transient expression of miR-510, supports the notion that growth factors, like TGF-beta, that induce EMT may play a role in the regulation of this and other miRNAs during breast cancer progression.
This and other studies have demonstrated the importance of PDEF regulation during breast cancer progression. We also show that miR-204 and miR-510 levels are elevated in breast cancer compared to non-tumor tissue. Until now the expression levels of miR-204 and miR-510 have not been specifically examined in breast cancer specimens, although a high through-put microarray study has been performed for miRNA profiling on breast cancer biopsies (37). While miR-204 levels were not specifically highlighted in this study, the datasets show a significant elevation of miR-204 in those breast cancer biopsy samples with a tumor cell percentage of 70% or more when compared to the normal controls. miR-510 levels were not evaluated in this study. Of interest, the genomic locations of these two miRNAs have been associated with cancer. Amplification of a non-coding region mapped to chromosome 9q21 (miR-204) has been associated with prostate cancer (38). Furthermore, amplification of Xq27 (miR-510) occurs during the process of cell transformation and tumorigenesis using breast cancer as a model system and in sporadic breast cancer (39, 40). Future studies directed towards elucidating the mechanism of activation of these miRNAs will provide valuable insight into the complex pathways involved in metastatic breast cancer progression. This is exemplified by the recent study that identified the metastatic specific induction of miRNA-10b through the action of the transcription factor Twist (41), another known regulator of EMT, emphasizing the fundamental role for miRNAs in cancer progression and their representation as a new class of tumor suppressors and oncogenes.
The function of the majority of miRNAs is still currently unknown, although a plethora of predicted targets exist. We show here for the first time a validated target, PDEF, for two miRNAs, miR-204 and miR-510, as well as functional consequences of their interaction. These studies support the model that miR-204 and miR-510 are potential oncogenes and that their elevated expression contributes to the loss of PDEF protein expression during breast cancer progression. It is likely that additional miRNAs are involved in the post-transcriptional regulation of PDEF and their identification will increase our understanding of the mechanisms that exist to regulate PDEF during breast cancer progression. Furthermore, future studies involved in the identification of additional targets for these miRNAs will provide greater insight into the complex biological pathways involved in metastatic progression.
Acknowledgments
We thank Richard Neve for the kind gift of cell lines. This work was supported in part by a grant from the National Institutes of Health (P01CA78582). The authors declare no competing financial interests.
References
- 1.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41(16):2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Turner DP, Moussa O, Sauane M, Fisher PB, Watson DK. Prostate-derived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer research. 2007;67(4):1618–25. doi: 10.1158/0008-5472.CAN-06-2913. [DOI] [PubMed] [Google Scholar]
- 3.Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer research. 2003;63(15):4626–31. [PubMed] [Google Scholar]
- 4.Gu X, Zerbini LF, Otu HH, et al. Reduced PDEF expression increases invasion and expression of mesenchymal genes in prostate cancer cells. Cancer research. 2007;67(9):4219–26. doi: 10.1158/0008-5472.CAN-06-3689. [DOI] [PubMed] [Google Scholar]
- 5.Nozawa M, Yomogida K, Kanno N, et al. Prostate-specific transcription factor hPSE is translated only in normal prostate epithelial cells. Cancer research. 2000;60(5):1348–52. [PubMed] [Google Scholar]
- 6.Gillanders WE, Mikhitarian K, Hebert R, et al. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Annals of surgery. 2004;239(6):828–37. doi: 10.1097/01.sla.0000128687.59439.d6. discussion 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitas M, Mikhitarian K, Hoover L, et al. Prostate-Specific Ets (PSE) factor: a novel marker for detection of metastatic breast cancer in axillary lymph nodes. British journal of cancer. 2002;86(6):899–904. doi: 10.1038/sj.bjc.6600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner DP, Findlay VJ, Moussa O, Watson DK. Defining ETS transcription regulatory networks and their contribution to breast cancer progression. Journal of cellular biochemistry. 2007;102(3):549–59. doi: 10.1002/jcb.21494. [DOI] [PubMed] [Google Scholar]
- 9.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43(10):1529–44. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky N. microRNA target predictions in animals. Nature genetics. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 12.Shilo S, Roy S, Khanna S, Sen CK. MicroRNA in cutaneous wound healing: a new paradigm. DNA and cell biology. 2007;26(4):227–37. doi: 10.1089/dna.2006.0568. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–29. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 14.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 15.Hornstein E, Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438(7068):671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & development. 2006;20(20):2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes & development. 2007;21(5):578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 19.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends in molecular medicine. 2006;12(12):580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown KA, Aakre ME, Gorska AE, et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6(3):R215–31. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilles C, Polette M, Birembaut P, Brunner N, Thompson EW. Expression of c-ets-1 mRNA is associated with an invasive, EMT-derived phenotype in breast carcinoma cell lines. Clinical & experimental metastasis. 1997;15(5):519–26. doi: 10.1023/a:1018427027270. [DOI] [PubMed] [Google Scholar]
- 28.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer research. 2002;62(6):1613–8. [PubMed] [Google Scholar]
- 29.Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. The International journal of developmental biology. 2004;48(5–6):365–75. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- 30.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 33.Ghadersohi A, Pan D, Fayazi Z, Hicks DG, Winston JS, Li F. Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibits breast cancer cell growth in vitro and xenograft tumor formation in vivo. Breast cancer research and treatment. 2007;102(1):19–30. doi: 10.1007/s10549-006-9314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′UTR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(23):9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 36.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells, tissues, organs. 2007;185(1–3):157–61. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 37.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Molecular cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer research. 1999;59(23):5975–9. [PubMed] [Google Scholar]
- 39.Balogh GA, Russo IH, Balsara BR, Russo J. Detection of chromosomal aberrations by comparative genomic hybridization during transformation of human breast epithelial cells in vitro. International journal of oncology. 2006;29(4):877–81. doi: 10.3892/ijo.29.4.877. [DOI] [PubMed] [Google Scholar]
- 40.Valladares A, Salamanca F, Madrigal-Bujaidar E, Arenas D. Identification of chromosomal changes with comparative genomic hybridization in sporadic breast cancer in Mexican women. Cancer genetics and cytogenetics. 2004;152(2):163–6. doi: 10.1016/j.cancergencyto.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 42.Oettgen P, Finger E, Sun Z, et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. The Journal of biological chemistry. 2000;275(2):1216–25. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]