Abstract
The nervous system maintains a delicate balance between excitation and inhibition, partly through the complex interplay between voltage-gated sodium and potassium ion channels. Because K+ channel blockade or gene deletion causes hyperexcitability, it is generally assumed that increases in K+ channel gene expression should reduce neuronal network excitability. We have tested this hypothesis by creating a transgenic mouse that expresses a Shaker-type K+ channel gene. Paradoxically, we find that addition of the extra K+ channel gene results in a hyperexcitable rather than a hypoexcitable phenotype. The presence of the transgene leads to a complex deregulation of endogenous Shaker genes in the adult central nervous system as well as an increase in network excitability that includes spontaneous cortical spike and wave discharges and a lower threshold for epileptiform bursting in isolated hippocampal slices. These data suggest that an increase in K+ channel gene dosage leads to dysregulation of normal K+ channel gene expression, and it may underlie a mechanism contributing to the pathogenesis of human aneuploidies such as Down syndrome.
Ion channel gene families typically encode membrane proteins that form multimers with similar, yet distinct, functional properties. One of the most extensive gene families is the voltage-gated K+ channel superfamily (1, 2). Members of this gene family are outward-rectifying K+ channels and can be classified into four subfamilies, Kv1, Kv2, Kv3, and Kv4 on the basis of amino acid identities. Kv1–4 K+ channels delimit the duration of the action potential and repetitive firing properties of excitable cells. Within this superfamily, functional diversity arises predominantly through subfamily-specific formation of homo- and heterotetrameric complexes of α subunits (3, 4). However, in the Kv1 gene subfamily, further heterogeneity occurs through the association of α transmembrane subunits with accessory cytoplasmic β subunits (5–7). These cytoplasmic β subunits can function both as chaperone proteins to direct localization of specific α subunits to the plasma membrane and as modulators of K+ channel inactivation (8, 9).
Insight into the role that K+ channels play in controlling neuronal excitability has come from studies of natural and induced mutations. Missense point mutations in the human voltage-gated K+ channel gene Kv1.1 are linked to episodic ataxia type 1 (10, 11). Expression of mutant Kv1.1 subunits with wild-type subunits in Xenopus oocytes results in a suppression of the corresponding K+ channel current and suggests that these missense mutations act through a dominant-negative effect (12). Introduction of a null mutation into the mouse Kv1.1 gene results in an epileptic phenotype characterized by frequent spontaneous tonic–clonic seizures leading to increased morbidity in young animals, as well as alteration of axonal action potential conduction in the sciatic nerve (13).
Precise spatiotemporal expression of each K+ channel subunit gene is also critical during brain development (14, 15). Overexpression of Kv1.1 mRNA in amphibian embryos leads to larger delayed-rectifier K+ channel currents, shorter action potentials, and a reduction in the number of morphologically differentiated neurons in culture (16). In the mammalian central nervous system (CNS), the regulatory mechanisms that coordinate K+ channel subunit gene expression and hence the stoichiometry of heteromeric ion channels are poorly understood. The complexity of Kv1 gene expression suggests that alterations in K+ channel gene copy number could result in multiple changes in the regulation of other K+ channel genes. Here we report a genetic approach to address this hypothesis through the overexpression of an Aplysia Shaker-type K+ channel gene (AKv1.1a) (17) in the murine CNS. We refer to this transgenic mouse as HypK.
Expression of the transgenic protein was detected in both neuronal cell somas and dendrites and resulted in a dysregulation of endogenous K+ channel Kv1 gene transcription and a paradoxical hyperexcitable phenotype in regions of the CNS corresponding to AKv1.1a expression. These data show that the presence of an extra K+ channel gene during CNS development alters the regulation of endogenous K+ channel genes, both within the Kv1α subfamily and between the interacting Kvβ subunit genes. These findings suggest that dysregulation of K+ channel gene expression may contribute to an underlying pathogenic mechanism for neurological disorders associated with altered gene dosage.
MATERIALS AND METHODS
Generation and Genotyping Transgenic Animals.
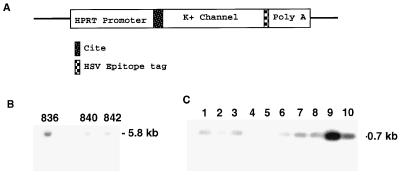
The 0.9-kbp (SalI–SacI) poly(A)+ DNA fragment and 2.2-kbp (EcoRI–SalI) K+ channel cDNA fragment were subcloned into plasmid pPP11 containing the human hypoxanthine phosphoribosyltransferase (HPRT) promoter. Prior to subcloning of the K+ channel cDNA, an idiotypic tag [derived from herpes simplex virus (HSV) glycoprotein D] was added to the 3′ terminus of the channel to aid in the localization of the transgene protein in situ. Expression of K+ current from the tagged K+ channel cDNA was recorded in an Xenopus oocyte expression system to confirm that the tag had no adverse effect on K+ channel function. A cap-independent translation enhancer sequence (CITE; Novagen) was included upstream of the ATG start codon for the K+ channel cDNA to increase the translation efficiency of the transgene. FVB mouse pronuclei were injected with a 6.5-kbp ScaI fragment prior to implantation into pseudopregnant females. Offspring were screened for the presence of the AKv1.1a sequence by using PCR and Southern blot analysis. Genomic DNA preparations from tails were characterized for transgene expression by Southern blot analysis using a 32P-labeled nick-translated probe complementary to the CITE sequence. The Southern blot was hybridized overnight at 55°C and washed at 65°C in 0.1× standard saline citrate (SSC). The PCR products were electrophoresed on a 0.8% agarose gel, prior to transfer onto a nylon membrane and hybridization with a 32P-labeled oligonucleotide probe internal to the PCR-amplified sequence. The Southern blot was washed at 55°C in 0.1× SSC and exposed to Kodak XAR film for 6 hr at room temperature. Three separate lines were identified, 836, 840, and 842, and they were maintained as hemizygotic through sibling crosses.
In Situ Hybridization and Immunocytochemistry.
In situ hybridization was performed as outlined by Wisden and Morris (18). Subunit-specific 45-mer antisense oligonucleotides were designed to nonhomologous regions of Kv1α and Kvβ K+ channel subunit cDNAs, as determined by amino acid alignments of mouse, rat, and Aplysia protein sequences. Horizontal 12-μm HypK mutant and age-matched control (+/+) brain sections were probed with subunit-specific Kv1α and Kv1β K+ channel antisense oligonucleotides (Genosys) that were 3′-end labeled by using terminal deoxynucleotidyltransferase (Promega) and deoxyadenosine 5′-[α-[35S]thio]triphosphate (1300 Ci/mmol; NEN; 1 Ci = 37 GBq) to a specific activity of 109 dpm/mg. Hybridization reactions were incubated overnight at 42°C, prior to washing in 0.3× SSC and 10 mM DTT, for 30 min at 55°C. Reverse images were printed directly from the x-ray film. Densitometry was performed with nih image software by observers blinded to the tissue genotype. Triplicate determinations of density were made in a precalibrated sample area within standardized regions of each brain section, and the ratio with respect to the adjacent white matter density was determined to normalize between sections. The means of these values for each genotype were then expressed as the ratio of mutant to wild-type control density levels for each subunit.
Immunocytochemistry was performed with a monoclonal antibody for HSV glycoprotein D (HSV-Tag, Novagen). Age-matched HypK adult transgenic and (+/+) FVB mice were anesthetized and perfused intra-aortically with 1× phosphate-buffered saline (PBS) followed by Accustain (Sigma) containing 4% formaldehyde. Brains were removed and fixed overnight in Accustain at 4°C, followed by cryoprotection in Accustain containing increasing concentrations (10–25%) sucrose, at 4°C. Horizontal sections (40 μm) were incubated at 25°C for 1 hr in Tris-buffered saline (TBS) containing 4% goat serum and 0.1% Triton X-100, rinsed three times in TBS containing 0.1% Triton X-100, and incubated in 0.3% hydrogen peroxide diluted in methanol for 30 min. The sections were washed three times in TBS containing 0.1% Triton X-100 prior to overnight incubation in TBS containing 0.1% Triton X-100, 2% normal goat serum, and 1 μg/ml HSV-Tag antibody. HSV-Tag immunoreactivity was detected by using an ABC Elite Mouse IgG kit (Vector Laboratories) and 3,3′-diaminobenzidine (DAB) substrate kit (Vector Laboratories) as outlined in the manufacturer’s protocols.
Cortical Electroencephalogram (EEG) Recordings.
Mice were anesthetized and bilateral cortical surface electrodes were implanted through cranial burr holes leading to a microminiature connector cemented to the skull. After several days of recovery, spontaneous electrocortical activity was recorded in the behaving animal for 60-min periods. Discharges were characterized by their frequency of appearance, duration, and spike frequency.
Hippocampal Slice Recordings.
Hippocampal slices (400 μm thick) were prepared from adult mice and recordings were carried out at 32–34°C under standard conditions in an interface chamber. The control solution contained 125 mM NaCl, 25 mM NaHCO3, 2 mM CaCl2, 1 mM MgCl2, 25 mM dextrose, and 2.5 mM KCl. Extracellular field potential recordings were made from area CA1, and only slices that exhibited >2-mV excitatory postsynaptic potential (EPSP) responses in normal saline and synchronous discharges in 10 mM K+ were used. Slices were serially exposed to various external K+ concentrations for 1-hr periods, and discharge frequency and duration were measured. Recordings were made from two to six slices per animal.
RESULTS
Generation of a Transgenic Shaker-Type K+ Channel Mouse.
Three lines of transgenic mice were generated expressing a construct containing the Aplysia AKv1.1a Shaker-type K+ channel cDNA, behind a fragment of the human HPRT promoter that drives CNS-specific expression (19) (Fig. 1). To allow unambiguous immunocytochemical identification of the transgenic protein, an 11-amino acid epitope tag from HSV glycoprotein D was added to the C terminus. The addition of the HSV epitopic marker resulted in the deletion of the AKv1.1a C-terminal ETDV sequence, which has been demonstrated in vitro to be necessary for subcellular specialization (20). The modified K+ channel cDNA was able to combine with mammalian Shaker subunits and form a functional channel in Xenopus oocytes. Transgenic mice showed no obvious behavioral or neurological deficits. Two strains were chosen for analysis, the high copy number (15 transgene copies) 836 strain and the lower copy number (4 transgene copies) 842 strain.
Figure 1.
HypK genotypic and mRNA characterization. (A) AKv1.1a K+ channel expression construct. (B) Southern blot analysis from genomic DNA tail preparations from founder mice shows highest copy number in 836 and 842 strains. (C) F2 mice carrying the transgene were identified by PCR analysis with forward and reverse oligonucleotide primers to CITE and 5′ Aplysia sequences, respectively.
Localization of the Transgenic Protein in the HypK Mouse.
The presence of AKv1.1a was detected by in situ hybridization with a subunit-specific antisense oligonucleotide probe. AKv1.1a mRNA was found in several brain areas, including neocortex, hippocampus, and cerebellum. Using a peptide antibody to the HSV sequence tag, we detected the strongest immunoreactivity in the cerebral cortex and hippocampus (Fig. 2 A and B). In both regions, pyramidal cell somas and dendrites were heavily stained, indicating that the invertebrate AKv1.1a protein was localized to subcellular compartments (Fig. 2C). Specific staining of interneurons was less evident, and no glial staining was seen.
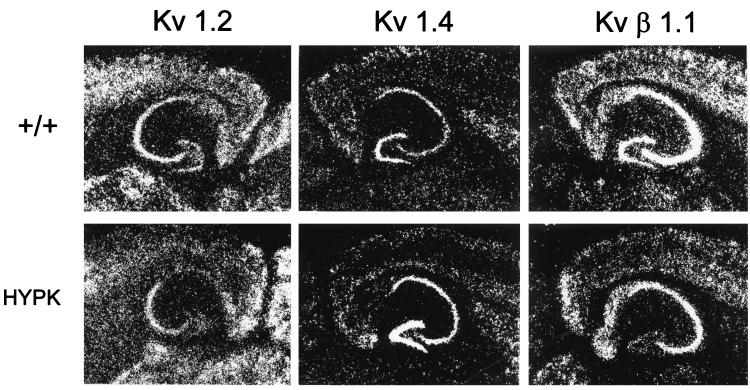
Figure 2.
Localization of AKv1.1a transgene protein in the adult HypK hippocampus. (A) Immunoreactive product for the AKv1.1a transgene protein was detected by using a monoclonal antibody to the HSV sequence in the hippocampus of an adult HypK mutant brain. (B) FVB control hippocampus was unstained under identical immunohistological conditions. (C) Immunoreactive staining for the AKv1.1a transgene protein was strongly positive in cell bodies and apical dendrites of CA1 pyramidal cells. Neocortex showed similar staining intensity. (Scale bars represent 100 μm for A and B and 3.5 μm for C.)
Analysis of Endogenous Kv1 α and β Subunit Gene Expression.
We tested the hypothesis that the presence of extra K+ channels during CNS development alters the regulation of endogenous neuronal K+ channel genes by using in situ hybridization analysis with antisense oligonucleotide probes that distinguish different native Shaker α (Kvα1.1–1.6) and β (Kvβ1 and -2) subunit mRNAs. Extensive subunit- and region-dependent alterations in adult Kv1α and Kvβ mRNA levels were observed in neocortex and hippocampus coincident with high AKv1.1α expression (Fig. 3 and Table 1). Kv1.2 showed a selective decrease within dentate granule cells. Kv1.5 and Kv1.6 subunits showed the most consistent widespread increases in mRNA expression, but the Kv1.5 transcript was clearly decreased in the hippocampal hilar cell subpopulation. Selective increases were also found for Kv1.4 mRNA in dentate granule cells, for Kv1.1 mRNA in neocortical layers II/III and hippocampal CA1 pyramidal cells, and for Kv1.3 mRNA in hippocampal CA1 and CA3 cells. In contrast to the Kv1α subunits, both Kvβ subunit mRNAs were down-regulated. Kvβ1 was reduced in dentate granule cells and neocortical layers II/III, while Kvβ2 was reduced in dentate granule and CA1 pyramidal cells. Neither the elevation of native Kv1α transcripts nor the decrease in expression of Kvβ mRNAs observed in the adult HypK CNS would be expected if a homeostatic regulatory mechanism functions to preserve subunit stoichiometry within K+ channel populations.
Figure 3.
Dysregulation of endogenous K+ channel gene expression in HypK transgenic mice. Representative autoradiograms of in situ hybridization of K+ channel subunit mRNA expression in adult FVB hippocampal control (+/+) (Upper) and HypK transgenic (Lower) mice. Kv1.2 mRNA expression is selectively down-regulated in the dentate gyrus of the HypK transgenic mouse, whereas Kv1.4 mRNA levels are up-regulated. Kvβ1 mRNA expression is down-regulated in the dentate gyrus and to a lesser degree in layers II/III of the overlying neocortex of the HypK transgenic mouse.
Table 1.
Regional K+ channel subunit expression changes in HYPK mutant brain
| Region | Normalized subunit expression
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Kv1.1 | Kv1.2 | Kv1.3 | Kv1.4 | Kv1.5 | Kv1.6 | Kvβ1.1 | Kvβ2 | |
| Dentate | 1.06 | 0.41* | 0.98 | 1.27* | 0.65* | 1.35* | 0.59* | 0.69* |
| Hilus | 1.1 | 1.03 | 1.16 | 0.97 | 1.00 | 1.63* | 0.89 | 1.00 |
| CA2/3 | 0.94 | 0.96 | 1.54* | 0.92 | 1.25* | 1.72* | 0.92 | 0.95 |
| CA1 | 1.3 | 0.93 | 1.33* | 0.99 | 1.55* | 1.62* | 0.9 | 0.73* |
| Cortex II/III | 1.32* | 0.78 | 1.12 | 1.00 | 1.48* | 2.18* | 0.75* | 1.05 |
| Cortex IV/VI | 1.08 | 1.05 | 1.1 | 0.84 | 1.34* | 1.7* | 0.98 | 1.12 |
Values represent ratio of the mean optical density from triplicate densitometric determinations of each specific hybridized probe in HypK brain sections divided by those from matched +/+ brain sections. Regulation of native mRNA levels by AKv1.1a transgene expression was neither region nor subunit dependent.
Levels in mutant brain altered by 25% or greater.
Analysis of Altered Network Excitability.
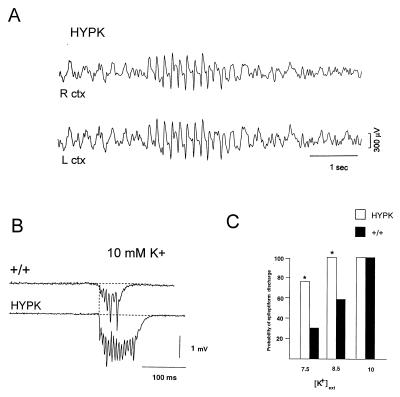
To determine whether the expression of the K+ channel transgene or up-regulation of native subunits might lead to reduced neuronal excitability, electroencephalograms were recorded from adult neocortex by using chronically implanted surface electrodes. Unexpectedly, the HypK mice showed spontaneous abnormal discharges (Fig. 4A), similar to those seen in genetic models of spike-wave epilepsy (21). Discharges were present in both strains examined, excluding the possibility of a de novo insertional mutation, and were more frequent in the high-copy number (836) strain. No abnormal seizure activity was observed in control FVB mice. We tested whether enhanced excitability might be present in hippocampal networks by examining in vitro slices. Latent network hyperexcitability was revealed by elevating extracellular potassium concentration ([K+]ext) in the bathing solution to produce network bursting. In slices bathed in supramaximal (10 mM) [K+]ext solutions, mutant slices demonstrated a small (≈20%) but significant increase in the duration of the synchronous discharge (+/+ = 97 ± 4 ms; HypK = 116 ± 7 ms: t test P > 0.05) (Fig. 4B), a feature seen in other spike-wave mutant models (22). The threshold for triggering synchronous network discharges was also decreased in HypK slices. At doses near the seizure threshold (7.5 and 8.5 mM) in wild-type slices, [K+]ext elevation raised the probability of bursting nearly 2-fold in HypK slices (bursting activity appeared in 11/36 (31% in 7.5 mM K+) and 17/29 (59% in 8.5 mM K+) of +/+ slices (n = 13 mice), compared with 29/38 (76% in 7.5 mM K+) and 25/25 (100% in 8.5 mM K+) slices from (836) HypK mice (n = 10 mice) (Fig. 4C).
Figure 4.
HypK mice exhibit spontaneous electroencephalogram (EEG) discharges in vivo and have a lower threshold for epileptiform discharge in hippocampal slices. Cortical EEG traces were recorded from the right (R) and left (L) hemispheres in mice from the HypK high transgene copy number line (836) (12 adults 2–3 months of age) and HypK low transgene copy number line (12 age-matched adults). (A) Spontaneous bilateral spike and wave seizure activity was routinely observed. In 6 control FVB (+/+) mice (not shown), abnormal synchronous activity was never present. (B) Extracellular field recordings from the CA3 pyramidal cell layer reveal prolongation of K+-induced network discharge in HypK hippocampus. Upper trace is from a control FVB slice in 10 mM [K+]ext and the lower trace is from the HypK high transgene copy number strain, 836. Discharges from the 836 HypK mutant strain were significantly prolonged (≈20%; P > 0.05, t test). (C) Lower threshold for network bursting in HypK hippocampal circuits. Histogram indicating the number of slices, expressed as a proportion of the total, that showed spontaneous epileptiform activity at each [K+]ext. The proportion of slices exhibiting spontaneous activity in both 7.5 mM and 8.5 mM [K+]ext was approximately 2-fold greater in transgenic mice compared with control (P > 0.05, sign test).
DISCUSSION
These data demonstrate the stable expression of an invertebrate K+ channel gene in the mammalian CNS. Shaker AKv1.1a immunoreactivity is localized to the soma and dendrites of neurons in the cerebral cortex and hippocampus of the HypK transgenic mouse. Two clear phenotypes are observed in response to AKv1.1a expression: (i) cellular changes in native K+ channel α and β subunit mRNA levels and (ii) network excitability increases in regions where both mRNA changes and transgenic K+ channel protein expression are most prominent.
AKv1.1a is an invertebrate voltage-gated K+ channel that is expressed in R15 and R2 neurons of the marine mollusk Aplysia. Transient overexpression of this gene in these ganglion cells results in a large increase in Shaker K+ channel current, inhibition of action potentials, and suppression of synaptic release (23). Since Kv1 invertebrate and vertebrate channels normally contribute to membrane potential repolarization, it was interesting to observe that overexpression of the AKv1.1a K+ channel gene in the HypK transgenic mouse results in a hyperexcitable network response.
Can the observed dysregulation of native Shaker α and β subunits in the HypK transgenic mouse directly explain the hyperexcitable phenotype? A numerical increase in most Kvα subunits would be inconsistent with enhanced excitability; similarly, since Kvβ1 promotes channel inactivation, the decreases of Kvβ subunits in several regions should prolong channel activity and thus accelerate membrane repolarization. One intriguing possibility is suggested by the recent finding that Kvβ2 may play a chaperone-like role in K+ channel protein trafficking, at least for Kv1.2 (24). Thus the reduced Kvβ2 mRNA levels observed in HypK brain might alter subcellular targeting and decrease the functional expression of K+ channel proteins in membranes, despite elevated levels of Kvα mRNAs. This spatial regulation could be especially important in modulating excitability in specific subcompartments of the neuron, for example in hippocampal dendrites, where low-voltage A-type currents dampen electrical propagation of Na+ and Ca2+ action currents (25). Decreases in native A currents or replacement by the higher threshold, longer inactivating AKv1.1a current would limit recovery from both subthreshold and action potential depolarization and interfere with dendritic integration, especially during increased firing activity.
Could the dysregulation of endogenous Kv1 gene expression be a direct result of seizure activity? Previously, changes in K+ channel gene regulation have been observed in a pentylenetetrazole-induced convulsive seizure model, where both Kv1.2 and Kv4.2 mRNAs were transiently down-regulated (26). Hence some or all of the changes in Kv1 α and β K+ channel mRNA levels seen in the HypK mouse could be due to acute seizure-driven changes in gene expression. The possibility that all changes in K+ channel mRNA levels might be an exclusive consequence of the epileptic phenotype rather than a contributor to it is unlikely, however, since in situ hybridization analysis of brain sections in an unrelated epileptic mutant, stargazer, with a >10-fold more severe spike-wave seizure phenotype, revealed no change in K+ channel mRNA levels (data not shown). Taken together, these observations suggest that at least some of the changes in endogenous Kv1 gene regulation in the HypK transgenic mouse are not seizure-driven but are instead a consequence of disrupting coordinated Kv1 gene expression during CNS development.
Since K+ channels play an important, but still incompletely characterized, role in nervous system development (14–16), changes in K+ currents resulting from expression of the AKv1.1a transgene could have profound effects on normal CNS development and excitability. These changes could prove to be heterogeneous in different brain pathways, depending on the types of endogenous K+ channels expressed. Single cell analysis will be required to demonstrate the functional expression of the Kv1.1α transgene within the dendrites, soma, and axon to determine the developmental impact of increased K+ channel currents on the integration of synaptic impulses, spike electrogenesis, and neurotransmitter release within an identified circuit.
Our data indicate that transgenic insertion strategies that modify ion channel gene dosage can lead to extensive cell-specific remodeling of endogenous ion channel gene expression within the CNS. The extent of secondary gene dysregulation adds an important dimension of developmental plasticity to the interpretation of gene function by transgenic mutational analysis, and to potential gene therapy strategies. This pattern of molecular plasticity might also contribute to the pathogenesis of deleterious neurological phenotypes in human gene duplication and aneuploidy syndromes (27), one of which (trisomy 21) is known to include two potassium channel genes within the critical region for Down syndrome (28, 29) and is associated with cognitive impairment and an increased incidence of seizures (30).
Acknowledgments
We thank Pragna Patel for the HPRT promoter construct. This work was supported by the American Epilepsy Society (M.L.S.), the National Institute on Drug Abuse (S.H.W.), National Institute of Neurological Disorders and Stroke, the National Institute of Child Health and Human Development (P.A.O. and J.L.N.), and the Blue Bird Circle Foundation for Pediatric Neurology.
ABBREVIATIONS
- CNS
central nervous system
- HypK
hyperexpressing K+ channel
- HPRT
hypoxanthine phosphoribosyltransferase
- HSV
herpes simplex virus
- [K+]ext
extracellular potassium concentration
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Roeper J, Pongs O. Curr Opin Neurobiol. 1996;6:338–341. doi: 10.1016/s0959-4388(96)80117-6. [DOI] [PubMed] [Google Scholar]
- 2.Sheng M, Jan Y N, Jan L. Prog Brain Res. 1995;105:87–93. doi: 10.1016/s0079-6123(08)63286-0. [DOI] [PubMed] [Google Scholar]
- 3.Isacoff E Y, Jan Y N, Jan L Y. Nature (London) 1990;345:475–476. doi: 10.1038/345530a0. [DOI] [PubMed] [Google Scholar]
- 4.Mackinnon R. Nature (London) 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- 5.Rettig J, Heinemann S H, Wunder F, Lorra C, Parcej D N, Dolly J O, Pongs O. Nature (London) 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 6.Heinemann S H, Rettig J, Wunder F, Pongs O. FEBS Lett. 1995;70:132–137. doi: 10.1016/0014-5793(95)01377-6. [DOI] [PubMed] [Google Scholar]
- 7.England S K, Uebele V N, Shear H, Kodali J, Bennett P B, Tamkun M M. Proc Natl Acad Sci USA. 1995;92:6309–6313. doi: 10.1073/pnas.92.14.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales M J, Castellino R C, Crews A L, Rasmussen R L, Strauss H C. J Biol Chem. 1995;270:6272–6277. doi: 10.1074/jbc.270.11.6272. [DOI] [PubMed] [Google Scholar]
- 9.Shi G, Nakahira K, Hammond S, Rhodes K J, Schecter L E, Trimmer J S. Neuron. 1996;16:843–852. doi: 10.1016/s0896-6273(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 10.Browne D L, Gancher S T, Nutt J G, Brunt E R P, Smith E A, Kramer P, Litt M. Nat Genet. 1994;8:136–140. doi: 10.1038/ng1094-136. [DOI] [PubMed] [Google Scholar]
- 11.Browne D L, Brunt E R P, Griggs R C, Nutt J G, Gancher S T, Smith E A, Litt M. Hum Mol Genet. 1995;4:1671–1672. doi: 10.1093/hmg/4.9.1671. [DOI] [PubMed] [Google Scholar]
- 12.Adelman J P, Bond C T, Pessia M, Maylie J. Neuron. 1995;15:1449–1454. doi: 10.1016/0896-6273(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 13.Smart S L, Lopantsev V, Zhang C L, Robbins C A, Wang H, Chiu S Y, Schwartzkroin R A, Messing A, Tempel B L. Neuron. 1998;20:809–819. doi: 10.1016/s0896-6273(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 14.Ribera A B, Spitzer N C. Ion Channels. 1992;3:1–38. doi: 10.1007/978-1-4615-3328-3_1. [DOI] [PubMed] [Google Scholar]
- 15.Catterall W A. Annu Rev Biochem. 1995;64:493–5316. doi: 10.1146/annurev.bi.64.070195.002425. [DOI] [PubMed] [Google Scholar]
- 16.Jones S M, Ribera A B. J Neurosci. 1994;14:2789–2799. doi: 10.1523/JNEUROSCI.14-05-02789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffinger P J, Furukawa Y, Zhao B, Dugan D, Kandel E R. J Neurosci. 1991;11:918–927. doi: 10.1523/JNEUROSCI.11-04-00918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisden W, Morris B, Hunt S P. In: Molecular Neurobiology—A Practical Approach. Chand J, Wheal H, editors. Oxford: Oxford Univ. Press; 1991. pp. 205–225. [Google Scholar]
- 19.Rincon-Limas D E, Geske R S, Xue J J, Hsu C Y, Overbeek P A, Patel P. J Neurosci Res. 1994;38:259–267. doi: 10.1002/jnr.490380304. [DOI] [PubMed] [Google Scholar]
- 20.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 21.Noebels J L, Sutherland M L, Nahm W K, DiPasquale E. Cold Spring Harbor Symp Quant Biol. 1997;61:319–326. [PubMed] [Google Scholar]
- 22.Noebels J L, Rutecki P. Brain Res. 1990;524:225–230. doi: 10.1016/0006-8993(90)90695-8. [DOI] [PubMed] [Google Scholar]
- 23.Kaang B-K, Pfaffinger P J, Grant S G N, Kandel E R, Furukawa Y. Proc Natl Acad Sci USA. 1992;89:1133–1137. doi: 10.1073/pnas.89.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes K J, Monaghan M M, Barrezuet N X, Nawoschik S, Bekele-Arcuri Z, Matos M F, Nakahira K, Schechter L E, Trimmer J S. J Neurosci. 1996;16:4846–4860. doi: 10.1523/JNEUROSCI.16-16-04846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman D A, Magee J C, Colbert C M, Johnston D. Nature (London) 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 26.Tsaur M L, Sheng M, Lowenstein D H, Jan Y N, Jan L Y. Neuron. 1992;8:1055–1067. doi: 10.1016/0896-6273(92)90127-y. [DOI] [PubMed] [Google Scholar]
- 27.Holtzman D M, Bayney R M, Li Y W, Khosrovi H, Berger C N, Epstein C J, Mobley W C. EMBO J. 1992;11:619–627. doi: 10.1002/j.1460-2075.1992.tb05094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malo M S, Srivastava K, Ingram V M. Gene. 1995;159:273–275. doi: 10.1016/0378-1119(95)00102-c. [DOI] [PubMed] [Google Scholar]
- 29.Ohira M, Seki N, Nagase T, Suzuki E, Nomura N, Ohara O, Hattori M, Sakaki Y, Eki T, Murakami Y, Saito T, Ichikawa H, Ohki M. Genome Res. 1997;7:47–58. doi: 10.1101/gr.7.1.47. [DOI] [PubMed] [Google Scholar]
- 30.Stafstrom C E. Am J Mental Retard. 1993;98:12–26. [PubMed] [Google Scholar]