Abstract
Functional redundancy in genomes arises from genes with overlapping functions, allowing phenotypes to persist after gene knockouts. Evolutionary redundancy or evolvability of a genome is one step removed, in that functional redundancy is absent but the genome has the potential to evolve to restore a lost phenotype. Exploring the extent to which this recovery alters gene networks can illuminate how functional gene interactions change through time. Here, the evolvability of lysis was studied in bacteriophage T7, revealing hidden functional interactions. Lysis is the destruction of host cell wall and membranes that releases progeny and is therefore essential for phage propagation. In most phages, lysis is mediated by a two-component genetic module: a muralytic enzyme that degrades the bacterial cell wall (endolysin) and a holin that permeabilizes the inner membrane to allow the endolysin access to the cell wall. T7 carries one known holin, one endolysin, and a second muralytic enzyme that plays little role in lysis by wild-type phage. If the primary endolysin is deleted, the second muralytic enzyme evolves to restore normal lysis after selection for faster growth. Here, a second level of evolutionary redundancy was revealed. When the second muralytic enzyme was prevented from adapting in a genome lacking the primary endolysin, the phage reevolved lysis de novo in the absence of any known muralytic enzymes by changes in multiple genes outside the original lysis module. This second level of redundancy proved to be evolutionarily inferior to the first, and both result in a lower fitness and slower lysis than wild-type T7. Deletion of the holin gene delayed lysis time modestly; fitness was restored by compensatory substitutions in genes that lack known roles in lysis of the wild type.
Keywords: experimental evolution, lysis, T7 bacteriophage, modularity, genome evolution, adaptive evolution
Introduction
Genes appear to interact in subsets known as modules, often associated with phenotypes such as DNA metabolism, stress responses, or metabolism. Although some interactions may change over time (Tischler et al. 2008), it is rarely clear how modules arise, expand, contract, or diverge through evolution. One experimental approach to this question is to remove one or more genes affecting a phenotype and then allow the dismantled gene network to evolve and restore function. Will the modularity and other genetic properties of the newly evolved phenotype resemble that of the old? What will be the source of the genes that control the newly evolved phenotype? This approach should not only yield insights regarding the evolution of network modularity but also augment other methods for network analysis such as protein and gene interaction maps.
Here, we use lysis timing in bacteriophages (phages, viruses of bacteria) as a model system to address these questions. Lysis is an important phage life history trait that varies in timing between phages and is easily assayed. Phage progeny need to escape the bacterial hosts infected by their parents to spread to new hosts. For many phages, this escape is accomplished by lysis, which involves destroying the bacterial cell wall; the osmotic pressure of the cytoplasm then ruptures the membrane to release the entire contents of the cell. A modular, two-component genetic model of lysis is general to most large double-stranded phages thus far studied, although the specific control mechanisms vary (Young 1992; Wang et al. 2000; Xu et al. 2004, 2005). The general two-component pathway involves a phage-encoded holin and endolysin, which together are necessary and sufficient for lysis. The endolysin is an enzyme that degrades the cell wall. Left to itself, it accumulates within the cytoplasm and unable to cross the inner membrane to contact the cell wall. The holin accumulates in the inner membrane and then suddenly causes permeabilization. The resulting lesions allow the endolysin to access and destroy the cell wall. Although the molecular mechanisms of holin activity are not fully understood and may indeed differ between holins, they have the common property of entirely preventing endolysin from attacking the cell wall until a precisely timed release (Grundling et al. 2001). This saltatory behavior of holins allows the cell to continue phage production unabated until the moment of lysis (Josslin 1970; Wang 2006). Most phages carry only one known holin, although in many cases, holin function is affected by another class of genes known as antiholins (Young and Wang 2005).
The modular two-component lysis system appears to be conserved at a functional level despite multiple origins (Wang et al. 2000). The many nonhomologous variants raise the possibility that such a system can evolve de novo, a prospect explored here by experimentally evolving a bacteriophage T7 with deletions of its lysis genes. First, can T7 evolve lysis when all of its muralytic enzymes are blocked from lysis involvement? Such an outcome would represent a pathway as yet unknown in other phages and demonstrate major latent adaptive capacity for the life history trait of lysis. Second, is a newly evolved mechanism of lysis genetically modular? Third, is lysis by wild-type T7 as simple as generally thought, or do multiple genes share holin function?
T7 Lysis Biology
Gp17.5 is the only known holin in T7, although its activity has only been examined in the context of phage λ (Vukov et al. 2000), and some evidence points to the existence of additional T7 genes with holin activity. (T7 genes are named as a number. The gene is usually nonessential if it includes a decimal.) Surprisingly, nonsense mutants of gene 17.5 plate at normal efficiency in a nonsuppressing host, although lysis of liquid cultures is somewhat delayed in gene 17.5 nonsense mutants of the related phage T3 (Miyazaki et al. 1978). Another indication that T7 may contain additional holins is the partial failure of an optimality model to predict lysis time evolution (Heineman and Bull 2007).
In addition to the holin, T7 also encodes an N-acetylmuramyl-L-alanine amidase (Inouye et al. 1973), usually called lysozyme or gp3.5, that serves as the endolysin. Phages lacking lysozyme activity lyse late, and they leave mature phage virions trapped inside cells (Silberstein and Inouye 1975). Lysozyme has an additional regulatory function, binding to T7 RNA polymerase, and altering gene expression (Moffatt and Studier 1987; Zhang and Studier 1997, 2004). The two functions of T7 lysozyme can be separated, however, as gene 3.5 mutants have been isolated that affect only its muralytic activity. Unlike in most phages, the T7 genes for lysozyme and holin are not close to each other on the genome.
The internal core protein gp16, which is essential for morphogenesis of infective particles (Moak and Molineux 2004), also carries a domain with muralytic activity. The protein is ejected from the phage particle at the initiation of infection (Kemp et al. 2005), and its muralytic activity is required for genome entry under suboptimal growth conditions (Moak and Molineux 2000). Elimination of muralytic activity in gp16 by substitutions of the catalytic residue slows genome penetration of the infected cell but has little effect on lysis at the end of the phage development. However, when a phage lacking the lysozyme gene was allowed to evolve, substitutions in gene 16 restored rapid lysis (Heineman et al. 2005). Thus, gene 16 provides a first level of evolutionary redundancy in endolysin function for T7.
Materials and Methods
Cell and Phage Lines
IJ1126 [Escherichia coli K-12, F− recC22 sbcA5 endA Gal− thi supL Δ(mcrC-mrr)102:Tn10] was used for transfections of T7 genomic DNA. IJ1133 [E. coli K-12, F- ΔlacX74 thi Δ(mcrC-mrr)102::Tn10] was the primary host used for all adaptations and assays. It is referred throughout as the “normal” host when it bears no plasmid. A plasmid (pPK70) that carries gene 16 in the vector pWSK129 (Wang and Kushner 1991) was used to provide gp16 in trans to phages lacking gene 16. For simplicity, IJ1133 cells carrying pPK70 are referred to as “+gp16” hosts. Plasmid pAR4521, which provides gp3.5 (Zhang and Studier 2004), was used to complement T7 mutants lacking gene 3.5, and pTP298, which expresses λ phage genes R, Rz, and Rz1 (Rennell et al. 1991), was used to propagate the T7 gene 3.5 mutant AFK136. The mutant lysozyme of AKF136 (see below) binds T7 RNA polymerase normally but lacks endolysin activity (Zhang and Studier 2004).
Our wild-type T7 (T7+, GenBank accession number AY264774, Bull et al. 2003) has the same sequence as that of the original T7 (GenBank accession number V01146, Dunn and Studier 1983) except for a 1-bp insertion following base 1896 in the nonessential gene 0.6. The Dunn and Studier sequence is used to define the location of changes that arose here during evolution. Evolved lines are denoted by a subscript E; for example, T7+ adapted to our laboratory conditions is T7+E.
Deletions in T7 were generally created by growing phage on a host carrying a pUC18 containing an insert of T7 sequence, typically with 100–200 bp identity flanking the region to be deleted. For example, the insert used to create T7 Δ 17.5 was comprised of a polymerase chain reaction (PCR) product containing T7 bp 35718–36343 juxtaposed to bp 36553–36933; the plasmid pΔ17.5 thus contains parts of genes 17 and 18 but lacks gene 17.5 (holin, coded by T7 bp 36344–36547). T7 17am61, which carries a stop codon in the essential upstream gene 17, was plated on the amber suppressing IJ486 [E. coli B argF40 supE (pΔ17.5)]. Phages from a plaque, some of which were recombinant over the gene 17 amber mutation and the 17.5 deletion, were resuspended and selectively plated on the nonsuppressing strain IJ1133, on which only recombinants that had acquired wild-type gene 17 form plaques. The resulting phages were then assayed by PCR to test for the 17.5 deletion. Other T7 deletion mutants included T7 Δ 3.5, which lacks almost all of gene 3.5 (Zhang and Studier 2004); AFK136, in which codons 130–135 of gene 3.5 are deleted, specifically eliminating the lytic activity of gp3.5 while preserving T7 RNA polymerase binding (Zhang and Studier 2004), and T7 Δ 16, a complete deletion of gene 16 (Moak and Molineux 2000).
Genomic fragment exchanges between phages were used to combine multiple gene deletions or to isolate the phenotypic effects of particular changes in adapted lines. DNA from different strains was digested with restriction enzyme, fragments were isolated and sets of fragments were ligated to regenerate genomes, which were then transfected into competent IJ1126 cells. The phages constructed by this method were 1) T7 Δ 3.5,Δ 17.5, carrying both the 3.5 and 17.5 deletions as well as all changes (except one in gene 17 at position 34975) from T7 adapted to grow well in our environmental conditions (T7+61 from Heineman et al. [2005]; here referred to as T7+E); 2) T7 Δ16, Δ 17.5, carrying the gene 16 and 17.5 deletions; 3) AFK Δ16, carrying both the partial 3.5 deletion from AFK136 and the 16 deletion; and 4) various T7 Δ 17.5 and AFK Δ 16 phages also carrying changes that arose during adaptation. Recombinant phages are named by the genes in which they bear substitutions and deletions. For example, the phage identical to AFK Δ 16 except that it carries changes in genes 0.7, 1.6, and 1.8 that evolved in the AFK Δ 16 strain is named AFK Δ 16+g0.7+g1.6+g1.8. (All AFK Δ16 phages with a gene 1.8 change carry the gene 1.8-M1T substitution rather than the insertion, see supplementary table S1, Supplementary Material online.)
Serial Transfer
Conditions used to adapt phage lines were identical to those of Heineman et al. (2005). Briefly, phages were propagated by serial transfer using exponentially growing cells. Frozen stocks of cells were added to 10 ml Luria–Bertani at 37 °C and incubated for 1 h to a density of 1–2 × 108 cells/ml. At this time, 104–107 phages were added. After incubation for 20–60 min, 104–107 phages and/or infected cells were added to the next culture or the culture was treated with chloroform and stored. Chloroform-treated phage samples were used to resume propagation at a later time. Flasks were sometimes allowed to progress to lysis to facilitate recombination among phages and thereby decrease the effect of clonal interference, which can slow adaptation (Miralles et al. 1999).
The three phages adapted in this fashion were T7 Δ17.5 (7.5 h of propagation), T7 Δ3.5,Δ17.5 (52 h), and AFK Δ16 (59 h). Times represent the accumulated growth period. AFK Δ16 was adapted on IJ1133 +gp16 cells, which compensated for the otherwise lethal deletion of gene 16. The fitness of the adapted T7 Δ3.5,Δ17.5 and AFK Δ16 lines were no longer increasing rapidly by the end of adaptation, but it is not known whether phenotypic evolution was at an endpoint.
An earlier study used selective conditions identical to those used here to adapt various phages (Heineman et al. 2005). The evolved phages from that study, T7 Δ3.5E, AFK136E, and T7+E (originally referred to as T7Δ3.562, AFK13643, and T7+61 respectively) are used for comparison to those created in this work.
Assays of Phenotype
The transfer conditions impose directional selection on phage growth rate: those genotypes that increase their numbers the fastest in the culture conditions replace the others. To be meaningful, estimates of fitness must match the nature of selection. Fitness is measured as doublings/hour of the phage population, a measure proportional to r, “the intrinsic rate of increase” often used in ecology. The value of r or doublings/hour indicates how quickly the phage population expands and is directly comparable across different phages, regardless of their generation time and other life history parameters, but the comparison is meaningful only when the different phages are assayed under the same conditions. In keeping with this understanding, fitness assays employed a protocol nearly identical to that used while transferring, except that the multiplicity of infection (moi) was maintained below 0.1. For fitness assays, phages were transferred for an accumulated total of 100 min across 4–5 cultures, and titers were taken at 40 and 100 min. The initial 40 min allowed the phage population to begin expanding approximately exponentially (Heineman et al. 2005). Doublings/hour is calculated as [log2(Nt/N0)]/t, where Nt is the density of phages at time t, taking into account dilutions over the course of the assay.
Lysis assays were done as described in Heineman et al. (2005) and involved infecting cells grown as above with phage at an moi of about 5 (phage density of ∼5 × 108/ml). At this phage density, almost all cells are infected in less than a minute. The average lysis time can be approximated by fitting the change in cell turbidity (measured by a Klett–Summerson photoelectric colorimeter using a 540 nm green filter) to a cumulative normal distribution using an empirical least squares method (Heineman et al. 2005). Decline in cell turbidity is often used as a proxy for lysis (Abedon 1992; Zhang and Studier 2004), and we have previously found this measure of lysis time to adequately measure phage release in a variety of T7 phages, even in T7 lysis mutants (Heineman et al. 2005). Cultures infected with lysis-defective phages frequently do not lyse completely at the time of cell death but suffer a relatively small initial drop in turbidity followed by a long period of no change. The lysis “turbidity decline index” is calculated as the percent decline in turbidity from the turbidity maximum of the culture (which occurs shortly after phage addition) to the turbidity at the endpoint. High index values indicate relatively complete lysis (more complete clearing of the culture).
Burst size assays involved adding 107 phage to 109 cells and incubating for 5 min, followed by a 103 or 106 dilution, and further incubation to a final time of 21.5–25.5 min. Measurements made early in the assay used a 103 dilution while those later used the 106 dilution in order to prevent further adsorption more effectively. Comparisons between titers with and without chloroform at 5.5–7.5 min were used in order to determine the number of unabsorbed phages and thus the number of infected cells. Burst sizes are calculated as the number of phages released per infected cell.
Recombination Assays of Compensatory Evolution
Recombination assays distinguish changes that are strictly compensatory for genomic modifications from changes that are generally advantageous under the growth conditions (Heineman et al. 2005). An evolved phage was recombined with the wild type. Recombination was achieved by cross-streaking both phages on a plate to achieve widespread coinfection of the same host. When the original genomic modification is deleterious relative to the wild type, the wild-type background will prevail but will acquire any evolved changes that are generally beneficial. In one assay, two phages had adapted by different pathways, and recombination between them was used to assess epistatic interactions between the changes associated with each pathway.
Sequencing and Statistical Tests
Sequencing employed an automated ABI3100 and ABI BigDye mix (v.3.1). Sequencing used either PCR products or viral genome as template, and data were analyzed with DNAStar Lasergene Seqman II software (v.5.05). The entire genomes of T7 Δ17.5E, T7 Δ3.5,Δ17.5E, AFK Δ16E, and AFK136E were sequenced. T7 Δ17.5E, unlike the other evolved lines, was sequenced and assayed from an isolate, rather than from a population. Endpoint recombination assay populations were sequenced over the regions that contained differences between the recombined phages; similarly, intermediate populations of the AFK Δ16 adaptation were sequenced only over regions in which changes were present in the final phage. All genomic fragment exchanges were verified by sequence or PCR analysis. Unless indicated otherwise, all statistical comparisons were based on two-tailed t-tests.
Results
General Approach
Phages with deletions or partial deletions of genes implicated in lysis were constructed and characterized for their lysis phenotypes. Many were then adapted for growth rate improvement, which often was achieved by improvement in lysis efficiency and timing. The compensatory substitutions responsible for lysis changes were identified, in the hope of understanding the layers of redundancy and evolvability of lysis. The genes deleted included 3.5 (lysozyme), 16 (a virion core protein with muralytic activity), and 17.5 (holin).
Phenotypic Evolution of a Lysis-Defective Phage while Constraining the Primary Evolutionary Pathway
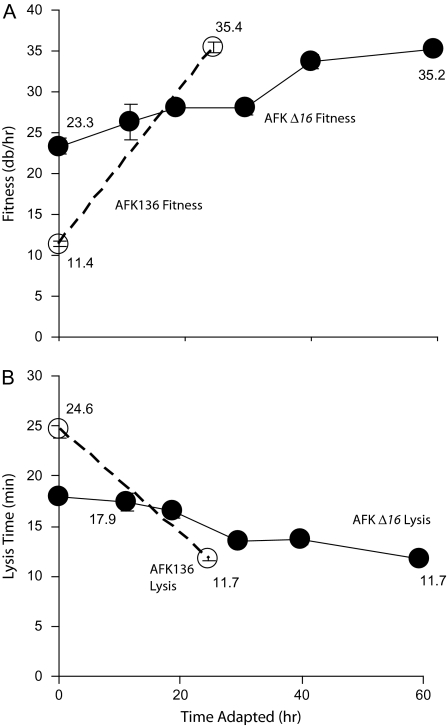
T7 AFK136 has a lysis-deficient endolysin (gp3.5). In prior work, this mutant evolved compensatory substitutions in the transglycosylase domain of gene 16, a gene that has only minor effects in lysis by wild-type T7. This adaptation is understandable because transglycosylases function as endolysins in other phages. Lysis time in AFK136 evolved from 24.6 to 11.7 min and fitness increased from 11.4 to 35.4 db/h, with most of the fitness increase attributable to substitutions in gene 16 (Heineman et al. 2005).
Recovery of AFK136 when gene 16 evolution is constrained is an obvious next step. AFK Δ16 productively infects cells carrying a plasmid encoding gene 16, but no evolution of gene 16 is possible because the wild-type gene is provided by the host rather than the phage genome. Thus, the main evolutionary pathway compensating for endolysin deficiency is blocked.
On the complementing (+gp16) host, the initial AFK Δ16 lyzed much later and had a lower fitness than wild-type T7 (figs. 1 and 2). The fitness of T7+ was relatively unaffected by whether the host provided gp16 or not (34.0 and 35.6 db/h, respectively); hence, the low fitness of AFK Δ16 was not primarily the result of the plasmid's presence in the host. Following adaptation of AFK Δ16 for 59 h, fitness increased and lysis time shortened (fig. 1). Fitness increase was gradual during serial transfer, without the steep initial rise that is frequently observed during adaptation to a novel environment. For comparison, unevolved AFK136 (on hosts not supplying gp16) had a lower fitness and a later time of lysis than the initial AFK Δ16 provided with gp16 in trans. This higher fitness of AFK Δ16 may be due to a beneficial effect of large amounts of host-produced gp16 to lysis when lysozyme is defective. Both AFK136E and AFK Δ16E attained similar phenotypic values after adaptation when the latter phage was provided with SP16 from host, but AFK136E did so much faster and from a later initial lysis time (fig. 1; Heineman et al. 2005), suggesting that constraining gene 16 evolution slows adaptation of lysis. Neither of the new pathways appears as effective as the original; from Heineman et al. (2005), T7+E has a fitness of 41.9 db/h (more than 6 db/h higher than either of the AFK adaptations) and a lysis time of 10.1 min (1.6 min faster).
FIG. 1.—
Evolution of AFK Δ16 (solid lines) on +gp16 hosts. (A) Fitness and (B) time to lysis. Data for AFK136 (dotted lines, open symbols) are from Heineman et al. (2005). Numbers are presented for the start and end of adaptation. Error bars, sometimes smaller than the data point, indicate ±one standard error.
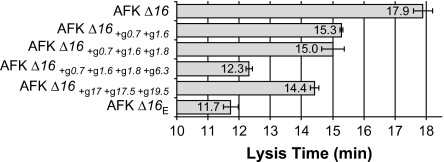
FIG. 2.—
Lysis times of various phages on hosts providing gp16. Error bars indicate ±one standard error.
A New Pathway for Lysis
The AFK Δ16E genome carried a number of changes, many in nonessential genes of unknown function (table 1). Genomes of intermediate populations were sequenced where substitutions were found in the evolved phage to determine when these changes arose. No changes were detected during the first 18.7 h of adaptation despite a significant fitness increase (from 23.4 to 27.9, P < 0.006). More comprehensive sequencing would be required to resolve the molecular basis of this change in fitness. For example, it could be that the fitter population was polymorphic for multiple changes, each too uncommon to be apparent in a consensus sequence.
Table 1.
Genetic Evolution of AFK Δ16 at Intermediate Times for Adaptation and after Recombination Assays
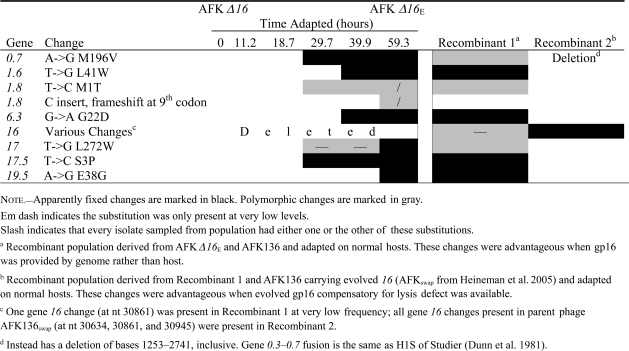 |
The effects of substitutions on lysis timing were assayed individually or in groups of 2–3 by genome fragment exchanges in the AFK Δ16 background (fig. 2). At least in combination, many of these changes affected lysis time. As might be anticipated, a change in 17.5 (holin) was observed, although the effect of this substitution alone was not assayed. The changes in gene 17 (tail fiber), gene 17.5, and gene 19.5 (see below) together resulted in much earlier lysis (from 17.9 to 14.4 min). All three gene products conceivably affect lysis time. The gene 0.7 and gene 1.6 substitutions together also have a major effect on lysis, hastening it from 17.9 min for the original unevolved phage to 15.3 min. Substitutions or deletions affecting gene 0.7 are common when T7 is serially transferred because gp0.7 is deleterious to phage development when host cells are growing in rich media (Studier 1979; Heineman et al. 2005; Springman et al. 2005). Gene 1.6 is nonessential; its function is unknown. Interestingly, gene 1.6 also evolved in two adaptations of lysozyme-deleted T7 (supplementary table S1, Supplementary Material online). Adding the substitution in gene 1.8 to the 0.7 and 1.6 mutations has little apparent effect, but the subsequent addition of the gene 6.3 change shortened lysis time from 15 to 12.3 min (P < 0.02; fig. 2). Little is known about gene 6.3. It is the last of the class II genes in the DNA metabolism cluster on the T7 genome and only codes for a 37 residue peptide. Gene 6.3 is also poorly conserved in otherwise closely related members of the T7 group. Interestingly, the same missense substitution in gene 6.3 is present in the gene 3.5 mutant T7 am13a (Molineux IJ, unpublished data), which was initially isolated nonselectively by an enzymatic assay after heavy mutagenesis of T7-infected cells (Studier 1972). Thus, although the evolved changes were analyzed in different backgrounds, large effect mutations occur in at least three different genes, and at least two of these are nonessential with no known function.
When AFK136 was adapted, gene 16 was the primary source of fitness improvement and faster lysis time. In the present experiment, gene 16 was not allowed to evolve, and (as noted above) a different molecular pathway resulted. The two adaptations thus offer two alternative pathways of evolution. Is one superior? To answer this question, we created a recombinant population carrying the mutations encoding both pathways for lysis and briefly adapted the progeny on normal hosts. If the pathways were compatible, the evolved phage would carry both sets of mutations; if not, the better pathway would spread. The fitnesses of both phages are too similar to suggest a superiority of one over the other (35.4 vs. 35.2; fig. 1), but a direct test is possible. For this comparison/competition, it was necessary that gene 16 be reintroduced into the AFK Δ16 background. One phage used to initiate the recombinant population thus carried the substitutions from AFK Δ16E that were still advantageous after the reintroduction of gene 16 into the genome (Recombinant 1; table 1). The second phage carried only the gene 16 changes that allowed lysis recovery during AFK136 adaptation (Heineman et al. 2005). The two phages were grown together to allow recombination and permit beneficial mutations to ascend in the population. This was thus a direct test of whether one evolutionary pathway of recovery was superior, or if both could be combined to increase fitness even more than either individually.
The phage resulting from this adaptation retained the gene 16 changes from the first recovery pathway, while losing all changes from AFK Δ16E (Recombinant 2; table 1). Thus, there was negative epistasis on fitness between the two sets of substitutions, and the primary pathway involving evolution of gene 16 was superior. The first layer of T7 evolutionary redundancy in lysis pathways is therefore more effective than the second.
The New Lysis Pathway Escapes gp16 Requirement
Gene 16 was prevented from evolving in the AFK Δ16 adaptation, but gp16 could nonetheless have been the primary enzymatic engine of lysis. This second possibility was suggested by the previously observed cryptic effect of gp16 on lysis (Heineman et al. 2005). Given that the wild-type gp16 can partially compensate for lysozyme deletion, the phage could have evolved to increase expression of the plasmid-borne gene 16 (cf. Bull et al. 2001), or it could have evolved to localize gp16 more effectively at the membrane in order to trigger lysis.
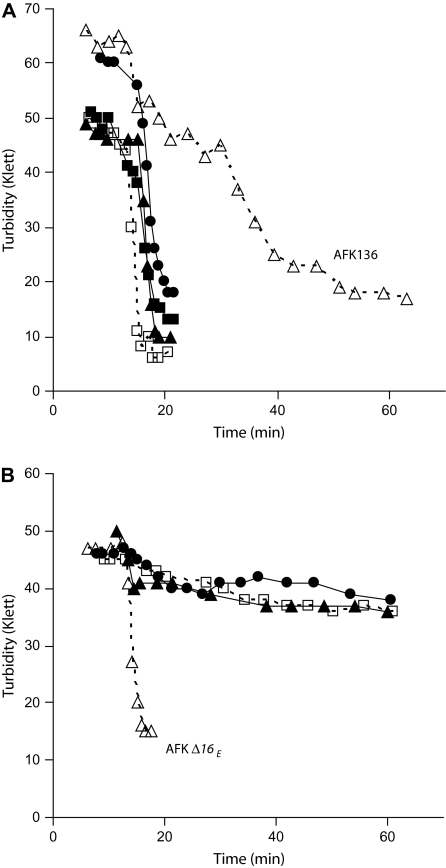
Deletion of gene 16 is an otherwise wild-type phage delayed lysis of infected, non-complementing bacteria by 2.1 minutes, to 15.9 min instead of 13.8 min (P < 0.0001, fig. 3). This delay was also seen with a 16 amber mutant (Heineman RH, unpublished data) and is therefore not due to the deletion per se. However, no lysis delay was noted after infection by 16am under different conditions of infection (Studier 1969) and any effect of gp16 on lysis when gp3.5 is present is therefore limited. Despite the delay observed, lysis by T7 Δ16 on non-complementing hosts was complete; culture turbidity declined by ∼85%, similar to decreases caused by wild-type (table 2, fig. 4A).
Table 2.
Turbidity Decline Indices
| Phage Line | Mean (%) |
| T7+ | 85 |
| T7 Δ16 | 84 |
| T7 Δ16,Δ17.5 | 69 |
| T7 Δ17.5 | 78 |
| AFK136 | 69 |
| AFK Δ16 | 14 |
| AFK Δ16+g0.7+g1.6 | 25 |
| AFK Δ16+g0.7+g1.6+g1.8 | 24 |
| AFK Δ16+g0.7+g1.6+g1.8+g6.3 | 21 |
| AFK Δ16+g17+g17.5+g19.5 | 12 |
| AFK Δ16E | 63 |
NOTE.—Higher percentages indicate more complete lysis/clearing of cell cultures. All values are from at least two measurements on normal (IJ1133) hosts.
FIG. 4.—
Representative turbidity lysis curves (measured in Klett units) on normal hosts after infection at an moi of 5. (A) Lysis of T7+ (open square), T7 Δ16 (closed triangle), T7 Δ17.5 (closed square), T7 Δ16,Δ17.5 (closed circle), and AFK136 (open triangle). (B) Lysis of AFK Δ16 (open square), AFK Δ16+g0.7+g1.6+g1.8+g6.3 (closed triangle), AFK Δ16+g17+g17.5+g19.5 (closed circle), and AFK Δ16E (open triangle).
However, gp16 is virtually essential for lysis when gp3.5 endolysin activity is absent; AFK Δ16 caused almost no lysis of cells lacking a gene 16 plasmid. The turbidity decline index was only ∼15%. (In contrast, those of AFK136 and T7 Δ16 were ∼70% and ∼85%, respectively; table 2, fig. 4.)
AFK Δ16E regained the ability to effectively lyse even cells that do not provide gp16; the ∼15% turbidity decline increased to ∼65% after adaptation (table 2, fig. 4B), indicating that some other gene or combination of genes acquired endolysin activity or its equivalent. None of the phages carrying subsets of the evolved substitutions lyzed cultures thoroughly (turbidity declines of less than 25%) despite the fact that all substitutions in AFK Δ16E were present between two of the phages created (AFK Δ16+g0.7+g1.6+g1.8+g6.3 and AFK Δ16+g17+g17.5+g19.5). Thus, substitutions in multiple genes (a minimum of one from each of the latter two phages) are required to permit effective lysis in the absence of both gp3.5 and gp16. Relative to the primary and secondary mechanisms of lysis, this tertiary pathway requires a larger set of functionally interacting genes and is thus a more complex module.
Genetic Analysis of Holin Activity, Another Component of Lysis
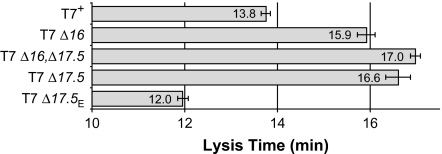
The phage holin is generally considered to control lysis time, and assays using phage λ as a reporter showed that T7 gp17.5 can act as a holin (Vukov et al. 2000). If λ lacks a holin, lysis is prevented entirely. In contrast, gp17.5 is not essential for T7 lysis. Consistent with earlier evidence from T3 (Miyazaki et al. 1978), cells infected by T7 Δ17.5 lyzed after 16.6 min, only 2.8 min later than lysis of T7+ (13.8 min, P < 0.0002; fig. 3). T7 Δ17.5 had a turbidity decline index of ∼80%, indistinguishable from the decline of ∼85% for T7+ (table 2, fig. 4A). This suggests that T7 may have another holin or membrane-degrading activity.
FIG. 3.—
Lysis time of various phages on normal hosts. Error bars indicate ±one standard error.
The idea that gp16 might function as a second holin was tested. Lysis was somewhat less complete for the double deletion mutant T7 Δ16,Δ17.5 than for either single mutant (∼70% decline in turbidity; table 2, fig. 4A). However, the double mutant lyzed at 17 min, only 1.1 min later than T7 Δ16 (P < 0.007) and only 0.4 min later than T7 Δ17.5 (P < 0.25; fig. 3). If both genes coded for holins, lysis should be delayed much more in the double deletion mutant. If gp17.5 and gp16 were the only two proteins with holin activity, one would further expect lysis by the double mutant to be profoundly delayed. These data indicate that gp16 is certainly not the only alternative to gp17.5 for holin activity, and they question whether gp16 has any holin activity.
Compensatory Evolution in 17.5 Deletion Mutants to Investigate Cryptic Holins
We considered whether T7 Δ17.5 could recover from its lysis delay and, if so, whether the sites of compensatory evolution might reveal additional holins. T7 Δ17.5 was serially transferred for 7.5 h to yield T7 Δ17.5E, which lyzed at 12.0 min, 4.6 min earlier than its immediate ancestor (P < 0.00002; fig. 3). Thus, the lack of gene 17.5 does not prevent the evolution to more rapid lysis, although some of this increase could stem from evolution of faster genome entry and earlier initiation of gene expression. Four changes were observed in T7 Δ17.5E (Supplementary Material online). Only the change in 19.5 was found to be strongly compensatory for the 17.5 deletion by a recombination test. Interestingly, a T7 carrying this substitution lyses 2 min later than a phage that lacks the gene 19.5 change (the two phages tested carry another mutation that has no independent effect on lysis; see Supplementary Material online). Gp19.5 is a nonessential protein of incompletely understood function. However, a potential role in lysis is not entirely unexpected. A phage with a deletion of gene 19.5 (along with the adjacent M-hairpin) lyses late and mutant phages make small plaques (Kim and Chung 1996), although much of this effect may be caused by the deletion of the hairpin (Kim et al. 1997).
One possibility is that the effect of the 19.5 substitution on lysis changes direction in different genetic backgrounds, indicating strong interactions with other genes or networks. Prior work on holins has not observed such interactions—holins have similar effects on lysis time even when transferred into profoundly different phages (Vukov et al. 2000). A second possibility is that, regardless of genetic background, the 19.5 change delays lysis as a side effect of delaying membrane permeabilization. Slow lysis due to a gene 17.5 deletion does not appear to increase burst size by itself (Heineman, RH, unpublished data). The lack of a burst size increase in a Δ17.5 phage may be due to holin activity of another gene such as gene 19.5, in which case the 19.5 mutation in a Δ17.5 background could permit larger bursts. However, the burst size of T7 Δ17.5 carrying the gene 19.5 substitution was rather variable, and the increase observed when the gene 19.5 change was introduced (from a burst of 168 to 238) is not significant (P = 0.24 by two-tailed t-test). Nevertheless, the data do not rule out a role of gp19.5 in membrane permeabilization at the end of an infection cycle, and even if it has a different function, it is evident that more genes are playing active roles in the lysis module than has been heretofore predicted. We also note that changes in gene 19.5 were also part of the secondary evolutionary pathway for lysis recovery seen in the AFK Δ16 adaptation.
Discussion
This study examined the levels of evolvability of lysis in bacteriophage T7 by removing individual genes and allowing the phage to recover by adaptation. Most phages, including T7, are thought to use a two-component genetic module to lyse their hosts: an endolysin to break open the cell wall and a holin to let the endolysin past the inner membrane. Thus, the removal of single genes is predicted to have profound and defined effects on lysis and fitness. Furthermore, as it was known from prior work that the evolutionary response to deletion of the phage endolysin gene was mutation in an enzyme whose normal role was outside of lysis, the design here prevented that recovery mechanism. The phage was able to recover from these perturbations, evolving to restore apparently normal lysis.
A priori, the reevolution of a phenotype whose primary genetic basis has been abolished is expected to occur by mutations in genes of similar function to that of the original (e.g., Hall 2003). Although lysis is generally considered an essential phenotype for a lytic phage, the deletion of genes underlying lysis is not as likely to stop replication entirely as is deletion of genes for other essential phenotypes. Furthermore, it is expected that there will be many possible pathways to de novo lysis. Lytic phages carry many genes that are lethal to the host, and a significant proportion of progeny phage is expected to be able to eventually escape a dead host, even if the timing of that escape is not ideal. Thus, a wide range of phage genes may be capable of making a small contribution to progeny release in the absence of normal lysis mechanisms. Long-term adaptation of improved lysis capability might then be expected to involve many genes, especially those interacting with host membranes, although there is no reason to expect that lysis should be capable of a recovery that closely approaches the characteristics of normal lysis.
One broad context for this work is the evolution of genetic modularity. Genetic interaction maps indicate that genomes are modular, with distinct groups of genes interacting to form phenotypes (Tong et al. 2004). Although these modules are often assumed to be lasting coalitions, they change over evolutionary time (Tischler et al. 2008). The reevolution of lysis in a short-term experiment where a strong selection was imposed thus offers insights regarding how modules can evolve in the long term. Although it might have been anticipated that the evolutionary restructuring of lysis would have come from genes with functions related to known lysis activities, even if outside the normal lysis module (as observed in the first layer of evolvability), our observations revealed a different outcome.
The De Novo Origin of Lysis Phenotypes
T7 coding for a defective lysozyme exhibits delayed lysis and low fitness, yet in prior work, both phenotypes greatly recovered with short-term adaptation; the improvement was mostly due to changes in the muralytic domain of a protein not normally involved in lysis. That recovery mechanism may represent a reversion to an ancestral state in which there was one gene with muralytic activity serving two functions, as currently seen in the T7 relative SP6 (Dobbins et al. 2004; Scholl et al. 2004; Heineman et al. 2005). In any case, that pathway merely recreated the standard lysis module, with a holin and endolysin. We have now shown that, when this primary evolutionary pathway is blocked, efficient lysis can still evolve but not from genes with known or suspected activities related to lysis. Although the molecular basis of the new lysis activity is not clear, it appears to be a novel type of lysis mechanism for phages. Multiple genes working in combination are required for what was previously performed by either of the two muralytic enzymes alone. Moreover, all the changes involved in the secondary adaptive pathway were shown to be disadvantageous when the primary recovery pathway is available. There are thus negative epistatic interactions between the two recovery mechanisms. For this reason, it would likely not have been possible to discover the secondary adaptive pathway without the artificial constraint on the primary pathway. Even reasonably effective layers of latent interaction may be difficult to identify without in-depth studies.
Many of the changes seen in the secondary adaptive pathway are difficult to explain in molecular terms. No function has been ascribed to many of these genes, perhaps because more efficient processes have superseded their effects on phage physiology. By selecting for alternative pathways of phage growth, increasingly unpredictable redundancies in metabolic pathways (which may be outside easily observed modules) can be revealed.
The low genetic modularity of T7 lysis under the second pathway of evolvability may be expected of nonessential phenotypes such as changes in physiology or behavior in response to environmental variations. Even if those phenotypes are controlled by a few genes, there may be a wide variety of latent interactions that can recreate some semblance of the original phenotype over evolutionary time. Other phenotypes, such as the ability to break down entirely novel substrates with multiple complex intermediate steps or the evolution of a structural component such as a capsid or tail, may be difficult or impossible to evolve in a design like ours because there are no overlapping functions in the genome that can provide viable, even if poor, intermediates.
However, this study suggests a method for evolving complicated traits in other artificial or natural genomes that may overcome a lack of intermediates. Gene products provided in trans may allow an essential phenotype to persist at low efficiency while evolution operates to improve the phenotype by changes in other parts of the genome. Here, this in trans assistance permitted the spread of the multiple substitutions responsible for recovering lysis in the absence of any known endolysin. The recovery of independent lysis in this study suggests that genomes can indeed evolve to be “weaned” of their need for the exogenous products.
A direct parallel to our study is provided by experimental evolution of new functions in E. coli. When lacZ was removed, E. coli recovered the ability to use lactose (albeit inefficiently) by multiple changes affecting a gene with no known role in normal lactose degradation (Hall 2003). In addition, when E. coli was aerobically adapted on a mix of citrate and glucose, it evolved to use citrate alone by multiple mutations, the first of which was not sufficient for citrate uptake (Blount et al. 2008). Our protocol differs from those examples in that it employs genetic manipulations rather than environmentally lenient conditions to achieve the selection.
Our analysis of the adaptation has neglected the underlying dynamics of mutation accumulation, although we attempted to use a large enough population size that adaptation would not be mutation limited. Assuming that any initially beneficial mutations happen at the same rate as other mutations in the genome, with a conservative estimate of 1 mutation per 300 replications (Drake et al. 1998), a culture of 109 infected cells (the approximate number at the end of each transfer) would yield each beneficial mutation hundreds of times over in a genome of 40 kb. Because recombination between genomes was permitted, the limiting step in these adaptations might then seem to be the time for beneficial mutations to ascend to high frequency and, in the case of strong, negative epistasis, the need for some mutations to reach high frequency before others can arise in a background that renders them favorable. However, the bottleneck size used here (typically ∼105) substantially reduces the chance that any individual beneficial mutation survives a transfer, introducing a strong element of chance in which mutations prevail (Wahl and Krakauer 2000), an effect we did not attempt to quantify.
There are a wide variety of different lysis systems, comprised of a structurally diverse group of proteins. As noted here, most dsDNA phages use a two-component lysis mechanism consisting of a holin and endolysin. The microvirid single-stranded phages instead encode a single lysis gene that achieves lysis by preventing the cell from synthesizing its cell wall so that the cell ruptures when it divides (Young et al. 2000). To a certain extent, these modules are interchangeable; transgenic viruses can be produced with partial or complete replacement of one module with another that still lyse cleanly (Vukov et al. 2000). However, when the dsDNA virus λ has its lysis module replaced by that of a microvirid, lysis is less efficient (Zheng et al. 2008). Similarly, in this study, the secondary adaptive pathway of lysis was less efficient than the first. It is possible that lysis modules evolve de novo relatively easily but that many of these modules are less effective.
Complexity of T7 Holin Activity
As noted above, the widely accepted general model of lysis in dsDNA phages involves a holin that acts with precise timing to permeabilize the inner membrane, allowing the previously accumulated endolysin to access and degrade the cell wall (Young 1992). Our study has revealed that holin activity in T7 is more complex than a single gene. There is only one documented holin in T7 (encoded by gene 17.5; Vukov et al. 2000), but the presence of a second holin or entity with holin-like activity is suggested by the observation that deletion of 17.5 delays lysis only slightly. In contrast, deletion of the holin in phage λ delays lysis profoundly (Bläsi et al. 1989).
Here we used an evolutionary approach to uncover the source of possible alternative holin activity in T7: evolve a genome lacking 17.5 and observe the sites of compensatory evolution. This approach revealed one candidate, gene 19.5. There is no function assigned to this gene, but it is predicted to encode a short, membrane-spanning protein (by TMHMM; Krogh et al. 2001; data not shown). A deletion mutant lacking gene 19.5 lyses late (Kim and Chung 1996), although the observed effect on lysis can perhaps be largely explained by the deletion also removing a nearby hairpin. However, Kim et al. (1997) have suggested that gene 19.5 acts to generate the hairpin, and if the hairpin triggers holin activity, 19.5 would be implicated in lysis indirectly. Gene 19.5 evolved during recoveries to both holin deletion and endolysin disruption. The substitution evolved in response to holin deletion (gp19.5-A24D) delays lysis and is predicted to destroy the transmembrane segment of the protein. The substitution evolved in response to endolysin disruption (gp19.5-E38G) is not predicted to affect the transmembrane domain of the protein, but in that phage, the gp17.5 holin is wild type, so less drastic changes in a secondary holin might be expected. Gene 19.5 is thus a worthwhile subject of further study of the mechanisms of T7-induced lysis.
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
R Springman constructed some of the phage lines and assisted with serial transfer in some cases. IN Wang, A Young, WR Harcombe, TE Keller, and others provided useful suggestions. National Institutes of Health (NIH) grants GM 57756 (J.J.B.) and NIH GM 32095 (I.J.M.) provided financial support. J.J.B. is also supported by the Miecher Regents Professorship at the University of Texas.
References
- Abedon ST. Lysis of lysis-inhibited bacteriophage T4-infected cells. J Bacteriol. 1992;174:8073–8080. doi: 10.1128/jb.174.24.8073-8080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsi U, Nam K, Hartz D, Gold L, Young R. Dual translational initiation sites control function of the λS gene. EMBO J. 1989;8:3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Molineux IJ. A general mechanism for viral resistance to suicide gene expression. J Mol Evol. 2001;53:47–54. doi: 10.1007/s002390010191. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Rokyta D, Molineux IJ. Experimental evolution yields hundreds of mutations in a functional viral genome. J Mol Evol. 2003;57:241–248. doi: 10.1007/s00239-003-2470-1. [DOI] [PubMed] [Google Scholar]
- Dobbins AT, George M, Jr, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J Bacteriol. 2004;186:1933–1944. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Elzinga M, Mark KK, Studier FW. Amino acid sequence of the gene 0.3 protein of bacteriophage T7 and nucleotide sequence of its mRNA. J Biol Chem. 1981;256:2579–2585. [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Grundling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci USA. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. The EBG system of E. coli: origin and evolution of a novel beta-galactosidase for the metabolism of lactose. Genetica. 2003;118:143–156. [PubMed] [Google Scholar]
- Heineman RH, Bull JJ. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evol Int J Org Evol. 2007;61:1695–1709. doi: 10.1111/j.1558-5646.2007.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman RH, Molineux IJ, Bull JJ. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J Mol Evol. 2005;61:181–191. doi: 10.1007/s00239-004-0304-4. [DOI] [PubMed] [Google Scholar]
- Inouye M, Arnheim N, Sternglanz R. Bacteriophage T7 lysozyme is an N-acetylmuramyl-L-alanine amidase. J Biol Chem. 1973;248:7247–7252. [PubMed] [Google Scholar]
- Josslin R. The lysis mechanism of phage T4: mutants affecting lysis. Virology. 1970;40:719–726. doi: 10.1016/0042-6822(70)90216-3. [DOI] [PubMed] [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Kim SH, Chung YB. Isolation of a mutant bacteriophage T7 deleted in nonessential genetic elements, gene 19.5 and m. Virology. 1996;216:20–25. doi: 10.1006/viro.1996.0030. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kim SH, Chung YB. Defects in concatemer processing of bacteriophage T7 DNA deleted in the M-hairpin region. Virology. 1997;236:37–46. doi: 10.1006/viro.1997.8715. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Miralles R, Gerrish PJ, Moya A, Elena SF. Clonal interference and the evolution of RNA viruses. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- Miyazaki JI, Ryo Y, Fujisawa H, Minagawa T. Mutation in bacteriophage T3 affecting host cell lysis. Virology. 1978;89:327–329. doi: 10.1016/0042-6822(78)90067-3. [DOI] [PubMed] [Google Scholar]
- Moak M, Molineux IJ. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol Microbiol. 2000;37:345–355. doi: 10.1046/j.1365-2958.2000.01995.x. [DOI] [PubMed] [Google Scholar]
- Moak M, Molineux IJ. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol. 2004;51:1169–1183. doi: 10.1046/j.1365-2958.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Studier FW. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987;49:221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J Mol Biol. 2004;335:1151–1171. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Silberstein S, Inouye M. Studies on the role of bacteriophage T7 lysozyme during phage infection. J Mol Biol. 1975;96:1–11. doi: 10.1016/0022-2836(75)90178-3. [DOI] [PubMed] [Google Scholar]
- Springman R, Badgett MR, Molineux IJ, Bull JJ. Gene order constrains adaptation in bacteriophage T7. Virology. 2005;341:141–152. doi: 10.1016/j.virol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Studier FW. Bacteriophage T7. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Studier FW. The genetics and physiology of bacteriaphage T7. Virology. 1969;39:562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Studier FW. Relationships among different strains of T7 and among T7-related bacteriophages. Virology. 1979;95:70–84. doi: 10.1016/0042-6822(79)90402-1. [DOI] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat Genet. 2008;40:390–391. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, et al. (42 co-authors) Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Vukov N, Scherer S, Hibbert E, Loessner MJ. Functional analysis of heterologous holin proteins in a λΔS genetic background. FEMS Microbiol Lett. 2000;184:179–186. doi: 10.1111/j.1574-6968.2000.tb09011.x. [DOI] [PubMed] [Google Scholar]
- Wahl LM, Krakauer DC. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics. 2000;156(3):1437–1448. doi: 10.1093/genetics/156.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN. Lysis timing and bacteriophage fitness. Genetics. 2006;172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- Xu M, Struck DK, Deaton J, Wang IN, Young R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci USA. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, Wang I. Phage Lysis. In: Calender R, editor. The bacteriophage. Oxford: Oxford University Press; 2005. pp. 104–125. [Google Scholar]
- Young I, Wang I, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- Zhang X, Studier FW. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J Mol Biol. 1997;269:10–27. doi: 10.1006/jmbi.1997.1016. [DOI] [PubMed] [Google Scholar]
- Zhang X, Studier FW. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J Mol Biol. 2004;340:707–730. doi: 10.1016/j.jmb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Dankenbring CA, Young R. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology. 2008;154:1710–1718. doi: 10.1099/mic.0.2008/016956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.