Abstract
Microscale topographical features have been known to affect cell behavior. An important target in this area is to integrate top down techniques with bottom up self-assembly to create three-dimensional (3D) patterned bioactive mimics of extracellular matrices. We report a novel approach toward this goal and demonstrate its use to study the behavior of human mesenchymal stem cells (hMSCs). By incorporating polymerizable acetylene groups in the hydrophobic segment of peptide amphiphiles (PAs), we were able to micro-pattern nanofiber gels of these bioactive materials. PAs containing the cell adhesive epitope arginine–glycine–aspartic acid–serine (RGDS) were allowed to self-assemble within microfabricated molds to create networks of either randomly oriented or aligned ~30 nm diameter nanofiber bundles that were shaped into topographical patterns containing holes, posts, or channels up to 8 μm in height and down to 5 μm in lateral dimensions. When topographical patterns contained nanofibers aligned through flow prior to gelation, the majority of hMSCs aligned in the direction of the nanofibers even in the presence of hole microtextures and more than a third of them maintained this alignment when encountering perpendicular channel microtextures. Interestingly, in topographical patterns with randomly oriented nanofibers, osteoblastic differentiation was enhanced on hole microtextures compared to all other surfaces.
1. Introduction
The behavior of cells depends strongly on the characteristics of their surrounding environment.1 In biological systems, the complex architecture from the extracellular matrix (ECM) controls many aspects of cell behavior. Both specific amino acid sequences and physical properties of the ECM, including topography, elasticity, and porosity, are known to direct cell fate and function.1–3 The bioactivity of these biochemical and physical signals can be modulated and optimized in vitro by controlling their spatial presentation at the nano- and micro-metre size scales. For example, the relative distance between cell adhesive epitopes can significantly enhance or diminish cell adhesion,4 proliferation,5 and differentiation.6 Similar manipulation of cell behaviors can be achieved through surface topographical features of specific size and geometry.7,8 In the context of synthetic biomaterials, tissue engineering and regenerative medicine strategies seek to stimulate and guide cell behavior in order to promote tissue and organ formation.9,10 In this context, the integration of nanoscale self-assembly of bioactive artificial matrices with three-dimensional (3D) micropatterning of topographical features is important to study and manipulate cell behavior both in vitro and in vivo.
Top-down processes (microfabrication techniques) have been used to develop well-defined topographical and biochemical patterns with micro- and nanoscale resolution to study cell behavior. The inherent precision of microfabrication allows small geometrical modifications of the surface features to better identify and quantify specific effects on cell adhesion,11 migration,12 proliferation,13,14 and differentiation.15,16 The majority of these studies have concentrated on understanding fundamental cellular mechanisms,17,18 growth and expansion of cell populations,19 drug discovery,20 and tissue engineering and regenerative medicine applications.21,22 Nonetheless, despite clear evidence that nano- and micro-topographies can affect and modulate cell and tissue behavior, the practicality of this approach has been hampered by the limited biocompatibility and lack of bioactivity of traditional microfabrication materials used to construct these devices.23,24 A number of approaches have been explored to overcome these limitations, including the development of techniques to topographically pattern more biocompatible/bioactive materials such as hyaluronic acid,25 chitosan,26 poly-caprolactone,27 poly(glycerol-sebacate),23 poly(ethylene glycol),28 or poly(lactic-co-glycolic acid) (PLGA).29 Furthermore, recent work also includes the incorporation of topographical patterns within 3D porous structures30,31 or attempts to combine topographical and chemical patterns.32,33 Strategies are still needed to create micropatterns of materials that also have nanoscale architecture and can display a broader range of molecular bioactivity.
Our laboratory has developed over the past few years self-assembling biomaterials, including networks of one-dimensional bioactive nanostructures based on peptide amphiphiles (PAs).34–39 The PAs can form nanofibers with a high aspect ratio that displays biological signals to control cell behavior both in vitro37–39 and in vivo.40,41 These nanofibers form gels that are highly bioactive, biocompatible, and biodegradable with great promise in both tissue engineering and regenerative medicine.38,40,41 Nanofiber-forming PA molecules are composed of three molecular segments, a hydrophobic alkyl tail that provides an important driving force for self-assembly through hydrophobic collapse in water once electrostatic repulsion among molecules is screened, a peptide segment with a strong propensity to form extended hydrogen bonded β-sheets which prevent aggregation of molecules into spherical micelles and favors instead the formation of fibrous aggregates, and a third segment displayed on nanofiber surfaces that can carry biological information. The resulting self-assembled PA nanofibers are 6–7 nm in diameter and microns in length and tend to aggregate into bundles. The architecture of these systems is therefore highly biomimetic of the fibrous elements commonly found in ECM such as collagen fibrils.
Integrating top down microfabrication with self-assembling PA systems offers a unique opportunity to combine in a single material and processing step, both physical and biomolecular elements that may control cell behavior. In this way, it may be possible to create bioactive scaffolds that combine biologically instructive nanoscale fibers with topographical features. In this work we report on a methodology and specific PA chemical structures amenable to this top down bottom up integration. Furthermore, we demonstrate the use of the strategy to study behavior of human mesenchymal stem cells in contact with an artificial matrix.
2. Materials and methods
Preparation of peptide amphiphiles
All amino acids and Rink MBHA resin were purchased from Novabiochem Corporation (San Diego, CA, USA). 10,12-Pentacosadiynoic acid was purchased from Fluka and Alfa Aesar (Ward Hill, MA, USA). n-Pentacosanoic acid was supplied by TCI America, Inc (Portland, OR, USA). All other reagents and solvents for peptide synthesis were purchased from Aldrich (St. Louis, MO, USA) or Mallinckrodt (Hazelwood, MO, USA) and used as provided. Peptide amphiphiles were manually synthesized by Fmoc solid phase peptide synthesis using orthogonal protecting strategies. Coupling of the first amino acid (Fmoc-Lys(Mtt)-OH) on to the resin was followed by deprotection of the Mtt group (5% Trifluoroacetic acid (TFA) cleaving cocktail in dichloromethane (DCM)) to couple the alkynyl tail (4: 4: 6 alkynyl tail/O-Benzotriazole-N, N, N′, N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU)/di(isopropyl)ethylamine (DIEA)). All subsequent couplings were done using HBTU (4 equiv), DIEA (6 equiv), and the appropriate Fmoc-protected amino acid (4 equiv). The PAs were cleaved from the resin using a cocktail comprised of TFA, triisopropylsilane (TIPS), and water (95: 2.5: 2.5). Compound purity was analyzed by an analytical reverse-phase high-performance liquid chromatography (RP-HPLC) on an Agilent HP 1050 system equipped with a Waters Atlantis C18 column (5 μm particle size, 150 × 4.6 mm or 250 × 4.6 mm). Purification by RP-HPLC was done using a preparative Varian HPLC system equipped with a Waters Atlantis C18 preparative column (5 μm particle size, 250 × 30.0 mm). Confirmation of mass and purity was accomplished through ESI (LCQ Advantage) and MALDI-TOF-MS (Voyager DE Pro). The purity of the final PA molecules was greater than 87%.
Microfabrication of PA topographical patterns
Mold fabrication
The first step to fabricate the PA topographical patterns combined microfabrication and soft lithographic techniques to create molds in which PA molecules can self-assemble into nanofibers. A 10 mm-thick layer of photoresist (SU-8 2010, MicroChem Corp., Newton MA, USA) was coated on top of a standard 3 inch diameter, (100)-oriented silicon wafer. Photolithography was used to transfer and develop the various patterns from a photomask to the coated silicon wafer. The topographically patterned silicon wafer was then coated with 1H,1H,2H,2H-perfluorodecyltrichlorosilane (Alfa Aesar, Ward Hill, MA, USA) to facilitate the release of the PDMS after casting. PDMS polymer and crosslinker (Sylgard 184, Dow Corning, Midland, MI, USA) were mixed in a ratio of 8: 1, degassed for 10 min, poured uniformly on top of the patterned silicon wafer, and cured for 1 h at 90 °C. PDMS replicas were released and used to confine PA solutions during self-assembly and generate precise micro- and macroscopic shapes of PA nanofiber networks.
PA material preparation
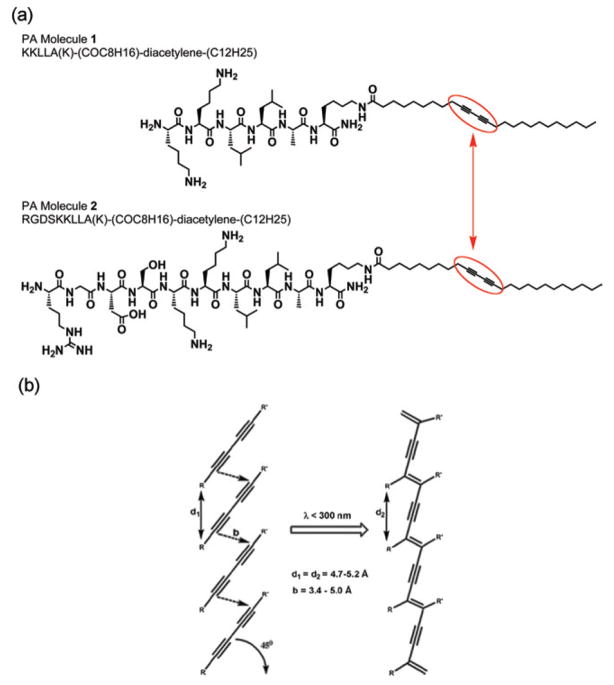
Both PA-molecule 1 and PA-molecule 2 (Fig. 1) were dissolved separately in deionized (DI) water to a concentration of 10 mM each. The two PA solutions were mixed to obtain solutions containing 20% PA-molecule 1 and 80% PA-molecule 2 wt.% prior to fabrication of the PA topographical patterns. The PA solution used to make topographical patterns with randomly oriented nanofibers was freshly made, while the one used to make topographical patterns comprising aligned PA nanofibers was aged for 72 h at room temperature prior to use. Aging of the PA solution promotes some nanofiber formation and increases the viscosity of the solution.
Fig. 1.
Peptide amphiphile (PA) molecules. (a) Molecular structure of the two PA molecules both comprising a photocrosslinkable diacetylene segment (surrounded in red). The smaller PA molecule serves as a filler to space out the bioactive PA molecule, which has the amino acid sequence arginine–glycine–aspartic acid–serine (RGDS) for integrin-based recognition and subsequent improved cell adhesion. (b) Illustration of the polymerization reaction of the diacetylene segments when UV irradiated.
PA topographical patterns
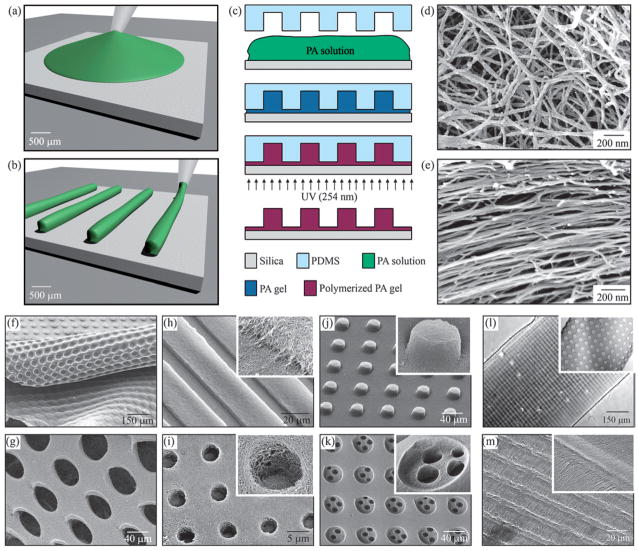
Two kinds of topographical patterns were fabricated. One type comprised a network of randomly oriented PA nanofibers, while the other was formed by aligned PA nanofibers. In both cases, silica slabs were used as substrates for the PA topographical patterns because they provide rigid surfaces that are transparent to 254 nm wavelength UV radiation. Prior to casting, the silica substrates were rendered hydrophilic by exposure to oxygen plasma using an uP80 Reactive Ion Etcher (Oxford Instruments, Oxfordshire, United Kingdom) at 150 W, 100 mTorr, for 45 s. For the topographical patterns consisting of random PA nanofibers (Fig. 2a, c, d), PDMS molds and 1 mm thick silica slabs (Esco Products, Oak Ridge, NJ, USA) were cut into 1 cm × 1 cm pieces. Then, 20 mL drops of the fresh PA solution were placed on top of the silica substrates, followed by placing of the PDMS molds over the PA. The silica–PA–PDMS mold configurations were then exposed to ammonium hydroxide (NH4OH) vapor in a sealed chamber (basic chamber) for 50 min. The base diffuses through the PA, screening the net charges of the PA molecules, and driving PA self-assembly between the silica slabs and the PDMS molds. Subsequently, a UV lamp (Spectronics Corporation, Westbury, NY, USA) was used to irradiate (2 min, 254 nm, 4 Watts) through the silica substrate to polymerize diacetylene groups. Covalent linking of PA molecules through a polydiacetylene backbone increases the mechanical integrity of gels. Finally, PDMS mold removal yields the PA topographical patterns. For the microtextures comprising aligned PA nanofibers (Fig. 2b, c, e), PDMS molds and 1 mm thick silica slabs were cut into 2 cm × 2 cm pieces. Then, 10 mL of the aged PA solution were slowly pipetted onto the silica substrate while dragging the pipette tip, creating a “noodle-shape” pattern of viscous PA solution. The PDMS mold was immediately placed on top of the PA and the same procedure of self-assembly, UV irradiation, and mold removal followed.
Fig. 2.
Fabrication techniques and resulting PA structures. (a–e) The fabrication process starts by either (a) dropping freshly dissolved PA for microtextures with randomly oriented nanofibers, or (b) dragging an aged PA solution for microtextures with aligned nanofibers on a silica substrate. (c) Then, a PDMS mold was used to cover the PA solution while allowing it to conform to the mold, self-assemble into nanofibers, and gel by exposure to ammonium hydroxide (NH4OH). The PA gel was then polymerized under UV irradiation and released from the mold to realize the PA microtextures. The process in (a, c) was used to achieve well-defined three-dimensional (3D) PA structures with (d) randomly oriented nanofibers including (f) removable layers with microtextures or (g) pores and surface microtextures such as (g) channels, (i) holes, (j) posts, and (k) two-level topographies with features down to 5 μm in size. On the other hand, following the process in (b, c), microtextures with (l, m) channels and holes were also achieved but with aligned nanofibers (inset in m).
Cell culture
Human mesenchymal stem cells (hMSCs) (Loanza, Walkersville, MD, USA) were expanded in mesenchymal stem cell growth media (PT-3238, Loanza, Walkersville, MD, USA). The PA substrates and control (glass) were sterilized by immersion in 70% ethanol for 30 min followed by thorough washing with 1× phosphate buffered saline (PBS) (HyClone, Logan, UT, USA). Cells were inoculated on day 0 at a seeding concentration of 10 000 cells/mL. At day 1, cell cultures investigated for osteogenic differentiation were transferred from the growth medium to an osteogenic medium (PT-3924, Loanza Walkersville, MD, USA) and cultured for an additional six days. Cells observed for morphology and alignment were fixed and analyzed after 24 h.
Cell culture analysis
Cells growing on topographical patterns comprising randomly oriented PA nanofibers were examined for effects on osteoblastic differentiation while those growing on topographical patterns formed with aligned PA nanofibers were analyzed to assess competitive effects of nano- and microtexture on cell morphology and alignment. Cell behaviors were investigated by RT-PCR, immunofluorescent staining, and brightfield, confocal, time-lapse, and scanning electron microscopy (SEM). In order to account for random variations between experiments and within a given experiment, statistical significance was defined at the 95% confidence interval using analysis of variance (ANOVA) test performed in Microsoft Excel (Microsoft Corp., Redmond, WA, USA).
Cell alignment
Cell alignment was quantified by photographing five randomly chosen fields of vision of hMSCs on the substrates comprising aligned PA nanofiber bundles and measuring the angle between the long axis of the cell processes and the long axis of the aligned nanofiber bundles (nanotexture). Results are presented as averages (± standard error) of these values. This type of alignment quantification technique has been used by other groups.42–44
RT-PCR
On Day 7, RT-PCR was performed to assess the effect of the PA topographical patterns on osteogenic differentiation. Briefly, cells were lysed with Trizol, followed by extraction and purification of the RNA. PCR was performed with an IQ Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using alkaline phosphatase (ALP) and osteopontin (OP) primers (Bio-search Technologies, Novato, CA, USA) as early and late markers of osteogenic differentiation, respectively.
Immunofluorescent staining
On day 7, immunofluorescent staining was used to qualitatively assess effects of PA topographical patterns on cell morphology and cell differentiation. For cell morphology, cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, rinsed three times with PBS, permeabilized for 4 min with 0.1 Triton X-100 (Sigma, St. Louis, MO, USA) in PBS, rinsed with PBS, blocked with 1% bovine serum albumin (BSA) (Gibco, Grand Island, NY, USA) in PBS, and rinsed again prior to staining of the actin cytoskeleton and cell nuclei. Specimens were then incubated with Rhodamine Phalloidin (Upstate Biotechnology, Charlottesville, VA, USA) at 1: 50 in PBS for 1 h at 25 °C, rinsed three times with PBS, and mounted with 6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI)-containing Vectashield (Vector Labs, Burlingame, CA, USA). For osteoblastic differentiation, cells were fixed as described above and stained for either Core Binding Factor Alpha-1 (CBFα-1), a critical transcription factor for osteoblastic differentiation, or osteopontin (OP). For CBFα-1 staining, fixed cells were incubated with anti-human CBFa-1 antibody (Alpha Diagnostics International, Inc, San Antonio, TX, USA) at 1: 400 diluted in 1% BSA 0.25% Triton X-100 in PBS for 2 h at 25 °C, followed by incubation with a secondary antibody, and rinsed with PBS. For OP staining, fixed cells were blocked with 0.1% Triton X-100 and 1% BSA in PBS, washed twice with PBS, and incubated with anti-human OP antibody (R&D Systems, Minneapolis, MN, USA) at 1: 50 overnight at 4 °C. Subsequently, cells were washed twice with PBS and incubated with anti-mouse IgG NL-637 secondary antibody (R&D systems, Minneapolis, MN) at 1: 200 for 60 min.
SEM fixation
Cells were fixed in a solution of 2% glutaraldehyde/3% sucrose in PBS at 4 °C. After 1 h, substrates were rinsed twice with PBS for 30 min at 4 °C and washed with deionized (DI) water for 5 min. Dehydration was performed by placing the substrates in 50% ethanol (in DI water) for 10 min and replacing it every 10 min while increasing the concentration to 60, 70, 80, 90, 95, and 100% ethanol. The liquid ethanol was removed while preserving the PA topographical patterns and cellular structures using critical point drying.
3. Results and discussion
Two PA molecules were designed for these studies (Fig. 1a). PA molecule 2 contains the bioactive cell adhesion epitope arginine–glycine–aspartic acid–serine (RGDS), and molecule 1 was non-bioactive and used to dilute the epitope on the nanofiber surface. A fixed dilution of 80% PA molecule 2 was observed to support cell adhesion and was therefore used to develop the microtexture fabrication process and in the cell experiments to isolate the effect of topography. Although patterns of similar PA monolayers,45 bilayers,46 and nanofibers47 have recently been reported, they have been limited to creating two-dimensional (2D) PA patterns. This is an important limitation because topographical patterns of microns in height are known to significantly affect cell behavior.7,21 While PA-based materials offer superb bioactive potential, the mechanical properties of the resulting PA gels have been an important limitation, which is in part due to the lack of covalent bonding within the fiber. PA nanofiber integrity relies on the formation of hydrogen bonds by amino acids promoting β-sheet configuration. Therefore, in order to achieve PA topographical features with microns in height, the PA molecules used here incorporate in their hydrophobic segment a photosensitive polymerizable diacetylene group that can covalently link the self-assembled molecules upon irradiation with UV light (254 nm) (Fig. 1). The design and synthesis of these PA molecules has been recently reported by our laboratory and polymerization of these systems has been found to enhance the mechanical properties of gels.48 Sequential control of the PA self-assembly followed by intrafiber polymerization of the diacetylene monomers permitted the fabrication of topographical patterns with ordered nano- and micron-size features in both vertical and horizontal directions. First, flowing of the PA solution (prior to self-assembly) within the microfabricated mold allowed the penetration and localization of PA molecules within the 3D space (inside the PDMS mold). Then, formation of the supramolecular nanofibers by self-assembling of the PA molecules provided the PA gel with a specific and well-defined 3D shape defined by the PDMS mold. Finally, UV irradiation of the PA gel resulted in conjugation of the diacetylene segments to form nanofibers with polydiacetylene backbones, which improved the mechanical integrity of the gels, permitted the removal of the PDMS mold, and consequently realized topographical patterns of bioactive PA gels. These topographical patterns are structurally hierarchical in that they consist of specific PA molecules that self-assemble into well-defined nanofibers, which in turn form a randomly oriented or aligned network of nanofibers that is shaped to form ordered 3D structures with micron-size features. Fig. 2 outlines the fabrication techniques used here to create patterned bioactive nanofiber gels.
In order to investigate the effect of physical guiding provided by topographies in these bioactive gels, two sets of experiments were conducted. First, PA topographical patterns with randomly oriented nanofibers were used to determine effects on hMSC differentiation into the osteoblastic phenotype. Then, an even higher level of topographical control was achieved by creating similar microtextures comprised of aligned PA nanofibers, which were used to investigate competitive effects between nano- and microtextures on hMSC alignment.
PA topographical patterns with randomly oriented nanofibers
The fabrication process illustrated in Fig. 2a, c, d was used to create PA topographical patterns made from randomly oriented PA nanofibers and comprised of features that were 8 μm high, ranging between 10 and 40 μm in lateral dimension. These substrates were tested in vitro using hMSCs cultured on PA surfaces with no microtextures (Smooth-PA), PA surfaces with 8 μm deep holes that were either 20 μm (20-PA) or 40 μm (40-PA) in diameter, and 8 μm high. We also tested surfaces containing 10 μm wide channels separated by 20 μm distances (CH-PA). In addition, commercially available tissue culture dishes (3513 Corning Incorporated, Corning, NY, USA) were used as controls (glass). Human MSCs were cultured under osteogenic conditions and analyzed using reverse transcription polymerase chain reaction (RT-PCR), immunofluorescent staining, bright-field, confocal, time-lapse, and scanning electron microscopy.
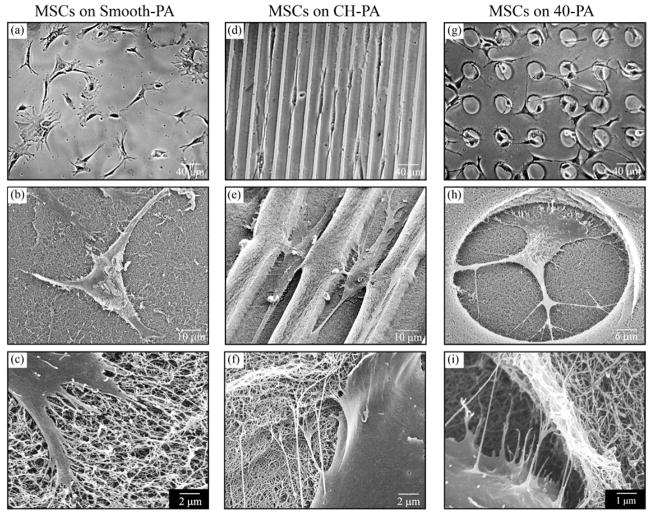
Cells were clearly observed to adhere to and interact with the RGDS-containing PA nanofibers, cell morphology varied on all tested substrates depending on their topographical features. This indicates that cells are recognizing both the biomolecular signaling provided by the RGDS epitope and the physical signaling (contact guidance) provided by the topographical patterns (Fig. 3). Furthermore, although cell contacts with the PA nanofibers were evident on all PA substrates, filopodia-nanofiber interactions were more noticeable on the vertical walls of the microtextures (Fig. 3f, i), indicating an effect of the vertical geometry provided by the bioactive PA topographies.
Fig. 3.
Morphological characteristics of cells on various PA substrates. hMSC morphology depended on the type of PA substrate on which they were growing. (a–c) Cells on Smooth-PA exhibited broad flattened shape with randomly oriented processes. (d–f) In contrast, hMSCs on CH-PA exhibited narrower cell bodies that aligned along the microchannel axis while (g–i) those growing on 40-PA tended to migrate and spread inside the 40 μm diameter holes. On all substrates, hMSCs interacted with the PA nanofiber bundles (c, f, i), which was especially evident along the vertical geometries of the channels (f) and holes (i).
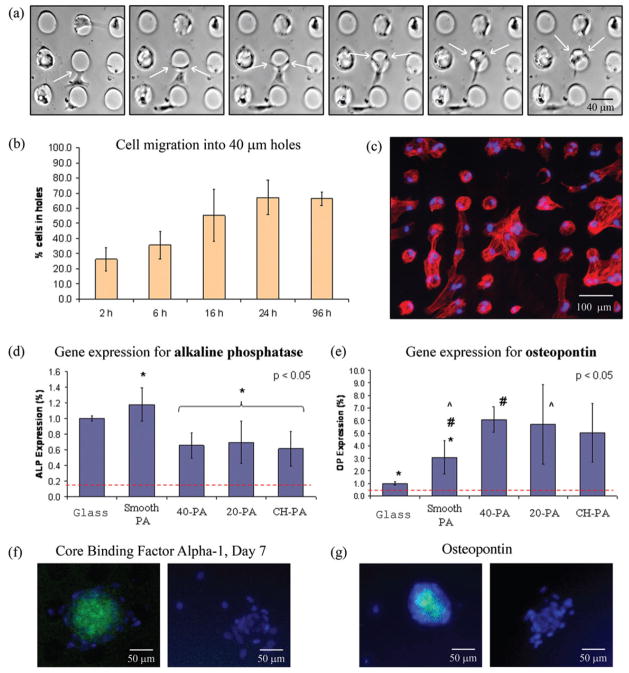
Cells growing on Smooth-PA and glass adopted a similar random morphology (Fig. 3a–c). On CH-PA, cells expressed an aligned morphology and actin cytoskeleton as well as migration along the channel direction (data not shown). This kind of behavior on microfabricated grooves or channels of similar size has been previously reported.42–44 Cells on 20-PA were observed to spread both over and inside the holes. A particularly interesting behavior was observed with cells growing on the 40-PA substrates. During the first hours after inoculation cells were present both inside and outside of the holes. After 2 h of cell inoculation, 26.3% (±7.7) of all cells were present inside of the 40 μm holes. This suggests that initial cell adhesion was not affected by the topographical features of the substrate. Once the cells adhered and spread, however, they were observed to interact with the edge of the 40 μm hole microtextures (Fig. 4a) and tended to migrate inside of them. Twenty-four hours post-seeding, 67.1% (±11.4) of the cells eventually migrated into the holes (Fig. 4b, c). One possibility for this observation is that although SEM observations revealed similar PA nanofiber distribution inside and outside of the holes, the thinner PA layer inside of the 40 μm holes might be a stiffer material than that on walls. It is a known fact that osteoblastic differentiation tends to occur in stiff matrices.49 However, cell contact with the vertical walls of the holes was very apparent (Fig. 3e, f, h, I and Fig. 4a) and suggests that their presence inside of the holes is at least partly a consequence of a preference to interact with the vertical geometries.7,14
Fig. 4.
Migration and osteoblastic differentiation of hMSCs on the 40-PA substrates. (a) Cells were observed to recognize the edge of the holes (white arrows) and migrate inside of them as evident by time-lapse microscopy (photographs represent a 70 min interval). (b) The number of cells inside the 40 μm holes increased from 26% two hours after inoculation to 67% after 24 h, (c) where they were observed to adhere and spread (blue = cell nuclei, red = actin cytoskeleton). (d) Osteoblastic differentiation was enhanced on the substrates comprising hole microtextures. At day 7, hMSCs on 20-PA and 40-PA expressed less alkaline phosphatase (ALP) (d) but significantly higher Osteopontin (OP) (e) than cells on Smooth-PA substrates, suggesting a more mature, differentiated cell. This result was confirmed by positive staining (green) for CBFα-1 (f) and OP (g) of cells on 40-PA, whereas no staining was observed on cells growing on Smooth-PA.
Cell differentiation was significantly affected by the type of substrate. When comparing the two types of smooth surfaces, cells on Smooth-PA expressed lower alkaline phosphatase (ALP) but significantly higher osteopontin (OP) gene expression than cells on glass (Fig. 5d). Furthermore, cells on the PA topographical patterns (20-PA, 40-PA, and CH-PA) expressed similar ALP mRNA levels, which were statistically lower with respect to those from cells cultured on Smooth-PA. However, cells on hole microtextures expressed significantly higher (p < 0.05) OP mRNA levels than those expressed by cells on Smooth-PA. ALP is an earlier marker of osteoblastic differentiation, while OP is a more mature indicator of differentiation.50 Therefore, cells on all PA substrates seemed to be transitioning faster from ALP mRNA expression into OP mRNA expression compared to cells on glass, demonstrating an enhanced differentiation effect of the RGDS-containing PA material. Furthermore, cells on hole microtextures have an even higher level of differentiation compared to those growing on Smooth-PA, demonstrating an enhanced differentiation effect of the 20 μm and 40 μm holes. In order to confirm that osteoblastic differentiation is highest on cells growing on holes, cells on Smooth-PA and 40-PA substrates were stained for both core binding factor a-1 (CBFα-1) and OP. At day 7, groups of cells growing on 40 μm holes were observed to stain (green) for both CBFα-1 and OP (Fig. 4f, g), whereas cells on Smooth-PA did not, qualitatively confirming the increased differentiation evident in the RT-PCR experiments. Differentiation of hMSCs is known to be triggered by an increased cell density.51 Cells growing on substrates comprising hole micro-textures tended to position their cell bodies inside the holes where they could interact with the wall of the holes in all 360° (Fig. 3h). We speculate that the increased differentiation of cells on hole microtextures may be a result of the cells sensing that they are enclosed, simulating the close contact that results from a high cell density, and starting to differentiate at an earlier stage prior to confluency. However, more detailed studies are needed to confirm such a hypothesis.
Fig. 5.
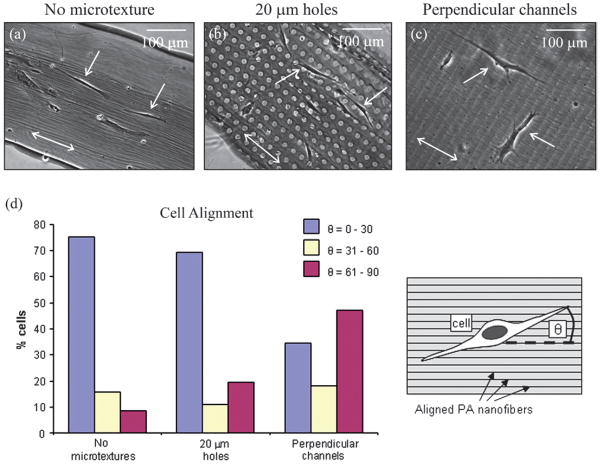
Cellular alignment on surface microtextures comprising aligned nanofiber bundles. (a–c) Representative images of cells (single-headed arrows) growing on substrates with aligned nanofibers bundles (direction of nanofibers represented by the double-headed arrow) with (a) no microtextures, (b) 20 μm diameter holes, or (c) 10 μm wide channels running perpendicular to the direction of the nanofiber bundles. (d) Quantification of cell alignment revealed that cells preferentially aligned along the long axis of the aligned nanofiber bundles (nanotexture) even in the presence of hole microtextures, and exhibited significantly different alignment to cells on substrates with microchannels perpendicular to the direction of the nanofibers. In the presence of these perpendicular microchannels, cells appeared to recognize both the nano- and microtexture, evident by aligning 45% of cells parallel to the long axis of the channels (60°–90°) and 35% of cells parallel to the nanofiber axis (0°–30°).
PA topographical patterns with aligned nanofibers
The fabrication process illustrated in Fig. 2b, c, e was used to create PA topographical patterns made from PA molecules that self-assemble into networks of aligned nanofiber bundles. Nanofiber alignment was achieved by injecting an aged PA solution under weak shear stresses immediately prior to gelation by exposure to ammonium hydroxide (NH4OH) vapor. Aging of PA solutions promotes nanofiber formation, which in turn increases the viscosity of the solution. By depositing the aged PA solution as illustrated in Fig. 2b and using a similar patterning technique to the one used to fabricate the topographical features with randomly oriented nanofibers, we were able to construct topographical micropatterns consisting of aligned PA nano-fibers. The resulting structures consisted of ~30 nm diameter aligned nanofiber bundles that act as nanoscale grooves, and ~4 μm deep holes and channels from lithographic micropatterning that ranged between 10 and 40 μm in lateral dimension (Fig. 2l, m and Fig. 5). Human MSCs were cultured on these substrates and their alignment quantified in order to determine the competitive effect of the nanoscale topography provided by the aligned bundles vs. the microscale topography. On the substrates without microtextures, hMSCs tended to align in the direction of nano-fiber orientation with a mean alignment of 23.2° ± 2.1°, suggesting that the cells are recognizing the texture created by aligned nanofiber bundles (Fig. 5a). Cell alignment with nano-scale grooves has been previously reported.52 Furthermore, cells growing on substrates comprising 20 μm (26.8° ± 5.0°) and 40 μm (26.8° ± 16.6°) diameter holes exhibited statistically similar alignment to those growing on substrates without microtextures. This behavior suggests that cells recognize the nanoscale texture (aligned nanofiber bundles) even in the presence of 20 μm and 40 μm diameter holes. Interestingly, cells growing on the substrates with microchannels that were perpendicular to the direction of the aligned nanofiber bundles exhibited statistically different alignment (49.4° ± 4.5°) than those growing on all the other substrates because they aligned either with the microchannels or with the bioactive nanofiber bundles (Fig. 5c). By defining relative cell alignment to the nanofiber bundles from 0° to 30° and to the microchannels from 60° to 90°, it can be observed that 45% of the cells preferred to align with microchannels while 35% preferred to align with nanofiber bundles even in the presence of the perpendicular microchannels (Fig. 5d). Although it is well known that cells recognize both nano- and microtextures, there is limited work comparing the competitive effects on cell behavior of topographical features of different size scales.53 By creating microtextures made from aligned PA nanofiber bundles, we were able to investigate the alignment response of hMSCs to topographical patterns at both the nano- and microscale while providing specific biological signals (RGDS). These experiments suggest that cytoskeleton organization, which determines cell shape, can respond to both bioactive textures of their matrix with size scales close to receptors and also to confinement by the matrix on cell size scales. Again, this is evident by the ability of the bioactive nanofibers to compete with micro-sized grooves in defining the alignment of cells.
4. Conclusions
By combining top down lithography and self-assembly of polymerizable molecules, we were able to create cell environments with microtextures and well-defined nanoscale bioactive features. The methodology developed enables the study of different types of bioactivity in soft materials combined with matrix architectures on the size scale of cells. The bioactive topographical patterns studied here were shown to significantly affect stem cell morphology, alignment, and differentiation. The hierarchical structures accessed by the simultaneous use of lithography and self-assembly can help craft bioactive soft matter to learn fundamental cell biology and use materials in biomedical devices and regenerative medicine.
Acknowledgments
Funding for this study was provided by the National Institutes of Health/National Institute of Dental & Craniofacial Research (NIH/NIDCR), Grant No. 5R01DE015920-3 and Baxter Healthcare International. This work made use of Central Facilities supported by the MRSEC program of the National Science Foundation (DMR-0520513) at the Materials Research Center of Northwestern University. Electron microscopy was performed in the Electron Probe Instrumentation Center (EPIC) facility of the NUANCE Center at Northwestern University, and is supported by NSF-NSEC, NSF-MRSEC, Keck Foundation, the State of Illinois, and Northwestern University. MALDI and ESI analysis were performed at the IMSERC Center at Northwestern University while confocal microscopy was performed in the Cell Imaging Facility at Northwestern University. We thank Mr Ben Myers, Dr William Russin, Mr Lennel Reynolds, and Dr Teng-Leong Chew for assistance with the various microscopy techniques. Conrado Aparicio would like to thank the Generalitat de Catalunya and the Technical University of Catalunya, Barcelona, Spain for fellowship support.
Footnotes
This paper is part of a Soft Matter theme issue on Self-Assembly. Guest editor: Bartosz Grzybowski.
References
- 1.Stevens MM, George JH. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 2.Ingber DE. Prog Biochem Biophys. 2008;97:163–179. doi: 10.1016/j.pbiomolbio.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirrell M, Kokkoli E, Biesalski M. Surf Sci. 2002;500:61–83. [Google Scholar]
- 4.Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. Chemphyschem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 5.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 6.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 7.Curtis A, Wilkinson C. Biomaterials. 1997;18:1573. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 8.Lim JY, Donahue HJ. Tissue Eng. 2007;13:1879. doi: 10.1089/ten.2006.0154. [DOI] [PubMed] [Google Scholar]
- 9.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci U S A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupp SI. MRS Bull. 2005;30:546–553. [Google Scholar]
- 11.Deutsch J, Motlagh D, Russell B, Desai TA. J Biomed Mater Res. 2000;53:267. doi: 10.1002/(sici)1097-4636(2000)53:3<267::aid-jbm12>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Mai J, Sun C, Li S, Zhang X. Biomed Microdevices. 2007;9:523. doi: 10.1007/s10544-007-9060-8. [DOI] [PubMed] [Google Scholar]
- 13.Mata A, Boehm C, Fleischman AJ, Muschler G, Roy S. J Biomed Mater Res. 2002;62:499. doi: 10.1002/jbm.10353. [DOI] [PubMed] [Google Scholar]
- 14.Mata A, Boehm C, Fleischman AJ, Muschler GF, Roy S. Int J Nanomed. 2007;2:389. [PMC free article] [PubMed] [Google Scholar]
- 15.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO. Nat Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 16.Yim EK, Pang SW, Leong KW. Exp Cell Res. 2007;313:1820. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100:1484. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkelmann M, Gold J, Hauert R, Kasemo B, Spencer ND, Brunette DM, Textor M. Biomaterials. 2003;24:1133. doi: 10.1016/s0142-9612(02)00449-0. [DOI] [PubMed] [Google Scholar]
- 19.Khademhosseini A, Ferreira L, Blumling J, 3rd, Yeh J, Karp MJ, Fukuda J, Langer R. Biomaterials. 2006;27:5968. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 20.Khetani SR, Bhatia SN. Nat Biotechnol. 2007 doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 21.Chehroudi B, Brunette DM. Biomaterials. 2002;23:229. doi: 10.1016/s0142-9612(01)00100-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Murakami H, Chehroudi B, Textor M, Brunette DM. Int J Oral Max Impl. 2006;21:354. [PubMed] [Google Scholar]
- 23.Bettinger CJ, Orrick B, Misra A, Langer R, Borenstein JT. Biomaterials. 2006;27:2558. doi: 10.1016/j.biomaterials.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 24.Kotzar G, Freas M, Abel P, Fleischman A, Roy S, Zorman C, Moran JM, Melzak J. Biomaterials. 2002;23:2737–2750. doi: 10.1016/s0142-9612(02)00007-8. [DOI] [PubMed] [Google Scholar]
- 25.Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, 3rd, Langer R, Burdick J. J Biomed Mater Res A. 2006;79:522. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda J, Khademhosseini A, Yeo Y, Yang X, Yeh J, Eng G, Blumling J, Wang CF, Kohane DS, Langer R. Biomaterials. 2006;27:5259. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S, Lee GY, Wong JY, Desai TA. Biomaterials. 2006;27:4775. doi: 10.1016/j.biomaterials.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 28.Suh KY, Seong J, Khademhosseini A, Laibinis PE, Langer R. Biomaterials. 2004;25:557. doi: 10.1016/s0142-9612(03)00543-x. [DOI] [PubMed] [Google Scholar]
- 29.Owen GR, Jackson J, Chehroudi B, Burt H, Brunette DM. Biomaterials. 2005;26:7447. doi: 10.1016/j.biomaterials.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 30.Stupp SI, Donners JM, Li LS, Mata A. MRS Bull. 2005;30:864. [Google Scholar]
- 31.Mata A, Kim EJ, Boehm A, Fleischman AJ, Muschler GF, Roy S. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2009.05.023. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charest JL, Eliason MT, Garcia AJ, King WP. Biomaterials. 2006;27:2487. doi: 10.1016/j.biomaterials.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Chen XD, Lenhert S, Hirtz M, Lu N, Fuchs H, Chi LF. Accounts Chem Res. 2007;40:393. doi: 10.1021/ar600019r. [DOI] [PubMed] [Google Scholar]
- 34.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 35.Klok HA, Hwang JJ, Hartgerink JD, Stupp SI. Macromolecules. 2002;35:6101–6111. [Google Scholar]
- 36.Hartgerink JD, Zubarev ER, Stupp SI. Curr Opin Solid St Mat. 2001;5:355–361. [Google Scholar]
- 37.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Biomaterials. 2007;28:4608. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 39.Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Acta Biomater. 2005;1:387. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Rajangam K, Behanna HA, Hui MJ, Han X, Hulvat JF, Lomasney JW, Stupp SI. Nano Lett. 2006;6:2086. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 41.Huang Z, Sargeant T, Hulvat JF, Mata A, Bringas P, Jr, Koh CY, Stupp SI, Snead ML. J Bone Miner Res. 2008;23:1995. doi: 10.1359/JBMR.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mata A, Boehm C, Fleischman AJ, Muschler G, Roy S. Biomed Microdevices. 2002;4:267. doi: 10.1023/a:1020950022074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walboomers XF, Croes HJE, Ginsel LA, Jansen JA. Biomaterials. 1998;19:1861. doi: 10.1016/s0142-9612(98)00093-3. [DOI] [PubMed] [Google Scholar]
- 44.Den Braber ET, Deruijter JE, Smits HTJ, Ginsel LA, Vonrecum AF, Jansen JA. J Biomed Mater Res. 1995;29:511. doi: 10.1002/jbm.820290411. [DOI] [PubMed] [Google Scholar]
- 45.Biesalski MA, Knaebel A, Tu R, Tirrell M. Biomaterials. 2006;27:1259. doi: 10.1016/j.biomaterials.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Stroumpoulis D, Zhang H, Rubalcava L, Gliem J, Tirrell M. Langmuir. 2007;23:3849. doi: 10.1021/la062375p. [DOI] [PubMed] [Google Scholar]
- 47.Hung AM, Stupp SI. Nano Lett. 2007;7:1165. doi: 10.1021/nl062835z. [DOI] [PubMed] [Google Scholar]
- 48.Hsu L, Cvetanovich GL, Stupp SI. J Am Chem Soc. 2008;130:3892–2899. doi: 10.1021/ja076553s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 50.Vats A, Bielby RC, Tolley NS, Nerem R, Polak JM. Lancet. 2005;366:592. doi: 10.1016/S0140-6736(05)66879-1. [DOI] [PubMed] [Google Scholar]
- 51.Caplan AI, Syftestad GT, Osdoby P. Clin Orthop Rel Res. 1983;174:243–263. [PubMed] [Google Scholar]
- 52.Teixeira AI, Nealey PF, Murphy CJ. J Biomed Mater Res A. 2004;71A:369. doi: 10.1002/jbm.a.30089. [DOI] [PubMed] [Google Scholar]
- 53.Tan J, Saltzman WM. Biomaterials. 2004;25:3593. doi: 10.1016/j.biomaterials.2003.10.034. [DOI] [PubMed] [Google Scholar]